A Metabolomics Approach to Increasing Chinese Hamster Ovary (CHO) Cell Productivity
Abstract
:1. Introduction
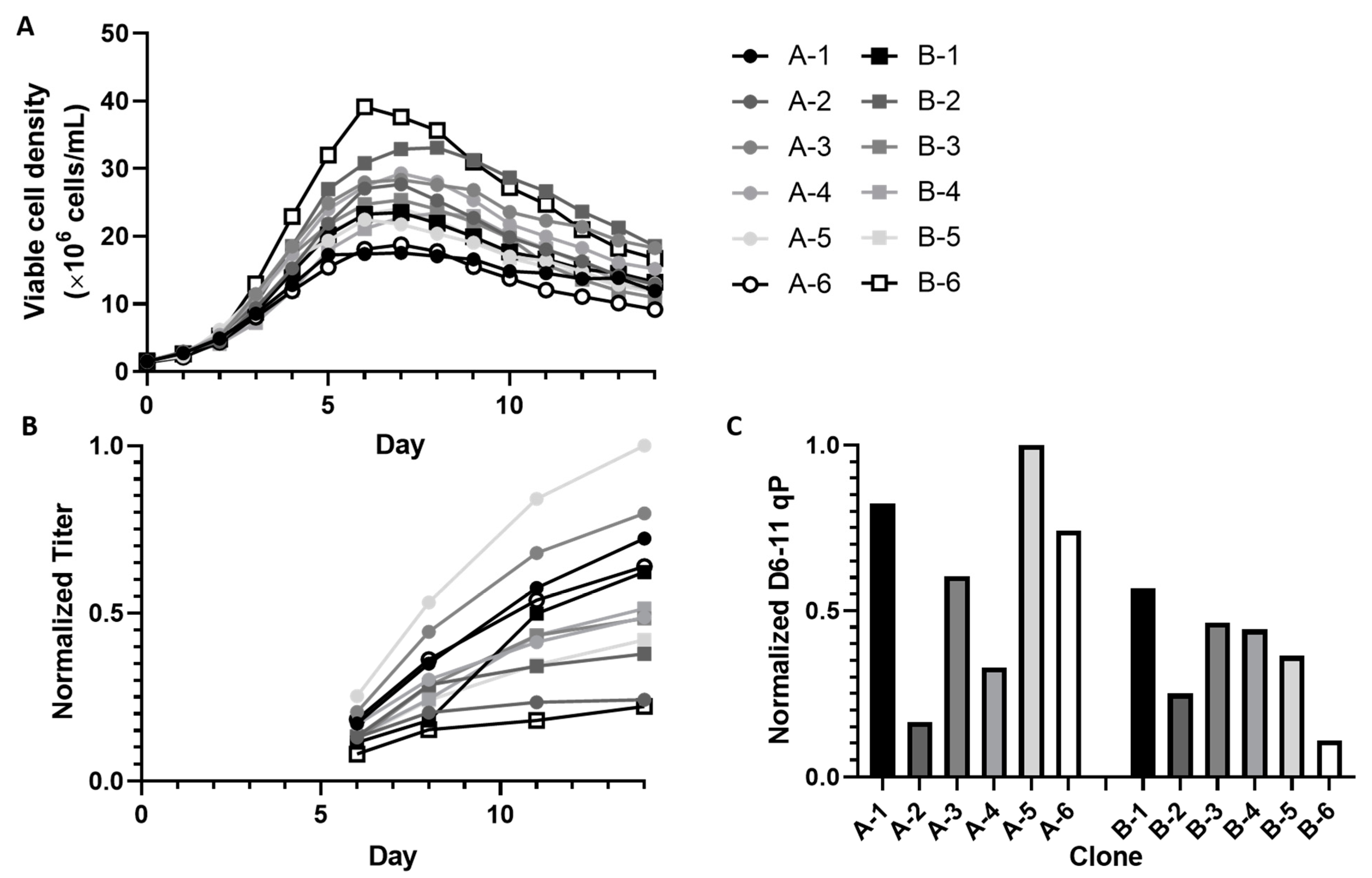
2. Results
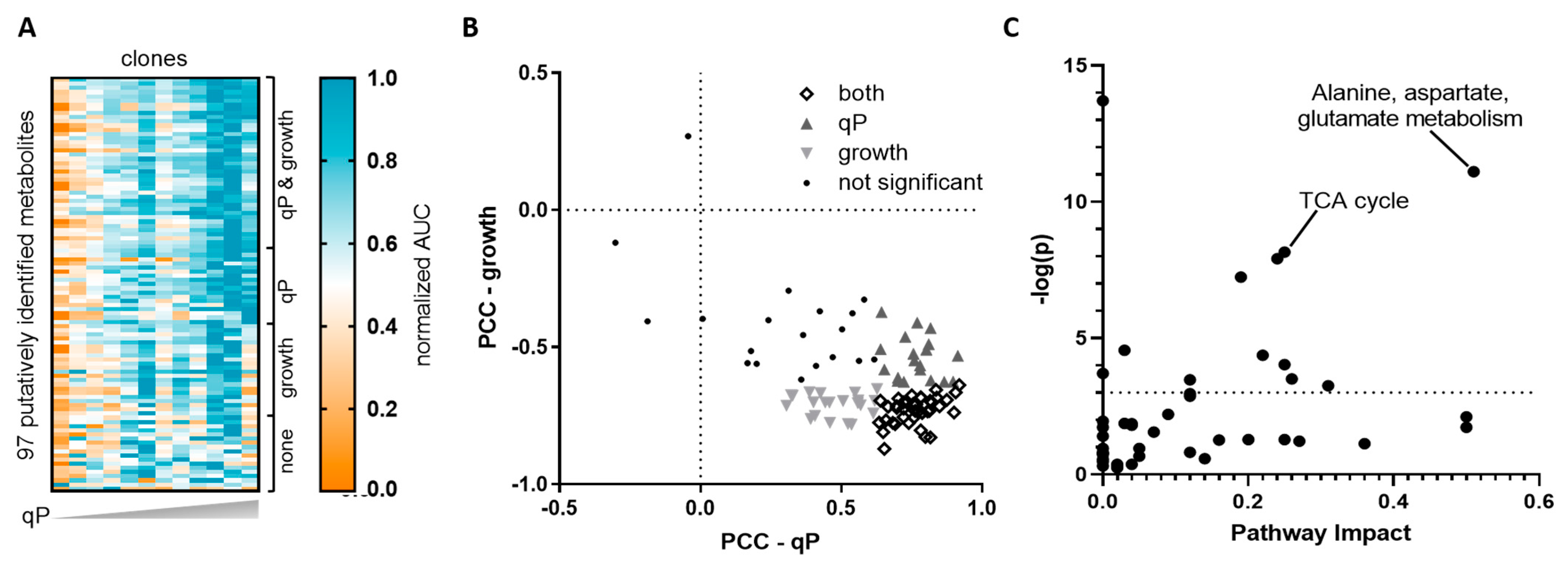
2.1. Untargeted Metabolomics Identified Endogenous CHO Cell Metabolites That Correlate with qP but Not Growth
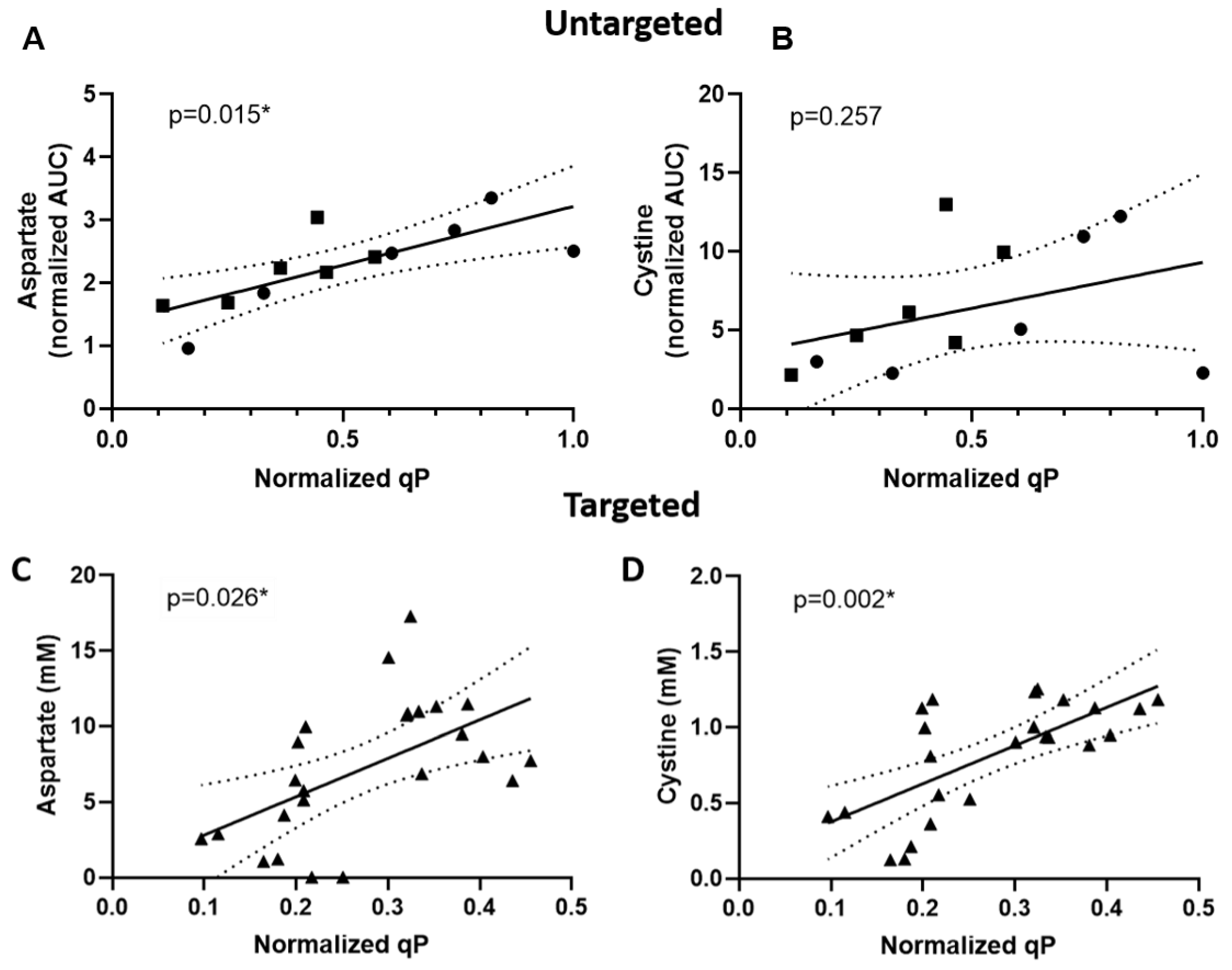
2.2. Targeted Analysis Confirmed Aspartate and Cystine as qP Specific Metabolite Indicators
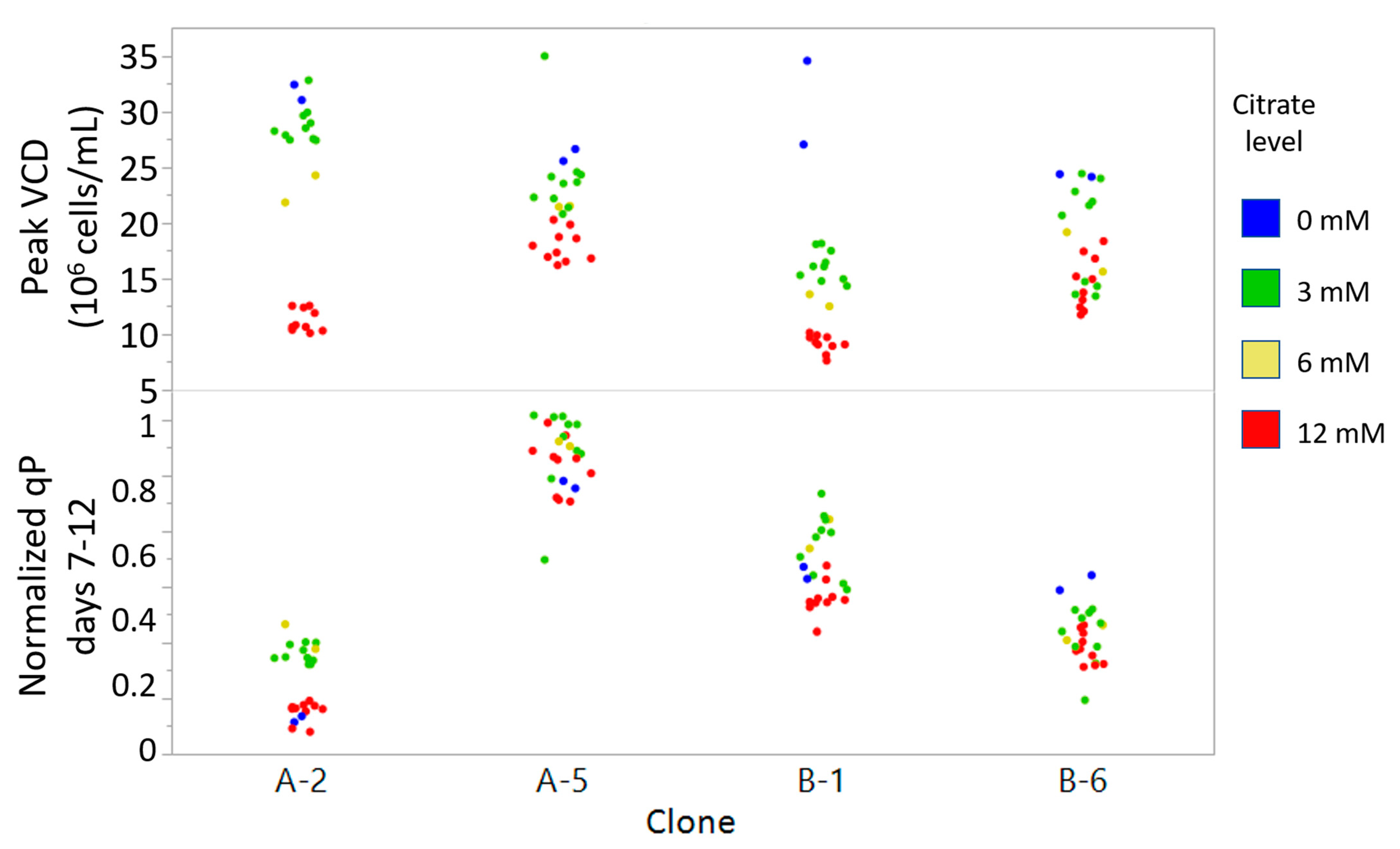
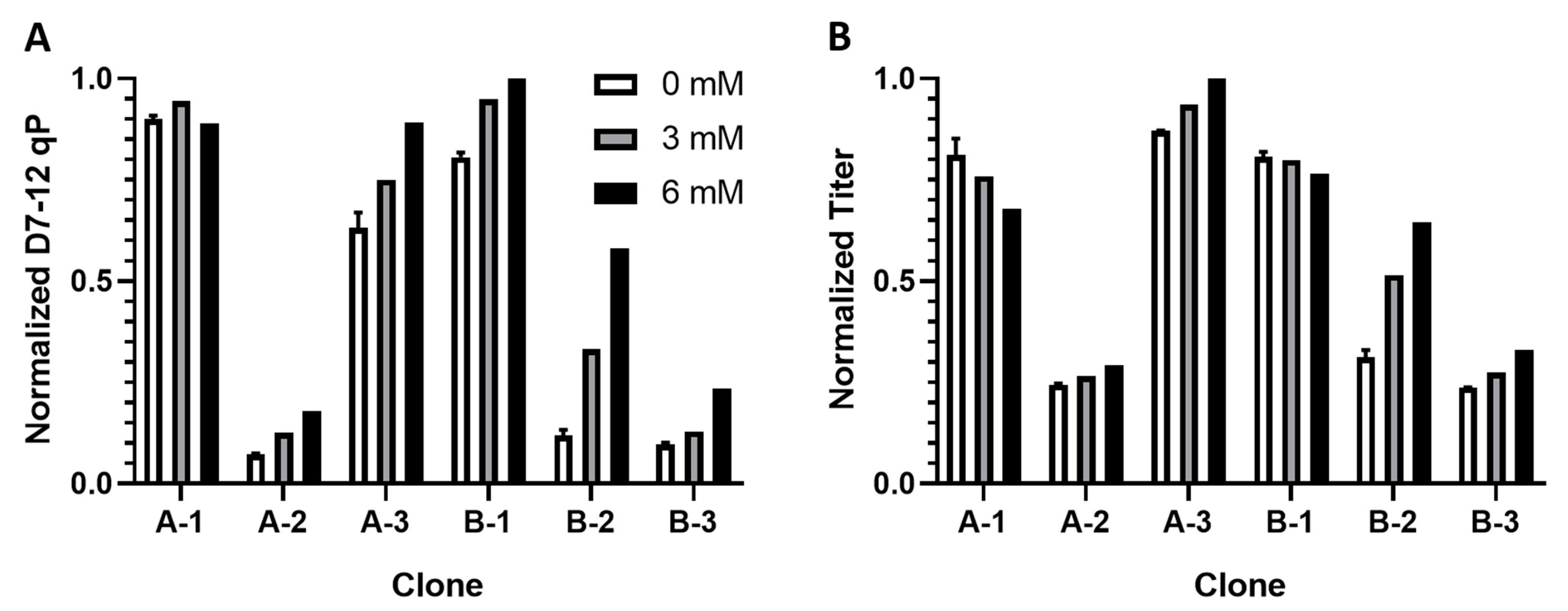
2.3. Medium Supplementation with qP Correlated Metabolite Improved qP and Titer
3. Discussion
3.1. Tradeoff between Cell Growth and mAb Production
3.2. Aspartic Acid and Cystine
3.3. Citrate Addition Improves qP and Titer
4. Materials and Methods
4.1. Cell Lines
4.2. Fed-Batch Cell Culture Experiments
4.3. Untargeted LC-MS
4.4. Feature Annotation
4.5. Data Analysis
4.6. Targeted LC-MS
4.7. Addback Experiments
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huang, Y.-M.; Hu, W.; Rustandi, E.; Chang, K.; Yusuf-Makagiansar, H.; Ryll, T. Maximizing productivity of CHO cell-based fed-batch culture using chemically defined media conditions and typical manufacturing equipment. Biotechnol. Prog. 2010, 26, 1400–1410. [Google Scholar] [CrossRef] [PubMed]
- Handlogten, M.W.; Lee-O’Brien, A.; Roy, G.; Levitskaya, S.V.; Venkat, R.; Singh, S.; Ahuja, S. Intracellular response to process optimization and impact on productivity and product aggregates for a high-titer CHO cell process. Biotechnol. Bioeng. 2018, 115, 126–138. [Google Scholar] [CrossRef] [PubMed]
- Takagi, Y.; Kikuchi, T.; Omasa, R.W.T. The enhancement of antibody concentration and achievement of high cell density CHO cell cultivation by adding nucleoside. Cytotechnology 2017, 69, 511–521. [Google Scholar] [CrossRef]
- Verhagen, N.; Teleki, A.; Heinrich, C.; Schilling, M.; Unsöld, A.; Takors, R. S-adenosylmethionine and methylthioadenosine boost cellular productivities of antibody forming Chinese Hamster ovary cells. Biotechnol. Bioeng. 2020, 117, 3239–3247. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Rehmann, M.S.; Xu, M.; Zheng, S.; Hill, C.; He, Q.; Borys, M.C.; Li, Z.J. Development of an intensified fed-batch production platform with doubled titers using N-1 perfusion seed for cell culture manufacturing. Bioresour. Bioprocess 2020, 7, 17. [Google Scholar] [CrossRef]
- Templeton, N.; Xu, S.; Roush, D.J.; Chen, H. 13C metabolic flux analysis identifies limitations to increasing specific productivity in fed-batch and perfusion. Metab. Eng. 2017, 44, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Stolfa, G.; Smonskey, M.T.; Boniface, R.; Hachmann, A.B.; Gulde, P.; Joshi, A.D.; Pierce, A.P.; Jacobia, S.J.; Campbell, A. CHO-Omics Review: The Impact of Current and Emerging Technologies on Chinese Hamster Ovary Based Bioproduction. Biotechnol. J. 2018, 13, e1700227. [Google Scholar] [CrossRef]
- Ang, K.S.; Hong, J.; Lakshmanan, M.; Lee, D.Y. Toward Integrated Multi-omics Analysis for Improving CHO Cell Bioprocessing. In Cell Culture Engineering; Lee, G.M., Kildegaard, H.F., Lee, S.Y., Nielsen, J., Stephanopoulos, G., Eds.; Wiley & Sons: New York, NY, USA, 2019; pp. 163–184. Available online: https://onlinelibrary.wiley.com/doi/abs/10.1002/9783527811410.ch7 (accessed on 20 July 2021).
- Morris, C.; Polanco, A.; Yongky, A.; Xu, J.; Huang, Z.; Zhao, J.; McFarland, S.; Park, S.; Warrack, B.; Reily, M.; et al. Bigdata analytics identifies metabolic inhibitors and promoters for productivity improvement and optimization of monoclonal antibody (mAb) production process. Bioresour. Bioprocess. 2020, 7, 31. [Google Scholar] [CrossRef]
- Huang, Z.; Yoon, S. Identifying metabolic features and engineering targets for productivity improvement in CHO cells by integrated transcriptomics and genome-scale metabolic model. Biochem. Eng. J. 2020, 159, 107624. [Google Scholar] [CrossRef]
- Chong, W.P.K.; Yusufi, F.N.K.; Lee, D.Y.; Reddy, S.G.; Wong, N.S.C.; Heng, C.K.; Yap, M.G.S.; Ho, Y.S. Metabolomics profiling of extracellular metabolites in recombinant Chinese Hamster Ovary fed-batch culture. Rapid Commun. Mass Spectrom. 2009, 23, 3763–3771. [Google Scholar] [CrossRef]
- Mohmad-Saberi, S.E.; Hashim, Y.Z.H.Y.; Mel, M.; Amid, A.; Ahmad-Raus, R.; Packeer-Mohamed, V. Metabolomics profiling of extracellular metabolites in CHO-K1 cells cultured in different types of growth media. Cytotechnology 2013, 65, 577–586. [Google Scholar] [CrossRef] [Green Version]
- Chong, W.P.K.; Yusufi, F.N.K.; Lee, D.-Y.; Reddy, S.G.; Wong, N.S.C.; KiatHeng, C.; Yap, M.G.S.; Ho, Y.S. Metabolomics-based identification of apoptosis-inducing metabolites in recombinant fed-batch CHO culture media. J. Biotechnol. 2011, 151, 218–224. [Google Scholar] [CrossRef]
- Mulukutla, B.C.; Kale, J.; Kalomeris, T.; Jacobs, M.; Hiller, G.W. Identification and control of novel growth inhibitors in fed-batch cultures of Chinese hamster ovary cells. Biotechnol. Bioeng. 2017, 114, 1779–1790. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Ray, S.; Dai, S.; Ivanov, A.R.; Abu-Absi, N.R.; Lewis, A.; Huang, Z.; Xing, Z.; Borys, M.C.; Li, Z.J.; et al. M Combined metabolomics and proteomics reveals hypoxia as a cause of lower productivity on scale-up to a 5000-liter CHO bioprocess. Biotechnol. J. 2016, 11, 1190–1200. [Google Scholar] [CrossRef] [PubMed]
- Chong, W.P.K.; Thng, S.H.; Hiu, A.P.; Lee, D.Y.; Chan, E.C.Y.; Ho, Y.S. LC-MS-based metabolic characterization of high monoclonal antibody-producing Chinese hamster ovary cells. Biotechnol. Bioeng. 2012, 109, 3103–3111. [Google Scholar] [CrossRef] [PubMed]
- Dean, J.; Reddy, P. Metabolic analysis of antibody producing CHO cells in fed-batch production. Biotechnol. Bioeng. 2013, 110, 1735–1747. [Google Scholar] [CrossRef] [PubMed]
- Templeton, N.; Smith, K.D.; McAtee-Pereira, A.G.; Dorai, H.; Betenbaugh, M.J.; Lang, S.E.; Young, J.D. Application of 13C flux analysis to identify high-productivity CHO metabolic phenotypes. Metab. Eng. 2017, 43, 218–225. [Google Scholar] [CrossRef]
- Huang, Z.; Xu, J.; Yongky, A.; Morris, C.S.; Polanco, A.L.; Reily, M.; Borys, M.C.; Li, Z.J.; Yoon, S. CHO cell productivity improvement by genome-scale modeling and pathway analysis: Application to feed supplements. Biochem. Eng. J. 2020, 160, 107638. [Google Scholar] [CrossRef]
- Qian, Y.; Sowa, S.W.; Aron, K.L.; Xu, P.; Langsdorf, E.; Warrack, B.; Aranibar, N.; Tremml, G.; Xu, J.; McVey, D.; et al. New insights into genetic instability of an industrial CHO cell line by orthogonal omics. Biochem. Eng. J. 2020, 164, 107799. [Google Scholar] [CrossRef]
- Alden, N.; Krishnan, S.; Porokhin, V.; Raju, R.; McElearney, K.; Gilbert, A.; Lee, K. Biologically Consistent Annotation of Metabolomics Data. Anal. Chem. 2017, 89, 13097–13104. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [Green Version]
- Pereira, S.; Kildegaard, H.F.; Andersen, M.R. Impact of CHO Metabolism on Cell Growth and Protein Production: An Overview of Toxic and Inhibiting Metabolites and Nutrients. Biotechnol. J. 2018, 13, e1700499. [Google Scholar] [CrossRef] [Green Version]
- Altamirano, C.; Cairó, J.J.; Gòdia, F. Decoupling cell growth and product formation in Chinese hamster ovary cells through metabolic control. Biotechnol. Bioeng. 2001, 76, 351–360. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [Green Version]
- Traustason, B. Amino Acid Requirements of the Chinese Hamster Ovary Cell Metabolism during Recombinant Protein Production. bioRxiv 2019, 796490. [Google Scholar] [CrossRef]
- Goos, P.; Jones, B. Optimal Design of Experiments: A Case Study Approach; John Wiley & Sons: Hoboken, NJ, USA, 2011. [Google Scholar]
- Lao, M.-S.; Toth, D. Effects of Ammonium and Lactate on Growth and Metabolism of a Recombinant Chinese Hamster Ovary Cell Culture. Biotechnol. Prog. 1997, 13, 688–691. [Google Scholar] [CrossRef] [PubMed]
- Toussaint, C.; Henry, O.; Durocher, Y. Metabolic engineering of CHO cells to alter lactate metabolism during fed-batch cultures. J. Biotechnol. 2016, 217, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Freund, N.W.; Croughan, M.S. A Simple Method to Reduce both Lactic Acid and Ammonium Production in Industrial Animal Cell Culture. Int. J. Mol. Sci. 2018, 19, 385. [Google Scholar] [CrossRef] [Green Version]
- Noh, S.M.; Park, J.H.; Lim, M.S.; Kim, J.W.; Lee, G.M. Reduction of ammonia and lactate through the coupling of glutamine synthetase selection and downregulation of lactate dehydrogenase-A in CHO cells. Appl. Microbiol. Biotechnol. 2017, 101, 1035–1045. [Google Scholar] [CrossRef]
- Mulukutla, B.C.; Mitchell, J.; Geoffroy, P.; Harrington, C.; Krishnan, M.; Kalomeris, T.; Morris, C.; Zhang, L.; Pegman, P.; Hiller, G.W. Metabolic engineering of Chinese hamster ovary cells towards reduced biosynthesis and accumulation of novel growth inhibitors in fed-batch cultures. Metab. Eng. 2019, 54, 54–68. [Google Scholar] [CrossRef]
- Alden, N.; Raju, R.; McElearney, K.; Lambropoulos, J.; Kshirsagar, R.; Gilbert, A.; Lee, K. Using Metabolomics to Identify Cell Line-Independent Indicators of Growth Inhibition for Chinese Hamster Ovary Cell-based Bioprocesses. Metabolites 2020, 10, 199. [Google Scholar] [CrossRef] [PubMed]
- Bielser, J.-M.; Wolf, M.; Souquet, J.; Broly, H.; Morbidelli, M. Perfusion mammalian cell culture for recombinant protein manufacturing-A critical review. Biotechnol. Adv. 2018, 36, 1328–1340. [Google Scholar] [CrossRef] [PubMed]
- Warikoo, V.; Godawat, R.; Brower, K.; Jain, S.; Cummings, D.; Simons, E.; Johnson, T.; Walther, J.; Yu, M.; Wright, B.; et al. Integrated continuous production of recombinant therapeutic proteins. Biotechnol. Bioeng. 2012, 109, 3018–3029. [Google Scholar] [CrossRef]
- Martínez, V.S.; Dietmair, S.; Quek, L.E.; Hodson, M.P.; Gray, P.; Nielsen, L.K. Flux balance analysis of CHO cells before and after a metabolic switch from lactate production to consumption. Biotechnol. Bioeng. 2013, 110, 660–666. [Google Scholar] [CrossRef]
- Fussenegger, M.; Schlatter, S.; Dätwyler, D.; Mazur, X.; Bailey, J.E. Controlled proliferation by multigene metabolic engineering enhances the productivity of Chinese hamster ovary cells. Nat. Biotechnol. 1998, 16, 468–472. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Gammell, P.; Clynes, M. Proliferation control strategies to improve productivity and survival during CHO based production culture: A summary of recent methods employed and the effects of proliferation control in product secreting CHO cell lines. Cytotechnology 2007, 53, 33–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larson, T.M.; Gawlitzek, M.; Evans, H.; Albers, U.; Cacia, J. Chemometric evaluation of on-line high-pressure liquid chromatography in mammalian cell cultures: Analysis of amino acids and glucose. Biotechnol. Bioeng. 2002, 77, 553–563. [Google Scholar] [CrossRef]
- Allen, E.L.; Ulanet, D.B.; Pirman, D.; Mahoney, C.E.; Coco, J.; Si, Y.; Chen, Y.; Huang, L.; Ren, J.; Choe, S.; et al. Differential Aspartate Usage Identifies a Subset of Cancer Cells Particularly Dependent on OGDH. Cell Rep. 2016, 17, 876–890. [Google Scholar] [CrossRef] [Green Version]
- Lasorsa, F.M.; Pinton, P.; Palmieri, L.; Fiermonte, G.; Rizzuto, R.; Palmieri, F. Recombinant expression of the Ca2+-sensitive aspartate/glutamate carrier increases mitochondrial ATP production in agonist-stimulated Chinese hamster ovary cells. J. Biol. Chem. 2003, 278, 38686–38692. [Google Scholar] [CrossRef] [Green Version]
- Chong, W.P.; Sim, L.C.; Wong, K.T.; Yap, M.G. Enhanced IFNgamma production in adenosine-treated CHO cells: A mechanistic study. Biotechnol. Prog. 2009, 25, 866–873. [Google Scholar] [CrossRef]
- Edros, R.; McDonnell, S.; Al-Rubeai, M. The relationship between mTOR signalling pathway and recombinant antibody productivity in CHO cell lines. BMC Biotechnol. 2014, 14, 15. [Google Scholar] [CrossRef] [Green Version]
- Young, J.D. Metabolic flux rewiring in mammalian cell cultures. Curr. Opin. Biotechnol. 2013, 24, 1108–1115. [Google Scholar] [CrossRef] [Green Version]
- Ali, A.S.; Raju, R.; Kshirsagar, R.; Ivanov, A.R.; Gilbert, A.; Zang, L.; Karger, B.L. Multi-Omics Study on the Impact of Cysteine Feed Level on Cell Viability and mAb Production in a CHO Bioprocess. Biotechnol. J. 2018, 14, e1800352. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.S.; Chen, R.; Raju, R.; Kshirsagar, R.; Gilbert, A.; Zang, L.; Karger, B.L.; Ivanov, A.R. Multi-Omics Reveals Impact of Cysteine Feed Concentration and Resulting Redox Imbalance on Cellular Energy Metabolism and Specific Productivity in CHO Cell Bioprocessing. Biotechnol. J. 2020, 15, 1900565. [Google Scholar] [CrossRef] [PubMed]
- Carinhas, N.; Duarte, T.M.; Barreiro, L.C.; Carrondo, M.J.; Alves, P.M.; Teixeira, A.P. Metabolic signatures of GS-CHO cell clones associated with butyrate treatment and culture phase transition. Biotechnol. Bioeng. 2013, 110, 3244–3257. [Google Scholar] [CrossRef]
- Sellick, C.A.; Croxford, A.S.; Maqsood, A.R.; Stephens, G.; Westerhoff, H.V.; Goodacre, R.; Dickson, A.J. Metabolite profiling of recombinant CHO cells: Designing tailored feeding regimes that enhance recombinant antibody production. Biotechnol. Bioeng. 2011, 108, 3025–3031. [Google Scholar] [CrossRef]
- Sellick, C.A.; Croxford, A.S.; Maqsood, A.R.; Stephens, G.M.; Westerhoff, H.V.; Goodacre, R.; Dickson, A.J. Metabolite profiling of CHO cells: Molecular reflections of bioprocessing effectiveness. Biotechnol. J. 2015, 10, 1434–1445. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Jiang, R.; Lin, H.; Xu, S. Feeding tricarboxylic acid cycle intermediates improves lactate consumption and antibody production in Chinese hamster ovary cell cultures. Biotechnol. Prog. 2020, e2975. [Google Scholar] [CrossRef]
- Gilbert, A.; McElearney, K.; Kshirsagar, R.; Sinacore, M.S.; Ryll, T. Investigation of metabolic variability observed in extended fed batch cell culture. Biotechnol. Prog. 2013, 29, 1519–1527. [Google Scholar] [CrossRef]
- Bai, Y.; Wu, C.; Zhao, J.; Liu, Y.H.; Ding, W.; Ling, W.L.W. Role of iron and sodium citrate in animal protein-free CHO cell culture medium on cell growth and monoclonal antibody production. Biotechnol. Prog. 2011, 27, 209–219. [Google Scholar] [CrossRef]
- Gupta, S.K.; Srivastava, S.K.; Sharma, A.; Nalage, V.H.; Salvi, D.; Kushwaha, H.; Chitnis, N.B.; Shukla, P. Metabolic engineering of CHO cells for the development of a robust protein production platform. PLoS ONE 2017, 12, e0181455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkens, C.A.; Gerdtzen, Z.P. Comparative metabolic analysis of CHO cell clones obtained through cell engineering, for IgG productivity, growth and cell longevity. PLoS ONE 2015, 10, e0119053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruttkies, C.; Schymanski, E.L.; Wolf, S.; Hollender, J.; Neumann, S. MetFrag relaunched: Incorporating strategies beyond in silico fragmentation. J. Cheminform. 2016, 8, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, F.; Greiner, R.; Wishart, D. Competitive fragmentation modeling of ESI-MS/MS spectra for putative metabolite identification. Metabolomics 2015, 11, 98–110. [Google Scholar] [CrossRef] [Green Version]
- NIST/EPA/NIH Mass Spectral Library NIST, Version 17. 6 June 2017. Available online: https://www.nist.gov/srd/nist-standard-reference-database-1a-v17 (accessed on 2 February 2018).
- Tautenhahn, R.; Cho, K.; Uritboonthai, W.; Zhu, Z.; Patti, G.J.; Siuzdak, G. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat. Biotechnol. 2012, 30, 826–828. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0—The Human Metabolome Database in 2013. Nucleic Acids Res. 2013, 41, D801–D807. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, G.; Aron, K.; Borys, M.; Li, Z.; Pendse, G.; Lee, K. A Metabolomics Approach to Increasing Chinese Hamster Ovary (CHO) Cell Productivity. Metabolites 2021, 11, 823. https://doi.org/10.3390/metabo11120823
Yao G, Aron K, Borys M, Li Z, Pendse G, Lee K. A Metabolomics Approach to Increasing Chinese Hamster Ovary (CHO) Cell Productivity. Metabolites. 2021; 11(12):823. https://doi.org/10.3390/metabo11120823
Chicago/Turabian StyleYao, Grace, Kathryn Aron, Michael Borys, Zhengjian Li, Girish Pendse, and Kyongbum Lee. 2021. "A Metabolomics Approach to Increasing Chinese Hamster Ovary (CHO) Cell Productivity" Metabolites 11, no. 12: 823. https://doi.org/10.3390/metabo11120823
APA StyleYao, G., Aron, K., Borys, M., Li, Z., Pendse, G., & Lee, K. (2021). A Metabolomics Approach to Increasing Chinese Hamster Ovary (CHO) Cell Productivity. Metabolites, 11(12), 823. https://doi.org/10.3390/metabo11120823







