A Global Metabolic Map Defines the Effects of a Si-Based Biostimulant on Tomato Plants under Normal and Saline Conditions
Abstract
:1. Introduction
2. Results and Discussion
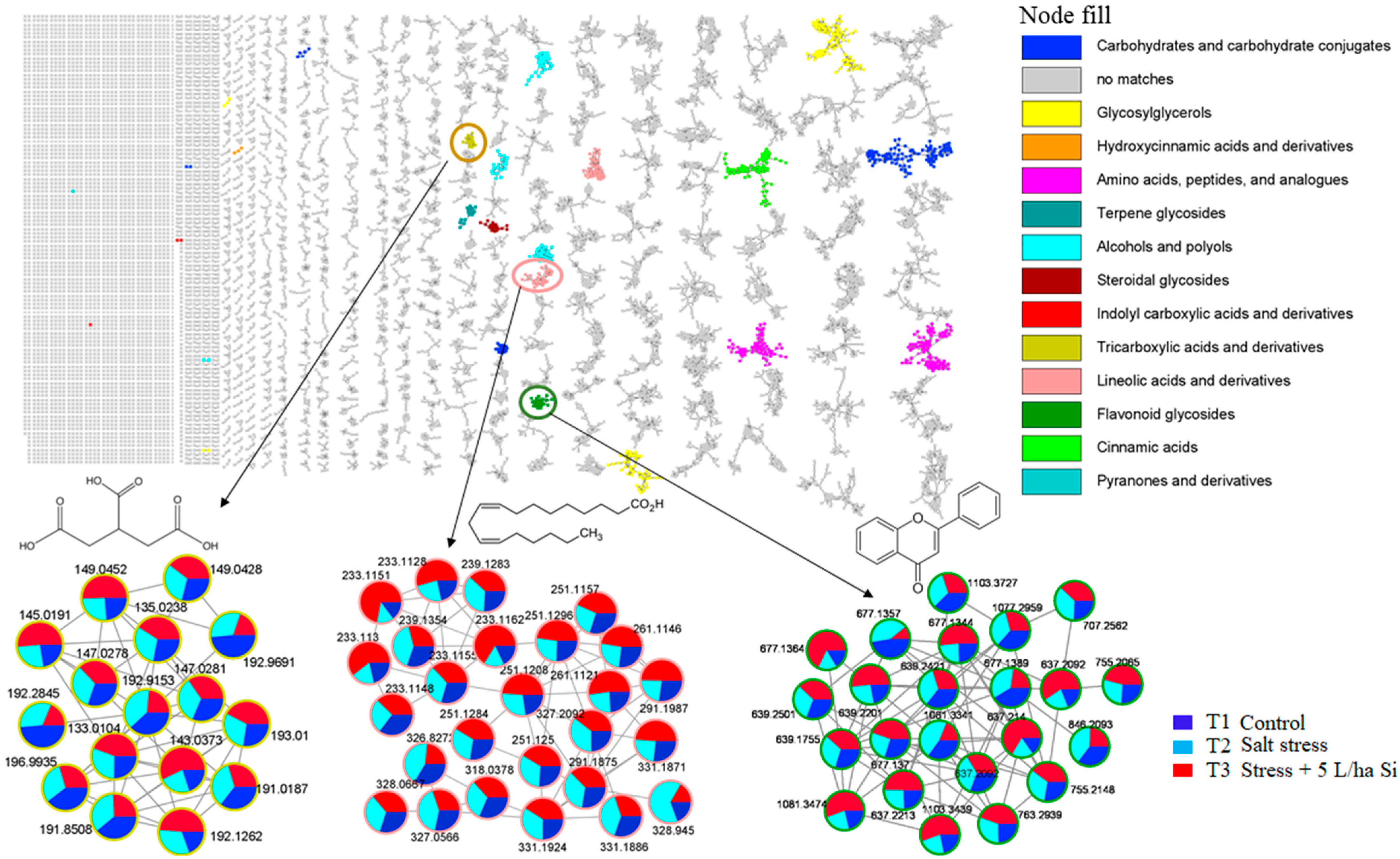
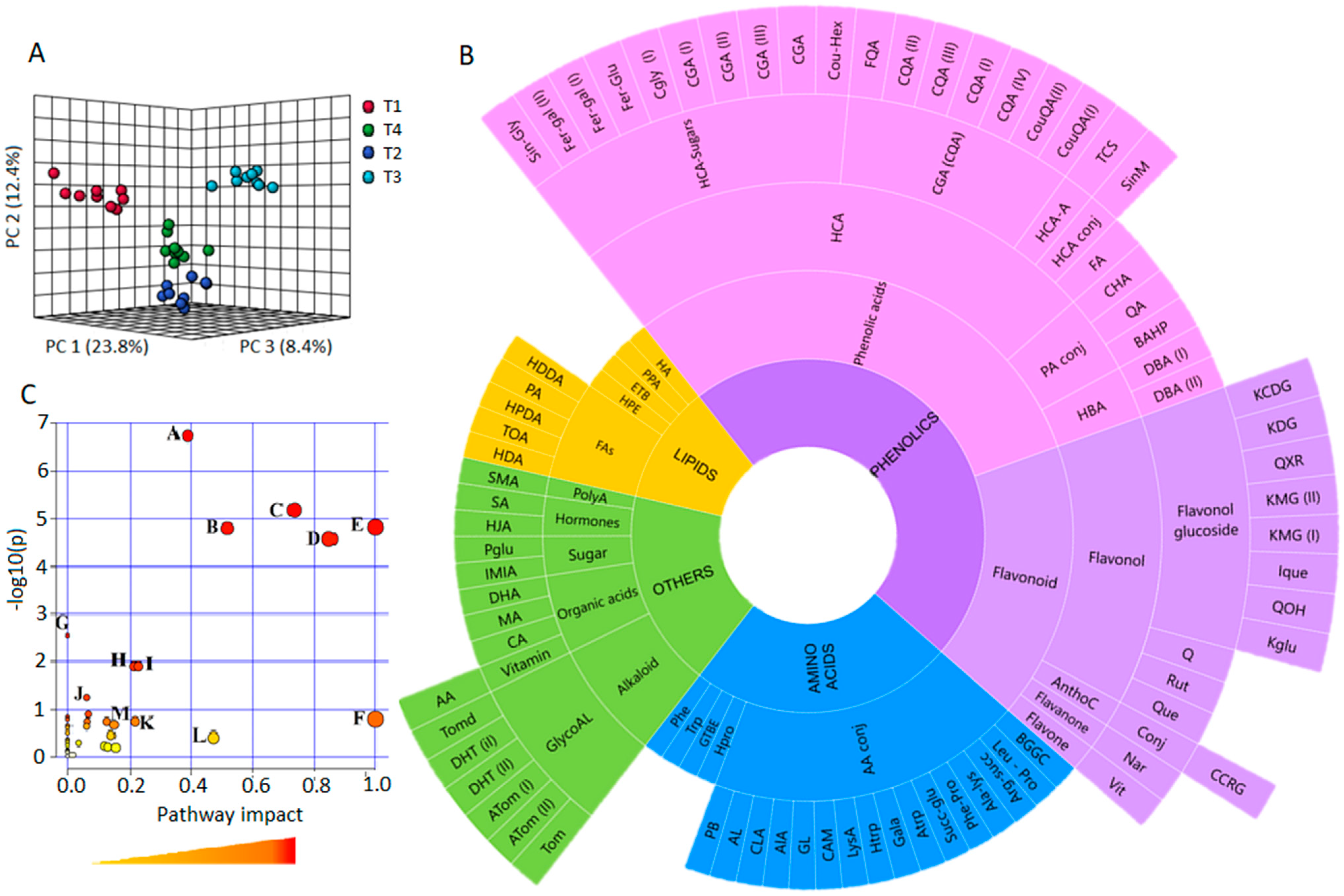
2.1. Metabolome Coverage and Metabolic Pathway/Network Context
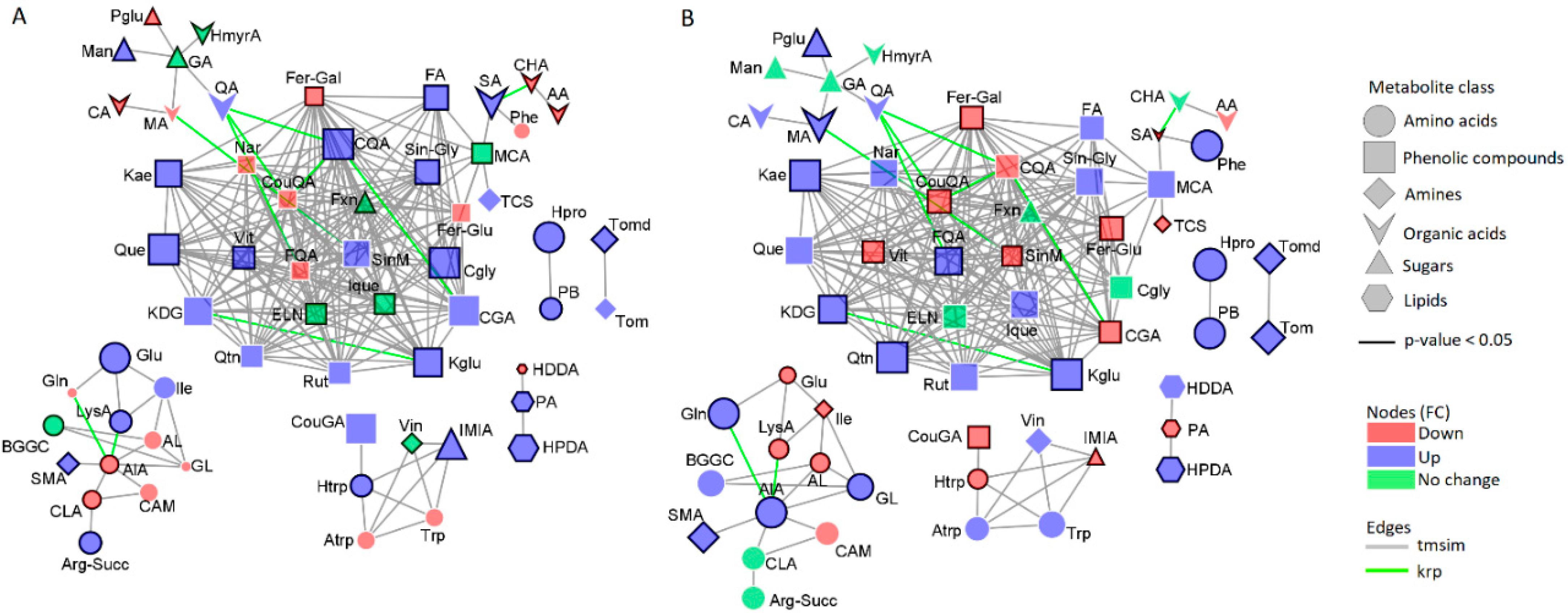
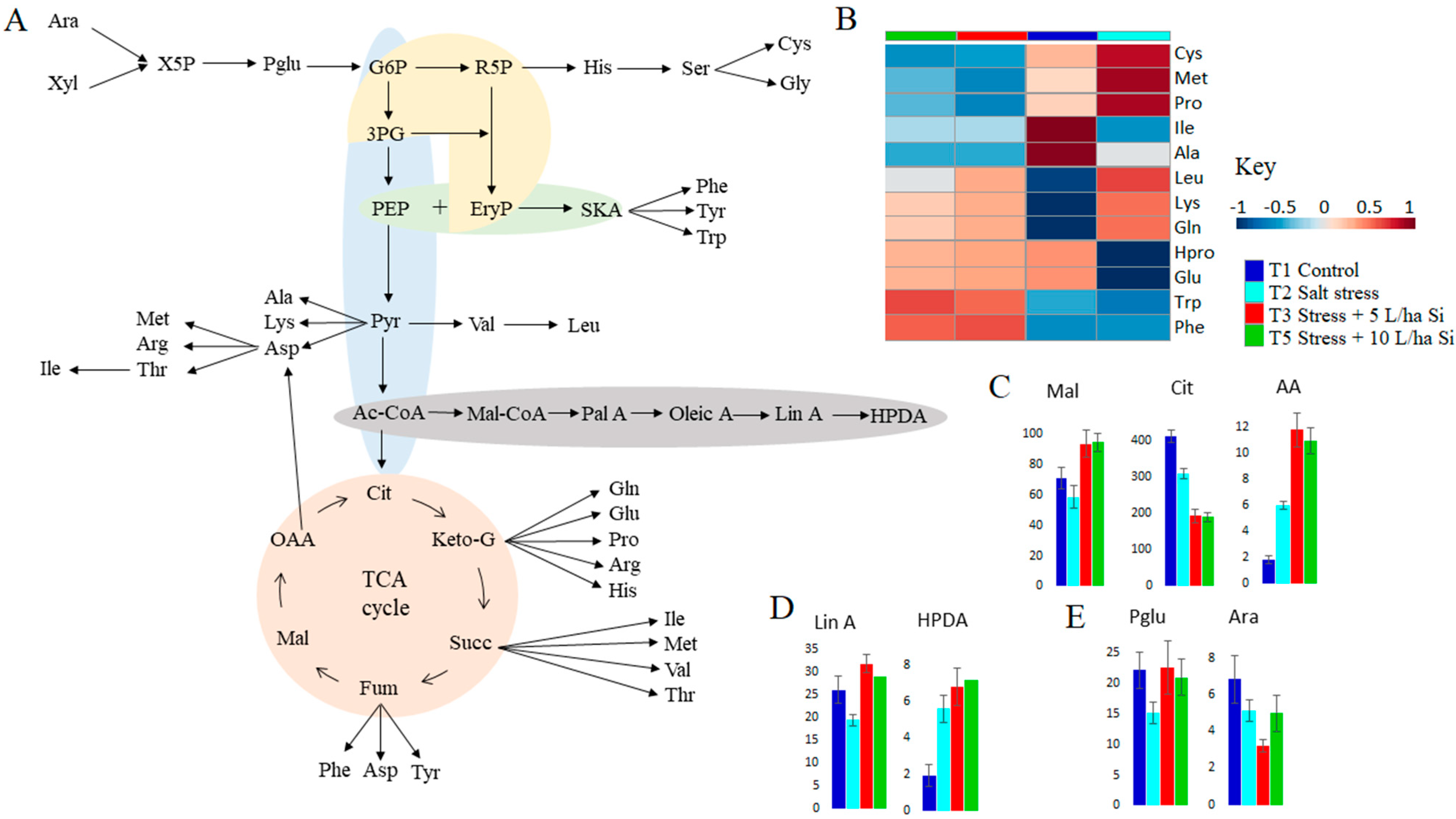
2.2. Primary Metabolism: Roles in Si-Induced Amelioration of Salt Stress Responses
2.2.1. Metabolic Reprogramming of Sugars and Organic Acids in Si Treated Salt Stressed Tomato Plants
2.2.2. Si Induced Modification of Fatty Acid Metabolism in Salt Stressed Tomato Plants
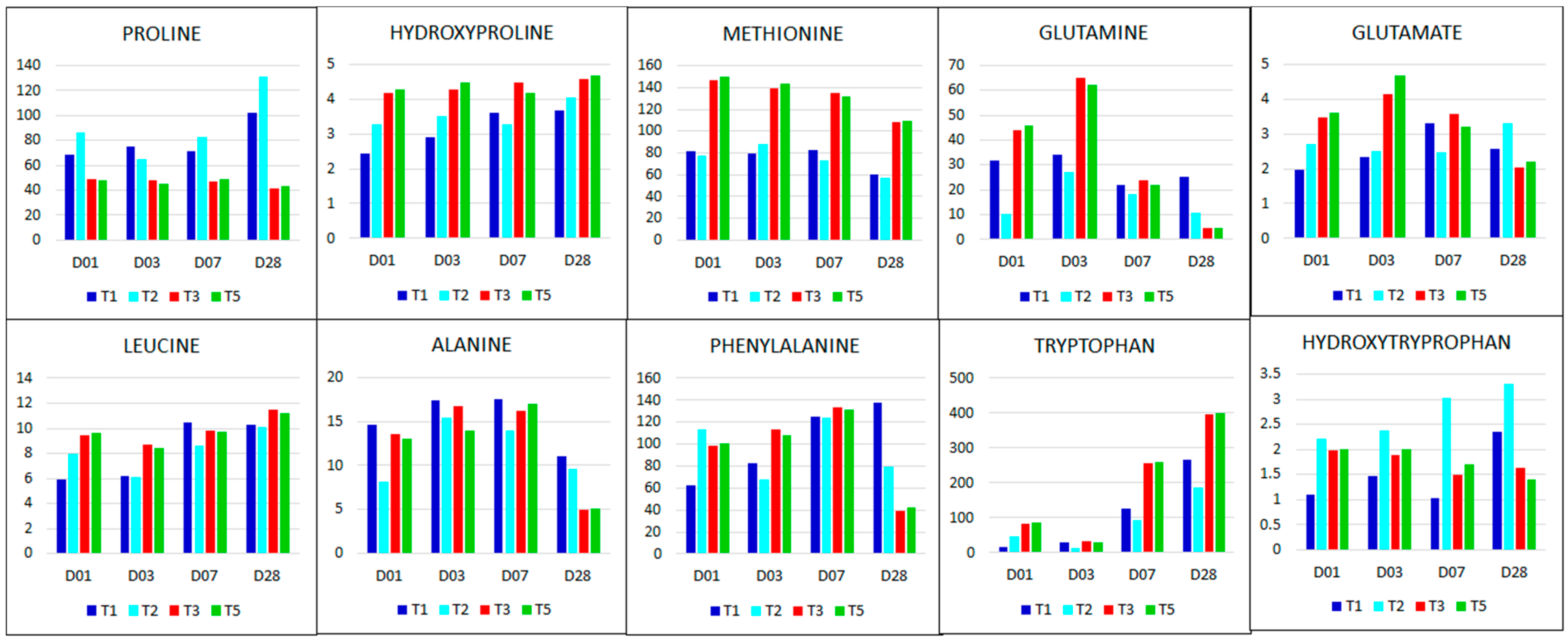
2.2.3. Reconfiguration of Amino Acid Metabolism in Response to Salt Stress and Si Treatment
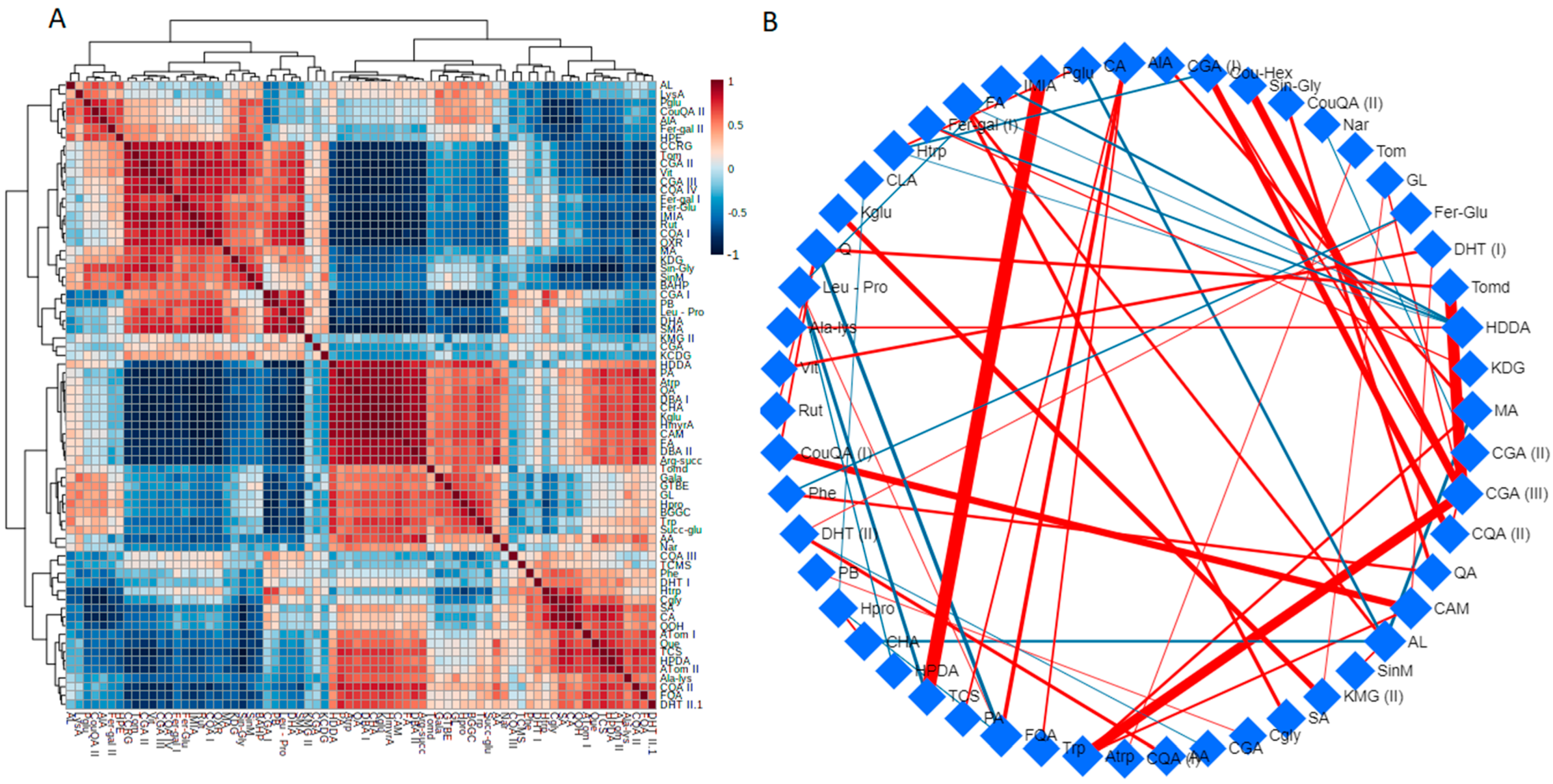
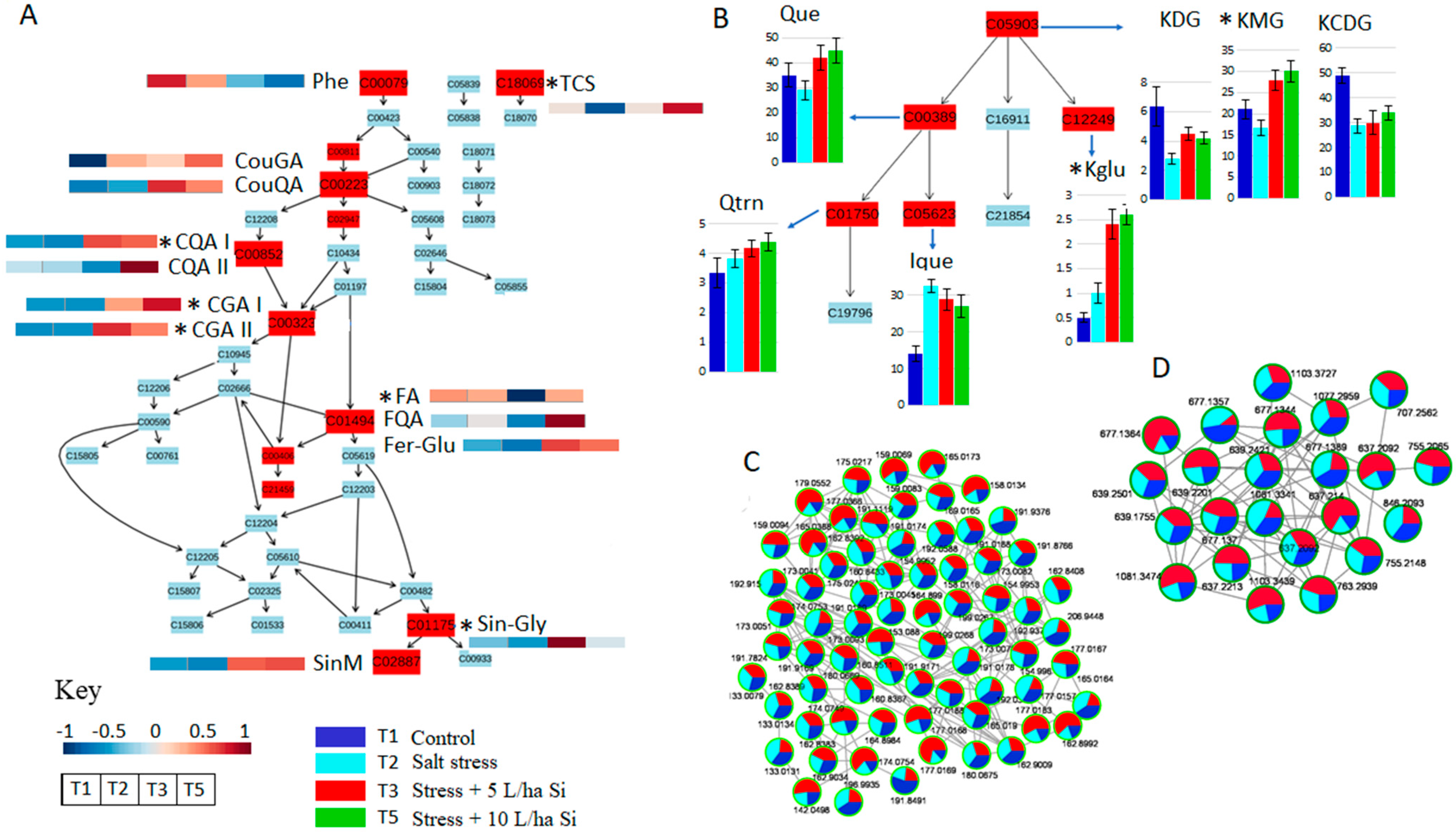
2.3. Si-Based Biostimulant Altered Secondary Metabolism towards the Salt-Stress Alleviation in Tomato Plants
2.3.1. Reconfiguration of Phenolics Metabolism in Response to Salt Stress
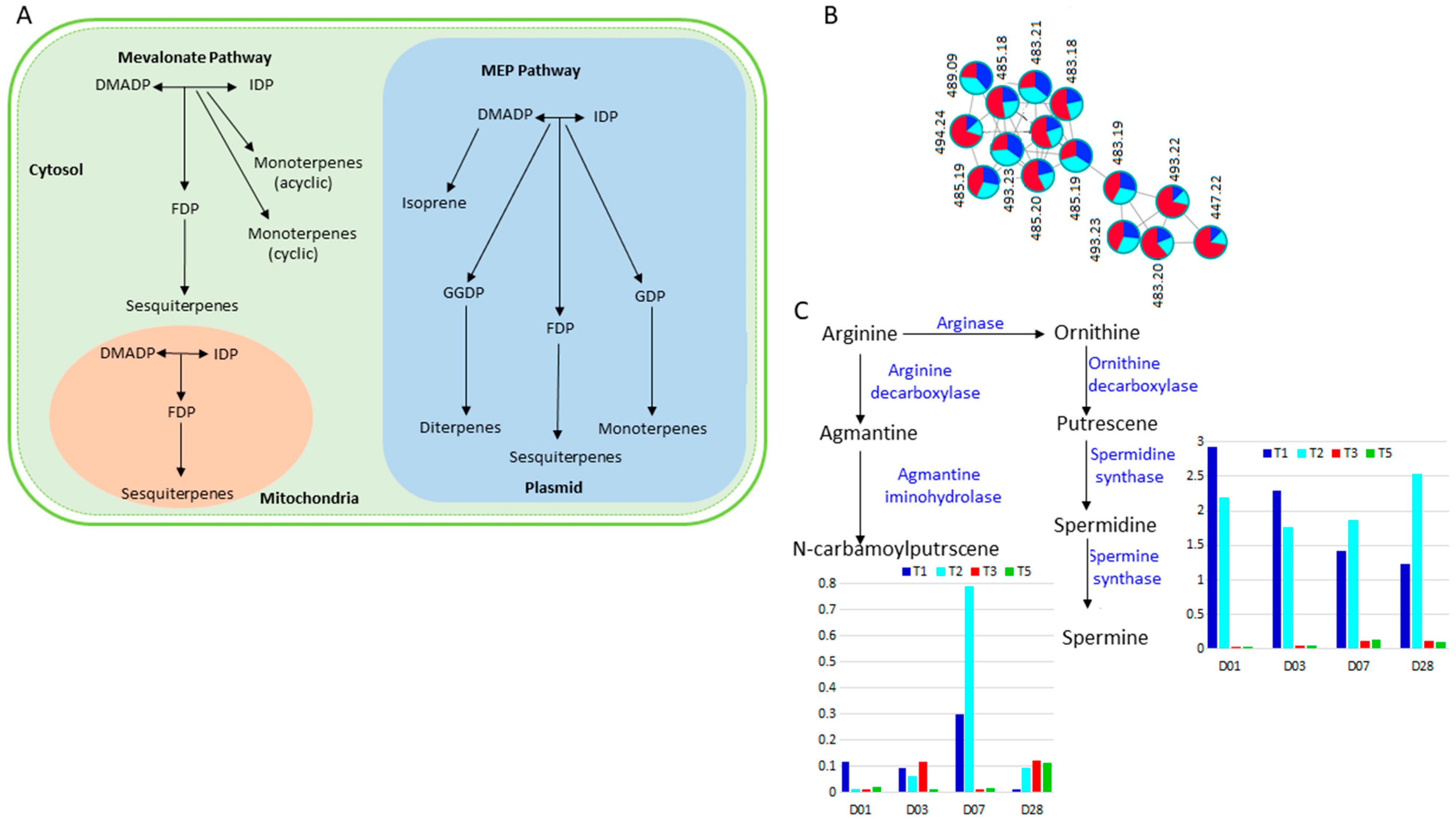
2.3.2. Terpenes and Polyamine Metabolism in Response to Si Treatment of Salt Stressed Plants
3. Materials and Methods
3.1. Plant Cultivation and Treatments
3.2. Harvesting Plant Materials and Metabolite Extraction
3.3. Data Acquisition Using Ultra-High-Performance Liquid Chromatography-High Definition Mass Spectrometry (UHPLC-HDMS)
3.4. Data Mining: Data Processing, Multivariate Data Exploration and Machine Learning-Based Classification
3.5. Molecular Networking
3.6. Metabolite Annotation and Biological Interpretation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, S.K.; Sohn, E.Y.; Hamayun, M.; Yoon, J.Y.; Lee, I.J. Effect of silicon on growth and salinity stress of soybean plant grown under hydroponic system. Agrofor. Syst. 2010, 80, 333–340. [Google Scholar] [CrossRef]
- Manivannan, A.; Soundararajan, P.; Muneer, S.; Ko, C.H.; Jeong, B.R. Silicon Mitigates Salinity Stress by Regulating the Physiology, Antioxidant Enzyme Activities, and Protein Expression in Capsicum annuum ‘Bugwang’. BioMed. Res. Int. 2016, 2016, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018, 60, 796–804. [Google Scholar] [CrossRef] [Green Version]
- Fürtauer, L.; Pschenitschnigg, A.; Scharkosi, H.; Weckwerth, W.; Nägele, T. Combined multivariate analysis and machine learning reveals a predictive module of metabolic stress response in Arabidopsis thaliana. Mol. Omics 2018, 14, 437–449. [Google Scholar] [CrossRef] [Green Version]
- Tebani, A.; Bekri, S. Paving the way to precision nutrition through metabolomics. Front. Nutr. 2019, 6, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Atkinson, N.J.; Lilley, C.J.; Urwin, P.E. Identification of genes involved in the response of arabidopsis to simultaneous biotic and abiotic stresses. Plant Physiol. 2013, 162, 2028–2041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tenhaken, R. Cell wall remodeling under abiotic stress. Front. Plant Sci. 2015, 5, 771. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.; Shahzad, B.; Kumar, V.; Kohli, S.K.; Sidhu, G.P.S.; Bali, A.S.; Handa, N.; Kapoor, D.; Bhardwaj, R.; Zheng, B. Phytohormones regulate accumulation of osmolytes under abiotic stress. Biomolecules 2019, 9, 285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Zelicourt, A.; Colcombet, J.; Hirt, H. The Role of MAPK Modules and ABA during Abiotic Stress Signaling. Trends Plant Sci. 2016, 21, 677–685. [Google Scholar] [CrossRef]
- Ahmad, P.; Prasad, M.N.V. (Eds.) Environmental Adaptations and Stress Tolerance of Plants in the Era of Climate Change; Springer: New York, NY, USA, 2012; ISBN 978-1-4614-0814-7. [Google Scholar]
- Brunetti, C.; George, R.M.; Tattini, M.; Field, K.; Davey, M.P. Metabolomics in plant environmental physiology. J. Exp. Bot. 2013, 64, 4011–4020. [Google Scholar] [CrossRef] [Green Version]
- Gupta, P.; De, B. Metabolomics analysis of rice responses to salinity stress revealed elevation of serotonin, and gentisic acid levels in leaves of tolerant varieties. Plant Signal. Behav. 2017, 12, e1335845. [Google Scholar] [CrossRef]
- Pandey, M.; Penna, S. Time course of physiological, biochemical, and gene expression changes under short-term salt stress in Brassica juncea L. Crop J. 2017, 5, 219–230. [Google Scholar] [CrossRef] [Green Version]
- Evelin, H.; Devi, T.S.; Gupta, S.; Kapoor, R. Mitigation of Salinity Stress in Plants by Arbuscular Mycorrhizal Symbiosis: Current Understanding and New Challenges. Front. Plant Sci. 2019, 10, 470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- du Jardin, P. Plant biostimulants: Definition, concept, main categories and regulation. Sci. Hortic. 2015, 196, 3–14. [Google Scholar] [CrossRef] [Green Version]
- Yakhin, O.I.; Lubyanov, A.A.; Yakhin, I.A.; Brown, P.H. Biostimulants in plant science: A global perspective. Front. Plant Sci. 2017, 7, 2049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricci, M.; Tilbury, L.; Daridon, B.; Sukalac, K. General principles to justify plant biostimulant claims. Front. Plant Sci. 2019, 10, 1–8. [Google Scholar] [CrossRef]
- Le Mire, G.; Nguyen, M.; Fassotte, B.; du Jardin, P.; Verheggen, F.; Delaplace, P.; Jijakli, H. Implementing plant biostimulants and biocontrol strategies in the agroecological management of cultivated ecosystems. Biotechnol. Agron. Soc. Environ. 2016, 20, 299–313. [Google Scholar] [CrossRef]
- Park, S.R.; Kim, H.S.; Lee, K.S.; Hwang, D.J.; Bae, S.C.; Ahn, I.P.; Lee, S.H.; Kim, S.T. Overexpression of rice NAC transcription factor OsNAC58 on increased resistance to bacterial leaf blight. J. Plant Biotechnol. 2017, 44, 149–155. [Google Scholar] [CrossRef] [Green Version]
- Van Oosten, M.J.; Pepe, O.; De Pascale, S.; Silletti, S.; Maggio, A. The role of biostimulants and bioeffectors as alleviators of abiotic stress in crop plants. Chem. Biol. Technol. Agric. 2017, 4, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Yuan, F.; Yang, H.; Xue, Y.; Kong, D.; Ye, R.; Li, C.; Zhang, J.; Theprungsirikul, L.; Shrift, T.; Krichilsky, B.; et al. OSCA1 mediates osmotic-stress-evoked Ca 2+ increases vital for osmosensing in Arabidopsis. Nature 2014, 514, 367–371. [Google Scholar] [CrossRef]
- Al Murad, M.; Khan, A.L.; Muneer, S. Silicon in horticultural crops: Cross-talk, signaling, and tolerance mechanism under Salinity Stress. Plants 2020, 9, 1–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Gong, H. Beneficial effects of silicon on salt and drought tolerance in plants. Agron. Sustain. Dev. 2014, 34, 455–472. [Google Scholar] [CrossRef] [Green Version]
- Fleming, T.R.; Fleming, C.C.; Levy, C.C.B.; Repiso, C.; Hennequart, F.; Nolasco, J.B.; Liu, F. Biostimulants enhance growth and drought tolerance in Arabidopsis thaliana and exhibit chemical priming action. Ann. Appl. Biol. 2019, 174, 153–165. [Google Scholar] [CrossRef]
- Tugizimana, F.; Engel, J.; Salek, R.; Dubery, I.; Piater, L.; Burgess, K. The Disruptive 4IR in the Life Sciences: Metabolomics. In The Disruptive Fourth Industrial Revolution; Doorsamy, W., Paul, B., Marwala, T., Eds.; Springer: Cham, Switzerland, 2020; Volume 674, ISBN 9783030482305. [Google Scholar]
- Miggiels, P.; Wouters, B.; van Westen, G.J.P.; Dubbelman, A.C.; Hankemeier, T. Novel technologies for metabolomics: More for less. TrAC Trends Anal. Chem. 2019, 120, 115323. [Google Scholar] [CrossRef]
- Rouphael, Y.; Colla, G. Editorial: Biostimulants in Agriculture. Front. Plant Sci. 2020, 11, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tugizimana, F.; Steenkamp, P.A.; Piater, L.A.; Labuschagne, N.; Dubery, I.A. Unravelling the metabolic reconfiguration of the post-challenge primed state in sorghum bicolor responding to colletotrichum sublineolum infection. Metabolites 2019, 9, 194. [Google Scholar] [CrossRef] [Green Version]
- Nothias, L.F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Aron, A.T.; Gentry, E.C.; McPhail, K.L.; Nothias, L.F.; Nothias-Esposito, M.; Bouslimani, A.; Petras, D.; Gauglitz, J.M.; Sikora, N.; Vargas, F.; et al. Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nat. Protoc. 2020, 15, 1954–1991. [Google Scholar] [CrossRef]
- Wang, M.; Jarmusch, A.K.; Vargas, F.; Aksenov, A.A.; Gauglitz, J.M.; Weldon, K.; Petras, D.; da Silva, R.; Quinn, R.; Melnik, A.V.; et al. Mass spectrometry searches using MASST. Nat. Biotechnol. 2020, 38, 23–26. [Google Scholar] [CrossRef]
- Watrous, J.; Roach, P.; Alexandrov, T.; Heath, B.S.; Yang, J.Y.; Kersten, R.D.; Van Der Voort, M.; Pogliano, K.; Gross, H.; Raaijmakers, J.M.; et al. Mass spectral molecular networking of living microbial colonies. Proc. Natl. Acad. Sci. USA 2012, 109, E1743–E1752. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oppong-Danquah, E.; Parrot, D.; Blümel, M.; Labes, A.; Tasdemir, D. Molecular networking-based metabolome and bioactivity analyses of marine-adapted fungi co-cultivated with phytopathogens. Front. Microbiol. 2018, 9, 1–20. [Google Scholar] [CrossRef]
- Hegazi, N.M.; Radwan, R.A.; Bakry, S.M.; Saad, H.H. Molecular networking aided metabolomic profiling of beet leaves using three extraction solvents and in relation to its anti-obesity effects. J. Adv. Res. 2020, 24, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Madsen, R.; Lundstedt, T.; Trygg, J. Chemometrics in metabolomics—A review in human disease diagnosis. Anal. Chim. Acta 2010, 659, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Duren, W.; Evans, C.R.; Burant, C.F.; Michailidis, G.; Karnovsky, A. Sparse network modeling and metscape-based visualization methods for the analysis of large-scale metabolomics data. Bioinformatics 2017, 33, 1545–1553. [Google Scholar] [CrossRef]
- Bartel, J.; Krumsiek, J.; Schramm, K.; Adamski, J.; Gieger, C.; Herder, C.; Carstensen, M.; Peters, A.; Rathmann, W.; Roden, M.; et al. The Human Blood Metabolome-Transcriptome Interface. PLoS Genet. 2015, 11, 1–32. [Google Scholar] [CrossRef]
- Kotze, H.L.; Armitage, E.G.; Sharkey, K.J.; Allwood, J.W.; Dunn, W.B.; Williams, K.J.; Goodacre, R. A novel untargeted metabolomics correlation-based network analysis incorporating human metabolic reconstructions. BMC Syst. Biol. 2013, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Rosato, A.; Tenori, L.; Cascante, M.; Carulla, P.R.D.A.; Dos Santos, V.A.M.; Saccenti, E. From correlation to causation: Analysis of metabolomics data using systems biology approaches. Metabolomics 2018, 14, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Schwahn, K.; Beleggia, R.; Omranian, N.; Nikoloski, Z. Stoichiometric correlation analysis: Principles of metabolic functionality from metabolomics data. Front. Plant Sci. 2017, 8, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Toubiana, D.; Puzis, R.; Wen, L.; Sikron, N.; Kurmanbayeva, A.; Soltabayeva, A.; Wilhelmi, M.D.M.R.; Sade, N.; Fait, A.; Sagi, M.; et al. Combined network analysis and machine learning allows the prediction of metabolic pathways from tomato metabolomics data. Commun. Biol. 2019, 2, 214. [Google Scholar] [CrossRef] [Green Version]
- Asensi-Fabado, M.A.; Amtmann, A.; Perrella, G. Plant responses to abiotic stress: The chromatin context of transcriptional regulation. Biochim. Et Biophys. Acta (BBA)-Gene Regul. Mech. 2017, 1860, 106–122. [Google Scholar] [CrossRef] [Green Version]
- Goodwin, C.R.; Covington, B.C.; Derewacz, D.K.; McNees, C.R.; Wikswo, J.P.; McLean, J.A.; Bachmann, B.O. Structuring microbial metabolic responses to multiplexed stimuli via self-organizing metabolomics maps. Chem. Biol. 2015, 22, 661–670. [Google Scholar] [CrossRef] [Green Version]
- Frew, A.; Weston, L.A.; Reynolds, O.L.; Gurr, G.M. The role of silicon in plant biology: A paradigm shift in research approach. Ann. Bot. 2018, 121, 1265–1273. [Google Scholar] [CrossRef] [Green Version]
- Isah, T. Stress and defense responses in plant secondary metabolites production. Biol. Res. 2019, 52, 39. [Google Scholar] [CrossRef] [Green Version]
- Erb, M.; Kliebenstein, D.J. Plant Secondary Metabolites as Defenses, Regulators, and Primary Metabolites: The Blurred Functional Trichotomy. Plant Physiol. 2020, 184, 39–52. [Google Scholar] [CrossRef]
- Rojas, C.M.; Senthil-Kumar, M.; Tzin, V.; Mysore, K.S. Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant Sci. 2014, 5, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Rapala-Kozik, M.; Kowalska, E.; Ostrowska, K. Modulation of thiamine metabolism in Zea mays seedlings under conditions of abiotic stress. J. Exp. Bot. 2008, 59, 4133–4143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corpas, F.J.; Barroso, J.B. NADPH-generating dehydrogenases: Their role in the mechanism of protection against nitro-oxidative stress induced by adverse environmental conditions. Front. Environ. Sci. 2014, 2, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Rizhsky, L.; Liang, H.; Mittler, R. The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol. 2002, 130, 1143–1151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karpiński, S.; Szechyńska-Hebda, M.; Wituszyńska, W.; Burdiak, P. Light acclimation, retrograde signalling, cell death and immune defences in plants. Plant Cell Environ. 2013, 36, 736–744. [Google Scholar] [CrossRef] [PubMed]
- Phoboo, S.; Sarkar, D.; Bhowmik, P.C.; Jha, P.K.; Shetty, K. Improving salinity resilience in Swertia chirayita clonal line with Lactobacillus plantarum. Can. J. Plant Sci. 2016, 96, 117–127. [Google Scholar] [CrossRef] [Green Version]
- Le Gall, G.; Colquhoun, I.J.; Davis, A.L.; Collins, G.J.; Verhoeyen, M.E. Metabolite profiling of tomato (Lycopersicon esculentum) using 1H NMR spectroscopy as a tool to detect potential unintended effects following a genetic modification. J. Agric. Food Chem. 2003, 51, 2447–2456. [Google Scholar] [CrossRef]
- Foyer, C.H. Reactive oxygen species, oxidative signaling and the regulation of photosynthesis. Environ. Exp. Bot. 2018, 154, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Ali, S.; Zandi, P.; Mehmood, A.; Ullah, S.; Ikram, M.; Ismail, M.A.S.; Babar, M.A. Role of sugars, amino acids and organic acids in improving plant abiotic stress tolerance. Pak. J. Bot. 2020, 52, 355–363. [Google Scholar] [CrossRef]
- Kövilein, A.; Kubisch, C.; Cai, L.; Ochsenreither, K. Malic acid production from renewables: A review. J. Chem. Technol. Biotechnol. 2020, 95, 513–526. [Google Scholar] [CrossRef] [Green Version]
- Schwachtje, J.; Fischer, A.; Erban, A.; Kopka, J. Primed primary metabolism in systemic leaves: A functional systems analysis. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Alseekh, S.; Fernie, A.R.; Luo, J. The Structure and Function of Major Plant Metabolite Modifications. Mol. Plant 2019, 12, 899–919. [Google Scholar] [CrossRef]
- Smirnoff, N. The function and metabolism of ascorbic acid in plants. Ann. Bot. 1996, 78, 661–669. [Google Scholar] [CrossRef] [Green Version]
- Das, K.; Roychoudhury, A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2014, 2, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Guo, R.; Shi, L.X.; Yan, C.; Zhong, X.; Gu, F.X.; Liu, Q.; Xia, X.; Li, H. Ionomic and metabolic responses to neutral salt or alkaline salt stresses in maize (Zea mays L.) seedlings. BMC Plant Biol. 2017, 17, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.; Long, M. A cytosolic NADP-malic enzyme gene from rice (Oryza sativa L.) confers salt tolerance in transgenic Arabidopsis. Biotechnol. Lett. 2007, 29, 1129–1134. [Google Scholar] [CrossRef] [PubMed]
- De Aragao, M.F.; Jolivet, Y.; Lima, M.G.S.; De Melo, D.F.; Dizengremel, P. NaCl-induced changes of NAD(P) malic enzyme activities in Eucalyptus citriodora leaves. Trees 1997, 12, 66–72. [Google Scholar] [CrossRef]
- Bybordi, A. Effect of Ascorbic Acid and Silicium on Photosynthesis, Antioxidant Enzyme Activity, and Fatty Acid Contents in Canola Exposure to Salt Stress. J. Integr. Agric. 2012, 11, 1610–1620. [Google Scholar] [CrossRef]
- Das, P.; Manna, I.; Sil, P.; Bandyopadhyay, M.; Biswas, A.K. Exogenous silicon alters organic acid production and enzymatic activity of TCA cycle in two NaCl stressed indica rice cultivars. Plant Physiol. Biochem. 2019, 136, 76–91. [Google Scholar] [CrossRef]
- Fougère, F.; Le Rudulier, D.; Streeter, J.G. Effects of salt stress on amino acid, organic acid, and carbohydrate composition of roots, bacteroids, and cytosol of alfalfa (Medicago sativa L.). Plant Physiol. 1991, 96, 1228–1236. [Google Scholar] [CrossRef] [Green Version]
- Yan, G.; Fan, X.; Peng, M.; Yin, C.; Xiao, Z.; Liang, Y. Silicon Improves Rice Salinity Resistance by Alleviating Ionic Toxicity and Osmotic Constraint in an Organ-Specific Pattern. Front. Plant Sci. 2020, 11, 1–12. [Google Scholar] [CrossRef]
- Ying, J.Z.; Shan, J.X.; Gao, J.P.; Zhu, M.Z.; Shi, M.; Lin, H.X. Identification of quantitative trait loci for lipid metabolism in rice seeds. Mol. Plant 2012, 5, 865–875. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Ma, D.; Zhang, Z.; Wang, S.; Du, S.; Deng, X.; Yin, L. Plant lipid remodeling in response to abiotic stresses. Environ. Exp. Bot. 2019, 165, 174–184. [Google Scholar] [CrossRef]
- Shaki, F.; Maboud, H.E.; Niknam, V. Effects of salicylic acid on hormonal cross talk, fatty acids profile, and ions homeostasis from salt-stressed safflower. J. Plant Interact. 2019, 14, 340–346. [Google Scholar] [CrossRef] [Green Version]
- Rasmussen, M.W.; Roux, M.; Petersen, M.; Mundy, J. MAP Kinase Cascades in Arabidopsis Innate Immunity. Front. Plant Sci. 2012, 3, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Asgher, M.; Khan, M.I.R.; Anjum, N.A.; Khan, N.A. Minimising toxicity of cadmium in plants—role of plant growth regulators. Protoplasma 2015, 252, 399–413. [Google Scholar] [CrossRef]
- Jagodzik, P.; Tajdel-Zielinska, M.; Ciesla, A.; Marczak, M.; Ludwikow, A. Mitogen-Activated Protein Kinase Cascades in Plant Hormone Signaling. Front. Plant Sci. 2018, 9, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Aziz, A.; Siti-Fairuz, M.; Abdullah, M.Z.; Ma, N.L.; Marziah, M. Fatty acid profile of salinity tolerant rice genotypes grown on saline soil. Malays. Appl. Biol. 2015, 44, 119–124. [Google Scholar]
- Abdel Latef, A.A.; Tran, L.S.P. Impacts of priming with silicon on the growth and tolerance of maize plants to alkaline stress. Front. Plant Sci. 2016, 7, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishna, R.; Karkute, S.G.; Ansari, W.A.; Jaiswal, D.K.; Verma, J.P.; Singh, M. Transgenic tomatoes for abiotic stress tolerance: Status and way ahead. 3 Biotech 2019, 9, 1–14. [Google Scholar] [CrossRef]
- Slama, I.; Abdelly, C.; Bouchereau, A.; Flowers, T.; Savouré, A. Diversity, distribution and roles of osmoprotective compounds accumulated in halophytes under abiotic stress. Ann. Bot. 2015, 115, 433–447. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Duan, G.; Li, C.; Liu, L.; Han, G.; Zhang, Y.; Wang, C. The Crosstalks Between Jasmonic Acid and Other Plant Hormone Signaling Highlight the Involvement of Jasmonic Acid as a Core Component in Plant Response to Biotic and Abiotic Stresses. Front. Plant Sci. 2019, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Madkour, M.A.; Smith, L.T.; Smith, G.M. Preferential osmolyte accumulation: A mechanism of osmotic stress adaptation in diazotrophic bacteria. Appl. Environ. Microbiol. 1990, 56, 2876–2881. [Google Scholar] [CrossRef] [Green Version]
- Hare, P.D.; Cress, W.A.; Van Staden, J. Dissecting the roles of osmolyte accumulation during stress. Plant Cell Environ. 1998, 21, 535–553. [Google Scholar] [CrossRef]
- García-Mauriño, S.; Monreal, J.A.; Alvarez, R.; Vidal, J.; Echevarría, C. Characterization of salt stress-enhanced phosphoenolpyruvate carboxylase kinase activity in leaves of Sorghum vulgare: Independence from osmotic stress, involvement of ion toxicity and significance of dark phosphorylation. Planta 2003, 216, 648–655. [Google Scholar] [CrossRef]
- Ferrario, M.; Brunelli, L.; Su, F.; Herpain, A.; Pastorelli, R. The systemic alterations of lipids, alanine-glucose cycle and inter-organ amino acid metabolism in swine model confirms the role of liver in early phase of septic shock. Front. Physiol. 2019, 10, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parthasarathy, A.; Savka, M.A.; Hudson, A.O. The synthesis and role of β-alanine in plants. Front. Plant Sci. 2019, 10, 921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rastogi, S.; Shah, S.; Kumar, R.; Vashisth, D.; Akhtar, M.Q.; Kumar, A.; Dwivedi, U.N.; Shasany, A.K. Ocimum metabolomics in response to abiotic stresses: Cold, flood, drought and salinity. PLoS ONE 2019, 14, e0210903. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zeng, X.; Xu, Q.; Mei, X.; Yuan, H.; Jiabu, D.; Sang, Z.; Nyima, T. Metabolite profiling in two contrasting Tibetan hulless barley cultivars revealed the core salt-responsive metabolome and key salt-tolerance biomarkers. AoB Plants 2019, 11, plz021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ballester, A.R.; Tikunov, Y.; Molthoff, J.; Grandillo, S.; Viquez-Zamora, M.; de Vos, R.; de Maagd, R.A.; van Heusden, S.; Bovy, A.G. Identification of loci affecting accumulation of secondary metabolites in tomato fruit of a Solanum lycopersicum × Solanum chmielewskii introgression line population. Front. Plant Sci. 2016, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Michaletti, A.; Naghavi, M.R.; Toorchi, M.; Zolla, L.; Rinalducci, S. Metabolomics and proteomics reveal drought-stress responses of leaf tissues from spring-wheat. Sci. Rep. 2018, 8, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Naikoo, M.I.; Dar, M.I.; Raghib, F.; Jaleel, H.; Ahmad, B.; Raina, A.; Khan, F.A.; Naushin, F. Role and regulation of plants phenolics in abiotic stress tolerance: An overview. Plant Signal. Mol. 2019, 157–168. [Google Scholar] [CrossRef]
- Tohge, T.; Fernie, A.R. Metabolomics-Inspired Insight into Developmental, Environmental and Genetic Aspects of Tomato Fruit Chemical Composition and Quality. Plant Cell Physiol. 2014, 56, 1681–1696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, J.; Yang, L.; Zhang, D.; Shi, J. Plant metabolomics: An indispensable system biology tool for plant science. Int. J. Mol. Sci. 2016, 17, 767. [Google Scholar] [CrossRef]
- Marcec, M.J.; Gilroy, S.; Poovaiah, B.W.; Tanaka, K. Mutual interplay of Ca2+ and ROS signaling in plant immune response. Plant Sci. 2019, 283, 343–354. [Google Scholar] [CrossRef]
- Etalo, D.; de Vos, R.C.; Joosten, M.H.A.J.; Hall, R. Spatially-resolved plant metabolomics: Some potentials and limitations of Laser-Ablation Electrospray Ionization (LAESI) Mass Spectrometry metabolite imaging. Plant Physiol. 2015, 169, 1424–1435. [Google Scholar] [CrossRef] [Green Version]
- Bouriche, H.; Kada, S.; Senator, A.; demirtas, I.; Ozen, T.; Toptanci, B.Ç.; Kizil, G.; Kizil, M. Phenolic Content and Biomolecule Oxidation Protective Activity of Globularia alypum Extracts. Braz. Arch. Biol. Technol. 2017, 60, 1–15. [Google Scholar] [CrossRef]
- Salehi, B.; Azzini, E.; Zucca, P.; Varoni, E.M.; Kumar, N.V.A.; Dini, L.; Panzarini, E.; Rajkovic, J.; Fokou, P.V.T.; Peluso, I.; et al. Plant-derived bioactives and oxidative stress-related disorders: A key trend towards healthy aging and longevity promotion. Appl. Sci. 2020, 10, 947. [Google Scholar] [CrossRef] [Green Version]
- Colaric, M.; Stampar, F.; Hudina, M. Changes in sugars and phenolics concentrations of Williams pear leaves during the growing season. Can. J. Plant Sci. 2006, 86, 1203–1208. [Google Scholar] [CrossRef]
- Diamond, S.; Rubin, B.E.; Shultzaberger, R.K.; Chen, Y.; Barber, C.D.; Golden, S.S. Redox crisis underlies conditional light-dark lethality in cyanobacterial mutants that lack the circadian regulator, RpaA. Proc. Natl. Acad. Sci. USA 2017, 114, E580–E589. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Zayed, O.; Zeng, F.; Liu, C.; Zhang, L.; Zhu, P.; Hsu, C.C.; Tuncil, Y.E.; Tao, W.A.; Carpita, N.C.; et al. Arabinose biosynthesis is critical for salt stress tolerance in Arabidopsis. New Phytol. 2019, 224, 274–290. [Google Scholar] [CrossRef]
- Sharma, A.; Kumar, V.; Yuan, H.; Kanwar, M.K.; Bhardwaj, R.; Thukral, A.K.; Zheng, B. Jasmonic acid seed treatment stimulates insecticide detoxification in Brassica juncea L. Front. Plant Sci. 2018, 871, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Maksymiec, W. Signaling responses in plants to heavy metal stress. Acta Physiol. Plant. 2007, 29, 177–187. [Google Scholar] [CrossRef]
- He, J.; Duan, Y.; Hua, D.; Fan, G.; Wang, L.; Liu, Y.; Chen, Z.; Han, L.; Qu, L.-J.; Gong, Z. DEXH Box RNA Helicase-Mediated Mitochondrial Reactive Oxygen Species Production in Arabidopsis Mediates Crosstalk between Abscisic Acid and Auxin Signaling. Plant Cell 2012, 24, 1815–1833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sampaio, B.L.; Edrada-Ebel, R.; Da Costa, F.B. Effect of the environment on the secondary metabolic profile of Tithonia diversifolia: A model for environmental metabolomics of plants. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.; Khan, A.L.; Muneer, S.; Kim, Y.H.; Al-Rawahi, A.; Al-Harrasi, A. Silicon and Salinity: Crosstalk in Crop-Mediated Stress Tolerance Mechanisms. Front. Plant Sci. 2019, 10, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Aras, S.; Keles, H.; Eşitken, A. Silicon nutrition counteracts salt-induced damage associated with changes in biochemical responses in apple. Bragantia 2020, 79, 1–7. [Google Scholar] [CrossRef]
- Ashour, H.A.; Abdel Wahab, M.; Mahmoud, A.W.M. Response of Jatropha integerrima Plants Irrigated with Different Levels of Saline Water to Nano Silicon and Gypsum. J. Agric. Stud. 2017, 5, 136. [Google Scholar] [CrossRef] [Green Version]
- Boncan, D.A.T.; Tsang, S.S.K.; Li, C.; Lee, I.H.T.; Lam, H.M.; Chan, T.F.; Hui, J.H.L. Terpenes and terpenoids in plants: Interactions with environment and insects. Int. J. Mol. Sci. 2020, 21, 7382. [Google Scholar] [CrossRef]
- Pazouki, L.; Niinemetst, U. Multi-substrate terpene synthases: Their occurrence and physiological significance. Front. Plant Sci. 2016, 7, 1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Odimegwu, J.I.; Odukoya, O.; Yadav, R.K.; Chanotiya, C.S.; Ogbonnia, S.; Sangwan, N.S. A new source of elemol rich essential oil and existence of multicellular oil glands in leaves of the dioscorea species. Sci. World J. 2013, 2013, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Romero-Aranda, M.R.; Jurado, O.; Cuartero, J. Silicon alleviates the deleterious salt effect on tomato plant growth by improving plant water status. J. Plant Physiol. 2006, 163, 847–855. [Google Scholar] [CrossRef]
- Ramasamy, J.; Kandasamy, R.; Palanisamy, S.; Nadesan, S. Optimization of Ultrasonic-Assisted Extraction of Flavonoids and Anti-oxidant Capacity from the Whole Plant of Andrographis echioides (L.) Nees by Response Surface Methodology and Chemical Composition Analysis. Pharmacogn. Mag. 2019, 15, 547–556. [Google Scholar] [CrossRef]
- Wang, S.; Liu, P.; Chen, D.; Yin, L.; Li, H.; Deng, X. Silicon enhanced salt tolerance by improving the root water uptake and decreasing the ion toxicity in cucumber. Front. Plant Sci. 2015, 6, 759. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.X.; Gong, H.J.; Yin, J.L. Role of silicon in mediating salt tolerance in plants: A Review. Plants 2019, 8, 147. [Google Scholar] [CrossRef] [Green Version]
- Cvikrová, M.; Gemperlová, L.; Martincová, O.; Vanková, R. Effect of drought and combined drought and heat stress on polyamine metabolism in proline-over-producing tobacco plants. Plant Physiol. Biochem. 2013, 73, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, M.; Ashraf, M. Gibberellic acid mediated induction of salt tolerance in wheat plants: Growth, ionic partitioning, photosynthesis, yield and hormonal homeostasis. Environ. Exp. Bot. 2013, 86, 76–85. [Google Scholar] [CrossRef]
- Paul, K.; Sorrentino, M.; Lucini, L.; Rouphael, Y.; Cardarelli, M.; Bonini, P.; Reynaud, H.; Canaguier, R.; Trtílek, M.; Panzarová, K.; et al. Understanding the biostimulant action of vegetal-derived protein hydrolysates by high-throughput plant phenotyping and metabolomics: A case study on tomato. Front. Plant Sci. 2019, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Tugizimana, F.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. A conversation on data mining strategies in LC-MS untargeted metabolomics: Pre-processing and pre-treatment steps. Metabolites 2016, 6, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faull, K.F.; Dooley, A.N.; Halgand, F.; Lorelei, D.; Norris, A.J.; Ryan, C.M.; Laganowsky, A.; Johnson, J.V.; Katz, J.E. An Introduction to the Basic Principles and Concepts of Mass Spectrometry. Compr. Anal. Chem. 2009, 52, 1–46. [Google Scholar] [CrossRef]
- Quan, N.T.; Anh, L.H.; Khang, D.T.; Tuyen, P.T.; Toan, N.P.; Minh, T.N.; The Minh, L.; Bach, D.T.; Thu Ha, P.T.; Elzaawely, A.A.; et al. Involvement of secondary metabolites in response to drought stress of rice (Oryza sativa L.). Agriculture 2016, 6, 23. [Google Scholar] [CrossRef] [Green Version]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; Vandergheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Hettne, K.M.; Williams, A.J.; van Mulligen, E.M.; Kleinjans, J.; Tkachenko, V.; Kors, J.A. Automatic vs. manual curation of a multi-source chemical dictionary: The impact on text mining. J. Cheminform 2010, 2, 3. [Google Scholar] [CrossRef] [Green Version]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [Green Version]

| Treatment (T) | Treatment Description | Application Rate (L/ha) |
|---|---|---|
| T1 | Control: No salt stress application, no biostimulant application | 0 |
| T2 | Control: Salt-stress application and no biostimulant application | 0 |
| T3 | Salt stress application and soil applied biostimulant | 5 |
| T4 | Salt stress application and foliar applied biostimulant | 5 |
| T5 | Salt stress application and soil applied biostimulant | 10 |
| T6 | Salt stress application and foliar applied biostimulant | 10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chele, K.H.; Steenkamp, P.; Piater, L.A.; Dubery, I.A.; Huyser, J.; Tugizimana, F. A Global Metabolic Map Defines the Effects of a Si-Based Biostimulant on Tomato Plants under Normal and Saline Conditions. Metabolites 2021, 11, 820. https://doi.org/10.3390/metabo11120820
Chele KH, Steenkamp P, Piater LA, Dubery IA, Huyser J, Tugizimana F. A Global Metabolic Map Defines the Effects of a Si-Based Biostimulant on Tomato Plants under Normal and Saline Conditions. Metabolites. 2021; 11(12):820. https://doi.org/10.3390/metabo11120820
Chicago/Turabian StyleChele, Kekeletso H., Paul Steenkamp, Lizelle A. Piater, Ian A. Dubery, Johan Huyser, and Fidele Tugizimana. 2021. "A Global Metabolic Map Defines the Effects of a Si-Based Biostimulant on Tomato Plants under Normal and Saline Conditions" Metabolites 11, no. 12: 820. https://doi.org/10.3390/metabo11120820
APA StyleChele, K. H., Steenkamp, P., Piater, L. A., Dubery, I. A., Huyser, J., & Tugizimana, F. (2021). A Global Metabolic Map Defines the Effects of a Si-Based Biostimulant on Tomato Plants under Normal and Saline Conditions. Metabolites, 11(12), 820. https://doi.org/10.3390/metabo11120820







