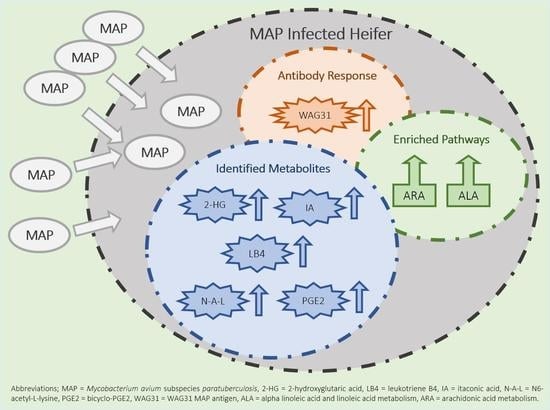
Metabolomic Changes in Naturally MAP-Infected Holstein–Friesian Heifers Indicate Immunologically Related Biochemical Reprogramming
Abstract
:1. Introduction
2. Results
3. Discussion
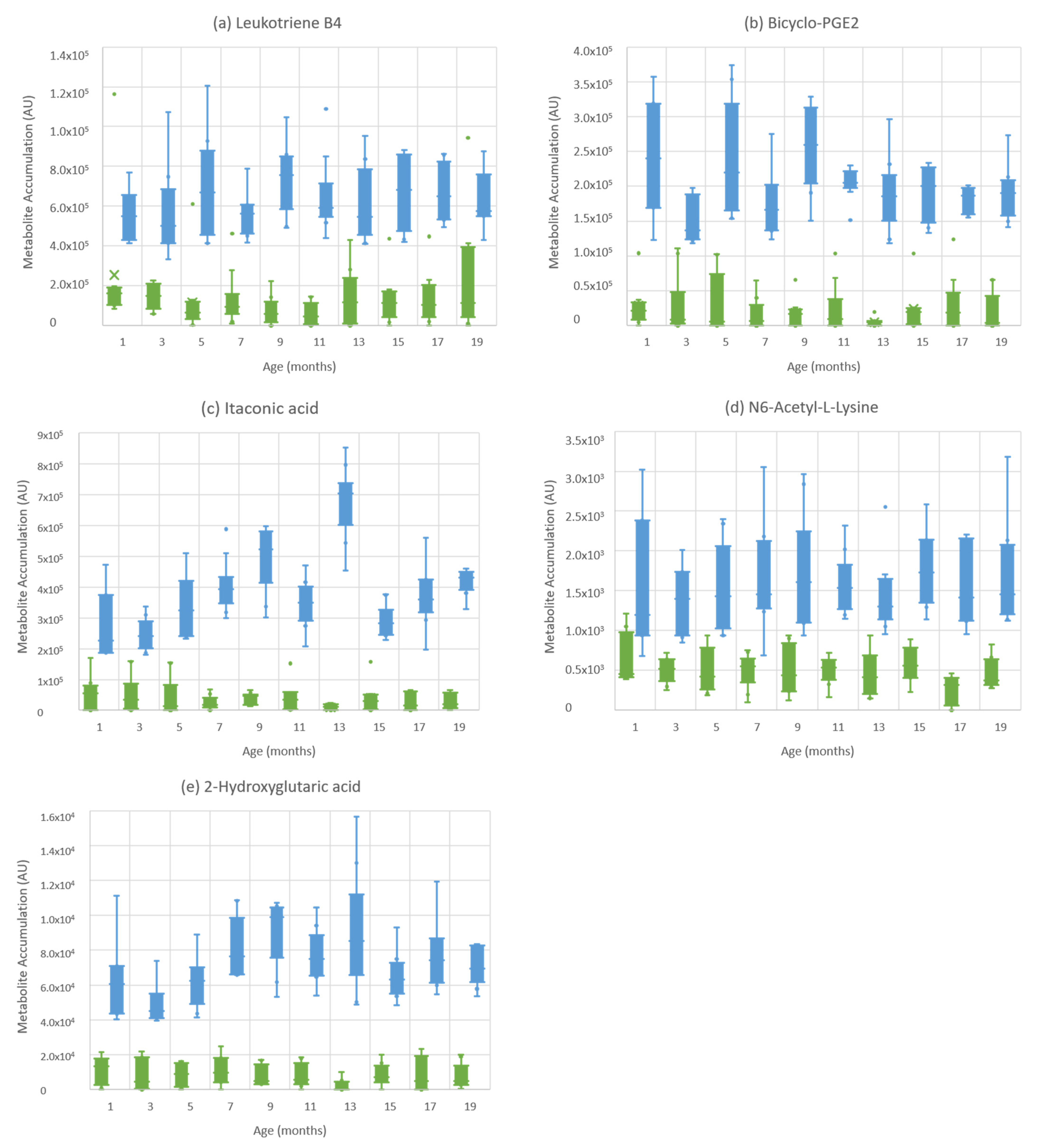
3.1. Early Changes in Bioenergetic Pathways Suggest Shifts towards Immunomodulatory Pathways
3.2. Differential Processing of Eicosanoids Are Features of Subclinically MAP-Infected Cattle
3.3. Key Metabolites Suggest Uraemic Changes That May Be Indicative of Th1–Th2 Switches
4. Materials and Methods
4.1. Animal Samples
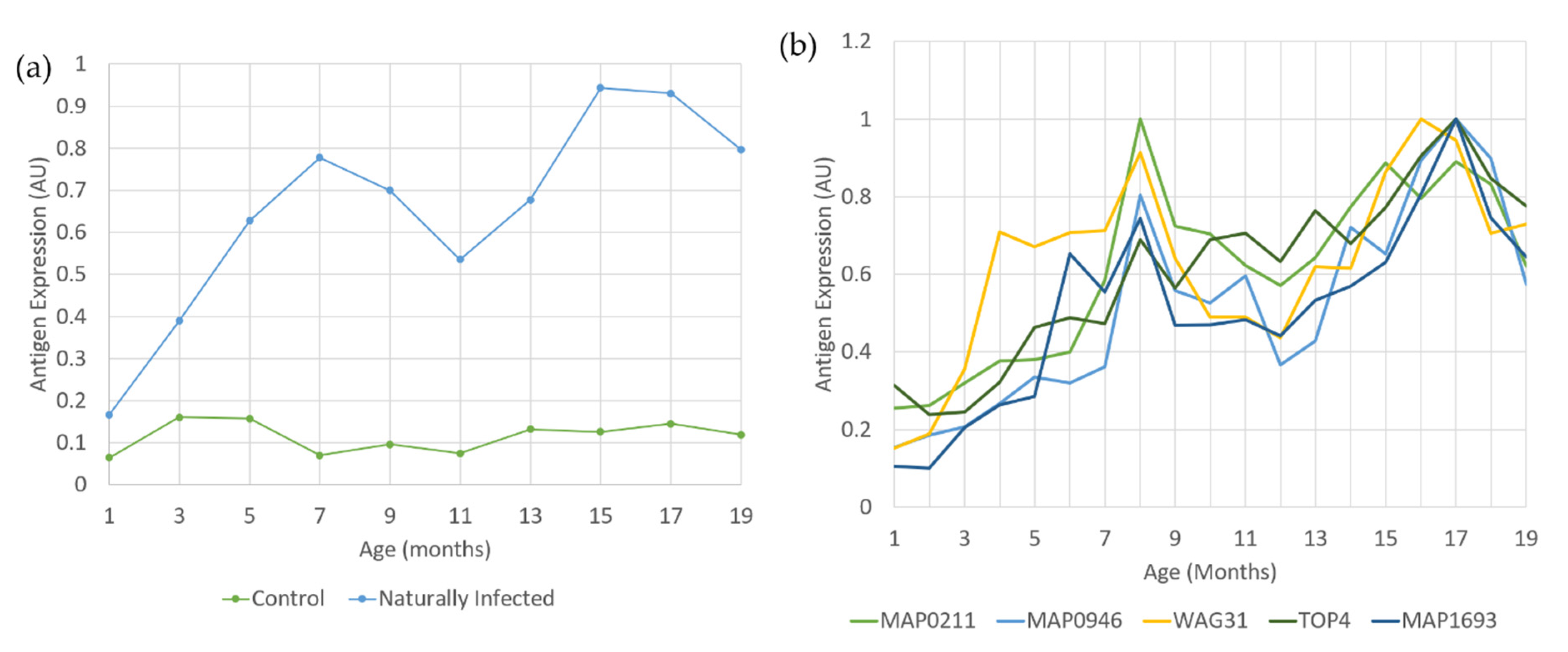
4.2. Antibody Tests
| Antigen. | Antigen Description (Uniprot for Recombinants) | |
|---|---|---|
| MAP_0210c | Ag1del1 | PirG; P36 or erp (Exported Repeated Protein) |
| MAP_0211 | - | Glf; UDP-galactopyranose mutase activity |
| MAP_0946 | - | Uncharacterized protein; DNA binding |
| MAP_1272 | - | NLPC_P60 domain-containing protein; integral membrane protein |
| MAP_1693c | - | Peptidyl-prolyl cis–trans isomerase |
| MAP_1889c | Antigen 84 | Wag31; Cell wall synthesis protein |
| MAP_2609 | Ag 7 | Uncharacterized protein; heme binding |
| MAP_2821 | - | Endoribonuclease L-PSP |
| MAP_2942c | Ag2, mpt53 | Mpt53; oxidoreductase activity |
| TOP4 | TOP4 | Mix of MAP_0210c, MAP_1272, MAP_1693c and MAP_2609 |
| - | ID-Vet | Commercially available absorbed ELISA (ID-Vet): ID Screen® Paratuberculosis Indirect |
| - | Pourquier | Commercially available absorbed ELISA (Pourquier): IDEXX® Pourquier Paratuberculosis ELISA Antibody |
4.3. Untargeted Metabolite Fingerprinting by Flow Infusion Electrospray Ionization-High-Resolution Mass Spectrometry (FIE–HRMS)
4.4. Statistical Analysis
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sweeney, R.W. Transmission of paratuberculosis. Vet. Clin. N. Am. Food Anim. Pract. 1996, 12, 305–312. [Google Scholar] [CrossRef]
- Whitlock, R.H.; Buergelt, C. Preclinical and clinical manifestations of paratuberculosis (including pathology). Vet. Clin. N. Am. Food Anim. Pract. 1996, 12, 345–356. [Google Scholar] [CrossRef]
- Sweeney, R.W. Pathogenesis of paratuberculosis. Vet. Clin. N. Am. Food Anim. Pract. 2011, 27, 537–546. [Google Scholar] [CrossRef]
- Stabel, J.R.; Bradner, L.; Robbie-Austerman, S.; Beitz, D.C. Clinical disease and stage of lactation influence shedding of Mycobacterium avium subspecies paratuberculosis into milk and colostrum of naturally infected dairy cows. J. Dairy Sci. 2014, 97, 6296–6304. [Google Scholar] [CrossRef] [Green Version]
- Salem, M.; Heydel, C.; El-Sayed, A.; Ahmed, S.A.; Zchock, M.; Baljer, G. Mycobacterium avium subspecies paratuberculosis: An insidious problem for the ruminant industry. Trop. Anim. Health Prod. 2013, 45, 351–366. [Google Scholar] [CrossRef]
- Barratt, A.S.; Arnoult, M.H.; Ahmadi, B.V.; Rich, K.M.; Gunn, G.J.; Stott, A.W. A framework for estimating society’s economic welfare following the introduction of an animal disease: The case of Johne’s disease. PLoS ONE 2018, 13, e0198436. [Google Scholar] [CrossRef]
- DEFRA. Available online: https://webarchive.nationalarchives.gov.uk/20110311110211/http://www.defra.gov.uk/foodfarm/farmanimal/diseases/atoz/documents/johnes-report0911.pdf (accessed on 24 June 2021).
- Olsen, I.; Sigurðardóttir, Ó.G.; Djønne, B. Paratuberculosis with special reference to cattle A review. Vet. Q. 2002, 24, 12–28. [Google Scholar] [CrossRef]
- Nielson, S.S.; Toft, N. Ante mortem diagnosis of paratuberculosis: A review of accuracies of ELISA, interferon-gamma assay and faecal culture techniques. Vet. Microbiol. 2008, 129, 217–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielson, S.S.; Toft, N. Age-specific characteristics of ELISA and fecal culture for purpose-specific testing for paratuberculosis. J. Dairy Sci. 2006, 89, 569–579. [Google Scholar] [CrossRef]
- Mortimer, R.A.R.; Barkema, H.W.; de Buck, J. Susceptibility to and diagnosis of Mycobacterium avium subspecies paratuberculosis infection in dairy calves: A review. Prev. Vet. Med. 2015, 121, 189–198. [Google Scholar] [CrossRef]
- Seth, M.; Lamont, E.A.; Janagama, H.K.; Widdel, A.; Vulchanova, L.; Stabel, J.R.; Waters, W.R.; Palmer, M.V.; Sreevatsan, S. Biomarker discovery in subclinical mycobacterial infections of cattle. PLoS ONE 2009, 4, e4578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, S.K.; Maclean, P.H.; Ganesh, S.; Shu, D.; Buddle, B.M.; Wedlock, D.N.; Heiser, A. Detection of microRNA in cattle serum and their potential use to diagnose severity of Johne’s disease. J. Dairy Sci. 2018, 101, 10259–10270. [Google Scholar] [CrossRef] [PubMed]
- Goldansaz, S.A.; Guo, A.C.; Sajed, T.; Steele, M.A.; Plastow, G.S.; Wishart, D.S. Livestock metabolomics and the livestock metabolome: A systematic review. PLoS ONE 2017, 12, e0177675. [Google Scholar] [CrossRef] [Green Version]
- Hollywood, K.; Brison, D.R.; Goodacre, R. Metabolomics: Current technologies and future trends. Proteomics 2006, 6, 4716–4723. [Google Scholar] [CrossRef]
- Overy, D.P.; Enot, D.P.; Tailliart, K.; Jenkins, H.; Parker, D.; Beckmann, M.; Draper, J. Explanatory signal interpretation and metabolite identification strategies for nominal mass FIE-MS metabolite fingerprints. Nat. Protoc. 2008, 3, 471–485. [Google Scholar] [CrossRef] [PubMed]
- Weiner, J., 3rd; Maertzdorf, J.; Sutherland, J.S.; Duffy, F.J.; Thompson, E.; Suilam, S.; McEwen, G.; Thiel, B.; Parida, S.K.; Zyla, J. Metabolite changes in blood predict the onset of tuberculosis. Nat. Commun. 2018, 9, 5208. [Google Scholar] [CrossRef] [Green Version]
- Mirsaeidi, M.; Banoei, M.M.; Winston, B.W.; Schraufnagel, D.E. Metabolomics: Applications and promise in Mycobacterial disease. Ann. Am. Thorac. Soc. 2015, 12, 1278–1287. [Google Scholar] [CrossRef] [Green Version]
- De Buck, J.; Shaykhutdinov, R.; Barkema, H.W.; Vogel, H.J. Metabolomic profiling in cattle experimentally infected with Mycobacterium avium subsp. paratuberculosis. PLoS ONE 2014, 9, e111872. [Google Scholar] [CrossRef]
- Tata, A.; Pallante, I.; Massaro, A.; Miano, B.; Bottazzzari, M.; Fiorini, P.; Dal Pra, M.; Paganini, L.; Stefani, A.; de Buck, J.; et al. Serum metabolomic profiles of paratuberculosis infected and infectious dairy cattle by ambient mass spectrometry. Front. Vet. Sci. 2021, 7, 625067. [Google Scholar] [CrossRef]
- Huda, A.; Jungersen, G.; Lind, P. Longitudinal study of interferon-gamma, serum antibody and milk antibody responses in cattle infected with Mycobacterium avium subsp. paratuberculosis. Vet. Microbiol. 2004, 104, 43–53. [Google Scholar] [CrossRef]
- Mortimer, R.A.R.; Barkema, H.W.; Orsel, K.; Wolf, R.; de Buck, J. Shedding patterns of dairy calves experimentally infected with Mycobacterium avium subspecies paratuberculosis. Vet. Res. 2014, 45, 71. [Google Scholar] [CrossRef] [Green Version]
- Mortier, R.A.R.; Barkema, H.W.; Negron, M.E.; Orsel, K.; Wolf, R.; de Buck, J. Antibody response early after experimental infection with Mycobacterium avium subspecies paratuberculosis in dairy calves. J. Dairy Sci. 2014, 97, 5558–5565. [Google Scholar] [CrossRef] [Green Version]
- Mortimer, R.A.R.; Barkema, H.W.; Wilson, T.A.; Sajobi, T.T.; Wolf, R.; de Buck, J. Dose-dependent interferon-gamma release in dairy calves experimentally infected with Mycobacterium avium subspecies paratuberculosis. Vet. Immunol. Immunopathol. 2014, 161, 205–210. [Google Scholar] [CrossRef]
- Swift, B.M.C.; Huxley, J.N.; Plain, K.M.; Begg, D.J.; de Silva, K.; Purdie, A.C.; Whittington, R.J.; Rees, C.E.D. Evaluation of the limitations and methods to improve rapid phage-based detection of viable Mycobacterium avium subsp. Paratuberculosis in the blood of experimentally infected cattle. BMC Vet. Res. 2016, 12, 115. [Google Scholar] [CrossRef]
- Linster, C.L.; Schaftingen, E.V.; Hanson, A.D. Metabolite damage and its repair or pre-emption. Nat. Chem. Biol. 2013, 9, 72–80. [Google Scholar] [CrossRef]
- Du, X.; Hu, H. The roles of 2-hydroxyglutarate. Front. Cell Dev. Biol. 2021, 9, 651317. [Google Scholar] [CrossRef] [PubMed]
- Gagné, L.M.; Boulay, K.; Topisirovic, I.; Huot, M.E.; Mallette, F.A. Oncogenic activities of IDH1/2 mutations: From epigenetics to cellular signaling. Trends Cell Biol. 2017, 27, 738–752. [Google Scholar] [CrossRef] [PubMed]
- Oldham, W.M.; Clish, C.B.; Yang, Y.; Loscalzo, J. Hypoxia-mediated increases in L-2-hydroxyglutarate coordinate the metabolic response to reductive stress. Cell Metab. 2015, 22, 291–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyrakis, P.A.; Palazon, A.; Macias, D.; Lee, K.L.; Phan, A.T.; Velica, P.; You, J.; Chia, G.S.; Sim, J.; Doedans, A.; et al. S-2-hydroxyglutarate regulates CD8+ T-lymphocyte fate. Nature 2016, 540, 236–241. [Google Scholar] [CrossRef]
- Michelucci, A.; Cordes, T.; Ghelfi, J.; Pailot, A.; Reiling, N.; Goldmann, O.; Binz, T.; Wegner, A.; Tallam, A.; Rausell, A.; et al. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc. Natl. Acad. Sci. USA 2013, 110, 7820–7825. [Google Scholar] [CrossRef] [Green Version]
- O’Neill, L.A.J.; Artyomov, M.N. Itaconate: The poster child of metabolic reprogramming in macrophage function. Nat. Rev. Immunol. 2019, 19, 273–281. [Google Scholar] [CrossRef]
- Malvisi, M.; Palazzo, F.; Morandi, N.; Lazzari, B.; Williams, J.L.; Pagnacco, G.; Minozzi, G. Responses of bovine innate immunity to Mycobacterium avium subsp. Paratuberculosis infection revealed by changes in gene expression and levels of microRNA. PLoS ONE 2016, 11, e0164461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubois, R.N.; Abramson, S.B.; Crofford, L.; Gupta, R.A.; Simon, L.S.; van de Putte, L.B.A.; Lipsky, P.E. Cyclooxygenase in biology and disease. FASEB J. 1998, 12, 1063–1073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goto, S.; Konnai, S.; Hirano, Y.; Kohara, J.; Okagawa, T.; Maekawa, N.; Sajiki, Y.; Watari, K.; Minato, E.; Kobayashi, A. Upregulation of PD-L1 expression by prostaglandin E2 and the enhancement of IFN-γ by anti-PD-L1 antibody combined with a COX-2 inhibitor in Mycoplasma bovis infection. Front. Vet. Sci. 2020, 7, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishimura, T.; Zhao, X.; Gan, H.; Koyasu, S.; Remold, H.G. The prostaglandin E2 receptor EP4 is integral to a positive feedback loop for prostaglandin E2 production in human macrophages infected with Mycobacterium tuberculosis. FASEB J. 2013, 27, 3827–3836. [Google Scholar] [CrossRef] [Green Version]
- Crooks, S.W.; Stockley, R.A. Leukotriene B4. Int. J. Biochem. Cell Biol. 1998, 30, 173–178. [Google Scholar] [CrossRef]
- Talahalli, R.; Zarini, S.; Sheibani, N.; Murphy, R.C.; Gubitosi-Klug, R.A. Increased synthesis of leukotrienes in the mouse model of diabetic retinopathy. Investig. Ophthalmol. Vis. Sci. 2013, 51, 1699–1708. [Google Scholar] [CrossRef]
- David, J.; Barkema, H.W.; Guan, L.L.; de Buck, J. Gene expression profiling of calves 6 and 9 months after inoculation with Mycobacterium avium subspecies paratuberculosis. Vet. Res. 2014, 45, 96. [Google Scholar] [CrossRef]
- Radzikowska, U.; Rinaldi, A.O.; Sözener, Z.C.; Karaguzel, D.; Wojcik, M.; Cypryk, K.; Akdis, M.; Akdis, C.A.; Sokolowska, M. The influence of dietary fatty acids on immune responses. Nutrients 2019, 11, 2990. [Google Scholar] [CrossRef] [Green Version]
- Marion-Letellier, R.; Butler, M.; Déchelotte, P.; Playford, R.J.; Ghosh, S. Comparison of cytokine modulation by natural peroxisome proliferator-activated receptor γ ligands with synthetic ligands in intestinal-like Caco-2 cells and human dendritic cells-potential for dietary modulation of peroxisome proliferator-activated receptor γ in intestinal inflammation. Am. J. Clin. Nutr. 2004, 87, 939–948. [Google Scholar] [CrossRef] [Green Version]
- Jaudszus, A.; Gruen, M.; Watzl, B.; Ness, C.; Roth, A.; Lochner, A.; Barz, D.; Grabriel, H.; Rothe, M.; Jahreis, G. Evaluation of suppressive and pro-resolving effects of EPA and DHA in human primary monocytes and T-helper cells. J. Lipid Res. 2013, 54, 923–935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verlengia, R.; Gorjao, R.; Kanunfre, C.C.; Bordin, S.; de Lima, T.M.; Martins, E.F. Effects of EPA and DHA on proliferation, cytokine production and gene expression in raji cells. Lipids 2004, 39, 857–864. [Google Scholar] [CrossRef]
- Hussain, T.; Shah, S.Z.A.; Zhao, D.; Sreevatsan, S.; Zhou, Z. The role of IL-10 Mycobacterium avium subsp. paratuberculosis infection. Cell Commun. Signal. 2016, 14, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jordao, L.; Lengeling, A.; Bordat, Y.; Boudou, F.; Gicquel, B.; Neyrolles, O.; Becker, P.D.; Guzman, C.A.; Griffiths, G.; Anes, E. Effects of omega-3 and -6 fatty acids on Mycobacterium tuberculosis in macrophages and in mice. Microbes and Infection. Microbes Infect. 2008, 10, 1379–1386. [Google Scholar] [CrossRef]
- Brosnan, J.T.; da Silva, R.P.; Brosnan, M.E. The metabolite burden of creatine synthesis. Amino Acids 2011, 40, 1325–1331. [Google Scholar] [CrossRef] [PubMed]
- Stein, I.M.; Cohen, B.D.; Kornhauser, R.S. Guanidinosuccinic acid in renal failure, experimental azotemia and inborn errors of the urea cycle. N. Engl. J. Med. 1969, 280, 926–930. [Google Scholar] [CrossRef]
- Cohen, B.D.; Stein, I.M.; Bonas, J.E. Guanidinosuccinic aciduria in uremia: A possible alternate pathway for urea synthesis. Am. J. Med. 1968, 45, 63–68. [Google Scholar] [CrossRef]
- Perez, G.; Rey, A.; Schiff, E. The biosynthesis of guanidinosuccinic acid by perfused rat liver. J. Clin. Investig. 1976, 57, 807–809. [Google Scholar] [CrossRef]
- Gryp, T.; Vanholder, R.; Vaneechoutte, M.; Glorieux, G. p-Cresyl Sulfate. Toxins 2017, 9, 52. [Google Scholar] [CrossRef] [Green Version]
- Smith, E.A.; Macfarlane, G.T. Enumeration of human colonic bacteria producing phenolic and indolic compounds: Effects of pH, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J. Appl. Bacteriol. 1996, 81, 288–302. [Google Scholar] [CrossRef]
- Smith, E.A.; Macfarlane, G.T. Dissimilatory amino acid metabolism in human colonic bacteria. Anaerobe 1997, 3, 327–337. [Google Scholar] [CrossRef]
- Vanholder, R.; De Smet, R.; Waterloos, M.-A.; Van Landschoot, N.; Vogeleere, P.; Hoste, E.; Ringoir, S. Mechanisms of uremic inhibition of phagocyte reactive species production: Characterization of the role of p-cresol. Kidney Int. 1995, 47, 510–517. [Google Scholar] [CrossRef] [Green Version]
- Faure, V.; Cerini, C.; Paul, P.; Berland, Y.; Dignat-George, F.; Brunet, P. The uremic solute p-cresol decreases leukocyte transendothelial migration in vitro. Int. Immunol. 2006, 18, 1453–1459. [Google Scholar] [CrossRef] [Green Version]
- Shiba, T.; Kawakami, K.; Sasaki, T.; Makino, I.; Kato, I.; Kobayashi, T.; Uchida, K.; Kaneko, K. Effects of intestinal bacteria-deprived p-cresyl sulfate on Th1-type immune response in vivo and in vitro. Toxicol. Pharmacol. 2014, 274, 191–199. [Google Scholar] [CrossRef]
- Hibbs, H.B.; Taintor, R.R.; Vavrin, Z. Macrophage cytotoxicity: Role for L-arginine deiminase and imino nitrogen oxidation to nitrite. Science 1987, 235, 473–476. [Google Scholar] [CrossRef]
- Rodriguez, P.C.; Zea, A.H.; DeSalvo, J.; Culotta, K.S.; Zabaleta, J.; Quiceno, D.G.; Ochoa, J.B.; Ochoa, A.C. L-Arginine consumption by macrophages modulates the expression of CD3 xi chain in T lymphocytes. J. Immunol. 2003, 171, 1232–1239. [Google Scholar] [CrossRef] [Green Version]
- Abumrad, N.N.; Barbul, A. The use of arginine in clinical practice. In Metabolic & Therapeutic Aspects of Amino Acids in Clinical Nutrition, 2nd ed.; Cynober, L.A., Ed.; CRC Press: Boca Raton, FL, USA, 2004; pp. 595–611. ISBN 0849313821. [Google Scholar]
- Bronte, V.; Zanovello, P. Regulation of immune responses by L-arginine metabolism. Nat. Rev. Immunol. 2005, 5, 641–654. [Google Scholar] [CrossRef] [PubMed]
- Magombedze, G.; Eda, S.; Koets, A. Can immune response mechanisms explain the faecal shedding patterns of cattle infected with Mycobacterium avium subspecies paratuberculosis. PLoS ONE 2016, 11, e0146844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smeed, J.A.; Watkins, C.A.; Rhind, S.M.; Hopkins, J. Differential cytokine gene expression profiles in the three pathological forms of sheep paratuberculosis. BMC Vet. Res. 2007, 3, 18. [Google Scholar] [CrossRef] [Green Version]
- Stabel, J.R. Transitions in immune responses to Mycobacterium paratuberculosis. Vet. Microbiol. 2000, 77, 465–473. [Google Scholar] [CrossRef]
- Wu, G.; Meininger, C.J. Regulation of nitric oxide synthesis by dietary factors. Annu. Rev. Nutr. 2002, 22, 61–86. [Google Scholar] [CrossRef]
- Closs, E.I.; Scheld, J.S.; Sharafi, M.; Forstermann, U. Substrate supply for nitric oxide synthase in macrophages and endothelial cells: Role of cationic amino acid transporters. Mol. Pharmacol. 2000, 57, 68–74. [Google Scholar]
- Willemsen, P.T.J.; Westerveen, J.; Dinkla, A.; Bakker, D.; van Zijderveld, F.G.; Thole, J.E.R. Secreted antigens of Mycobacterium avium subspecies paratuberculosis as prominent immune targets. Vet. Microbiol. 2006, 114, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Mon, M.L.; Viale, M.; Bachetti, G.; Pinedo, F.A.; Gioffre, A.; Traveria, G.; Willemsen, P.; Bakker, D.; Romano, M.I. Search for Mycobacterium avium subspecies paratuberculosis antigens for the diagnosis of paratuberculosis. Vet. Med. Int. 2012, 2012, 860362. [Google Scholar] [CrossRef] [Green Version]
- Beckmann, M.; Parker, D.; Enot, D.P.; Duval, E.; Draper, J. High-throughput, nontargeted metabolite fingerprinting using nominal mass flow injection electrospray mass spectrometry. Nat. Protoc. 2008, 3, 486–504. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Wishart, D.; Xai, J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef]
- Foroutan, A.; Fitzsimmons, C.; Mandal, R.; Piri-Moghadam, H.; Zheng, J.; Guo, A.C.; Li, C.; Guan, L.L.; Wishart, D.S. The bovine metabolome. Metabolites 2020, 10, 233. [Google Scholar] [CrossRef]
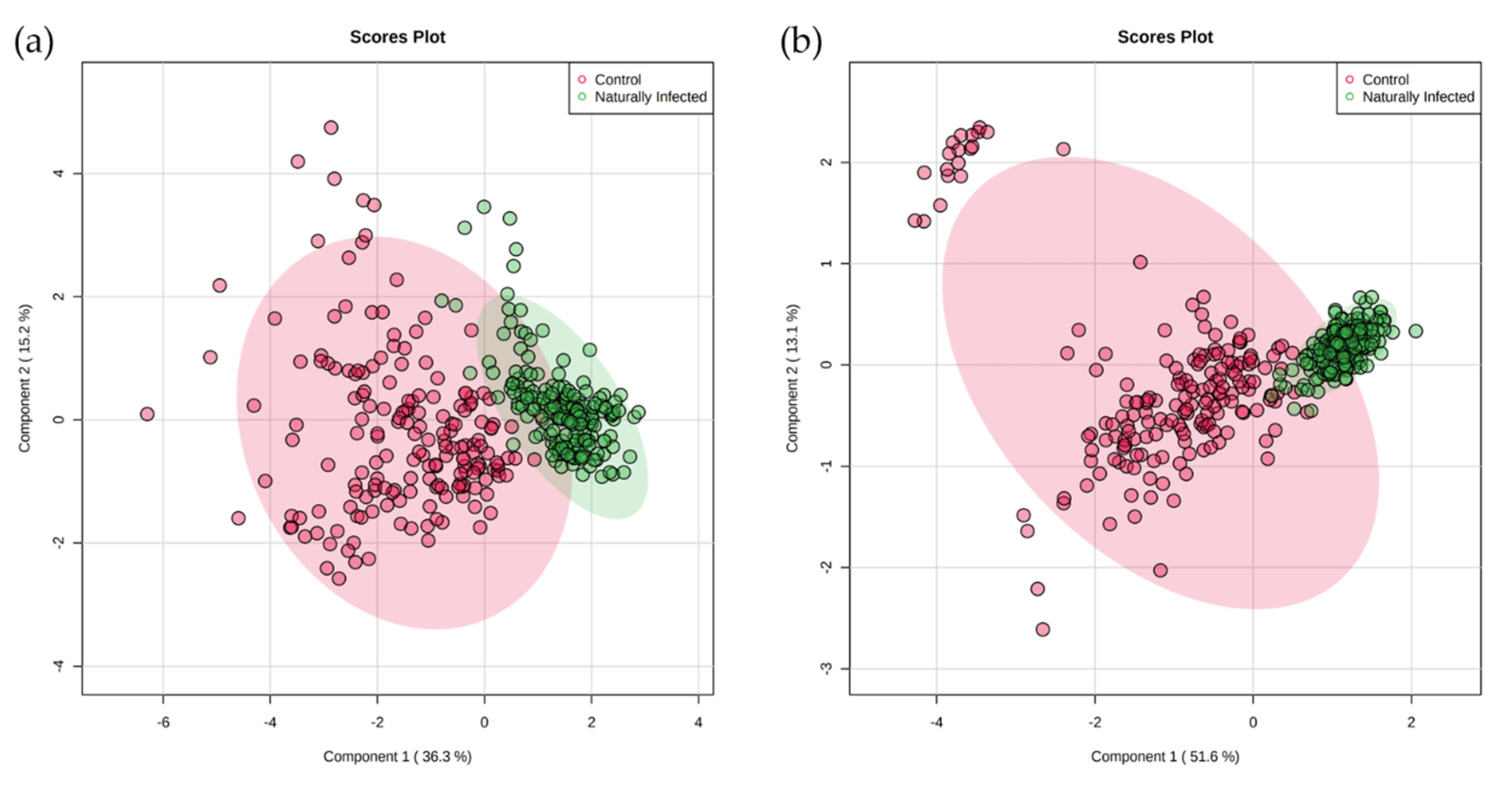



| Control | Naturally Infected | Class Error | |
|---|---|---|---|
| Control | 52 | 1 | 0.0189 |
| Naturally Infected | 0 | 60 | 0.0000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taylor, E.N.; Beckmann, M.; Villarreal-Ramos, B.; Vordermeier, H.-M.; Hewinson, G.; Rooke, D.; Mur, L.A.J.; Koets, A.P. Metabolomic Changes in Naturally MAP-Infected Holstein–Friesian Heifers Indicate Immunologically Related Biochemical Reprogramming. Metabolites 2021, 11, 727. https://doi.org/10.3390/metabo11110727
Taylor EN, Beckmann M, Villarreal-Ramos B, Vordermeier H-M, Hewinson G, Rooke D, Mur LAJ, Koets AP. Metabolomic Changes in Naturally MAP-Infected Holstein–Friesian Heifers Indicate Immunologically Related Biochemical Reprogramming. Metabolites. 2021; 11(11):727. https://doi.org/10.3390/metabo11110727
Chicago/Turabian StyleTaylor, Emma N., Manfred Beckmann, Bernardo Villarreal-Ramos, Hans-Martin Vordermeier, Glyn Hewinson, David Rooke, Luis A. J. Mur, and Ad P. Koets. 2021. "Metabolomic Changes in Naturally MAP-Infected Holstein–Friesian Heifers Indicate Immunologically Related Biochemical Reprogramming" Metabolites 11, no. 11: 727. https://doi.org/10.3390/metabo11110727