The Functionally Characterization of Putative Genes Involved in the Formation of Mannose in the Aplanospore Cell Wall of Haematococcus pluvialis (Volvocales, Chlorophyta)
Abstract
:1. Introduction
2. Results
2.1. Morphological Changes of Algal Cells in Responding to SAHL Stresses
2.2. The Genes Involved in the Development of the Cell Wall of H. pluvialis When Exposed to the SAHL Stresses
2.3. The Expression Profiles of Genes Involved in the Biosynthesis Pathways of Cell Wall
2.4. Correlations between Expression of the Cell Wall Genes and the Components
3. Discussion
4. Materials and Methods
4.1. H. pluvialis Strain and Culture Conditions
4.2. Observation of Microalgal Morphology by Transmission Electron Microscope (TEM)
4.3. The Measurement of the Components Comprised Cell Wall
4.3.1. The Measurement of Cellulose
4.3.2. The Measurement of Mannose, Glucose and Fructose
4.4. The Explore of the Cell Wall Related Genes
4.5. Real-Time Quantitative Reverse Transcriptase PCR
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hagen, C.; Siegmund, S.; Braune, W. Ultrastructural and chemical changes in the cell wall of Haematococcus pluvialis (Volvocales, Chlorophyta) during aplanospore formation. Eur. J. Phycol. 2002, 37, 217–226. [Google Scholar] [CrossRef]
- Gross, G.J.; Lockwood, S.F. Acute and chronic administration of disodium disuccinate astaxanthin (Cardax TM) produces marked cardioprotection in dog hearts. Mol. Cell. Biochem. 2005, 272, 221–227. [Google Scholar] [CrossRef]
- Wang, X.; Willen, R.; Wadstrom, T. Astaxanthin-rich algal meal and vitamin C inhibit Helicobacter pylori infection in BALB/cA mice. Antimicrob. Agents Chemother. 2000, 44, 2452–2457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Downie, C.G.; Aysegul, C.; Gultekin, Y.; Necdet, D.; Saffet, O.; Owen, E.G.; Berna, S.; Mutay, A. Effect of astaxanthin on hepatocellular injury following ischemia/reperfusion. Toxicology 2010, 267, 147–153. [Google Scholar]
- Chi-Huang, C.; Chung-Yin, C.; Jueen-Ya, C.; Yung, P.R.; Chiung-Huei, P. Astaxanthine secured apoptotic death of PC12 cells induced by beta-amyloid peptide 25–35: Its molecular action targets. J. Med. Food 2010, 13, 548–556. [Google Scholar] [CrossRef] [PubMed]
- Izumi-Nagai, K.; Nagai, N.; Ohgami, K.; Satofuka, S.; Ozawa, Y.; Tsubota, K.; Ohno, S.; Oike, Y.; Ishida, S. Inhibition of Choroidal Neovascularization with an anti-inflammatory carotenoid astaxanthin. Investig. Ophthalmol. Vis. Sci. 2008, 49, 1679–1685. [Google Scholar] [CrossRef] [PubMed]
- Pomb, C.B.; Winey, W.M.; Stone, P.J.; Silva, W.T. Dietary beta-carotene and astaxanthin but not canthaxanthin stimulate splenocyte function in mice. Anticancer. Res. 1999, 19, 5223–5227. [Google Scholar]
- Gwak, Y.H.Y.; Wang, B.; Kim, M.; Jeong, J.; Lee, C.G.; Hu, Q.; Han, D.; Jin, E. Comparative analyses of lipidomes and transcriptomes reveal a concerted action of multiple defensive systems against photooxidative stress in Haematococcus pluvialis. J. Exp. Bot. 2014, 65, 4317–4334. [Google Scholar] [CrossRef]
- Damiani, M.C.; Leonardi, P.I.; Pieroni, O.I.; Cáceres, E.J. Ultrastructure of the cyst wall of Haematococcus pluvialis (Chlorophyceae): Wall development and behaviour during cyst germination. Phycologia 2006, 45, 616–623. [Google Scholar] [CrossRef]
- Khan, M.I.; Fatma, M.; Per, T.S.; Anjum, N.A.; Khan, N.A. Salicylic acid-induced abiotic stress tolerance and underlying mechanisms in plants. Front. Plant Sci. 2015, 6, 462. [Google Scholar] [CrossRef] [Green Version]
- Meng, C.; Liang, C.; Su, Z.; Qin, S.; Tseng, C. There are two 5′-flanking regions of bkt encoding beta-carotene ketolase in Haematococcus pluvialis. Phycologia 2006, 45, 218–224. [Google Scholar] [CrossRef]
- Chun-Xiao, M.; Chang-Ying, T.; Peng, J.; Song, Q.; Cheng-Kui, T. Cloning and characterization of beta-carotene ketolase gene promoter in Haematococcus pluvialis. Acta Biochim. Biophys. Sin. 2005, 37, 270–275. [Google Scholar]
- Gao, Z.Q.; Meng, C.X. Regulatory Analysis of IPP Isomerase Gene in Haematococcus pluvialis. Mar. Bull. 2009, 11, 37–42. [Google Scholar]
- Gao, Z.Q.; Meng, C.X.; Ye, N.H. Three 5′-flanking regions of crtO encoding β-carotene oxygenase in Haematococcus pluvialis. Bull. Mar. Sci. 2010, 12, 59–64. [Google Scholar]
- Gao, Z.; Li, Y.; Wu, G.; Li, G.; Sun, H.; Deng, S.; Shen, Y.; Chen, G.; Zhang, R.; Meng, C.; et al. Transcriptome Analysis in Haematococcus pluvialis: Astaxanthin Induction by Salicylic Acid (SA) and Jasmonic Acid (JA). PLoS ONE 2015, 10, e0140609. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Miao, X.; Zhang, X.; Wu, G.; Guo, Y.; Wang, M.; Li, B.; Li, X.; Gao, Y.; Hu, S. Comparative fatty acid transcriptomic test and iTRAQ-based proteomic analysis in Haematococcus pluvialis upon salicylic acid (SA) and jasmonic acid (JA) inductions. Algal Res. 2016, 17, 277–284. [Google Scholar] [CrossRef]
- Gao, Z.; Meng, C.; Zhang, X.; Xu, D.; Zhao, Y.; Wang, Y.; Lv, H.; Liming, Y.; Chen, L.; Ye, N. Differential expression of carotenogenic genes, associated changes on astaxanthin production and photosynthesis features induced by JA in H. pluvialis. PLoS ONE 2012, 7, e42243. [Google Scholar] [CrossRef]
- Gao, Z.; Meng, C.; Zhang, X.; Xu, D.; Miao, X.; Wang, Y.; Yang, L.; Lv, H.; Chen, L.; Ye, N. Induction of salicylic acid (SA) on transcriptional expression of eight carotenoid genes and astaxanthin accumulation in Haematococcus pluvialis. Enzym. Microb. Technol. 2012, 51, 225–230. [Google Scholar] [CrossRef]
- Zhao, Y.; Hou, Y.; Chai, W.; Liu, Z.; Wang, X.; He, C.; Hu, Z.; Chen, S.; Wang, W.; Chen, F. Transcriptome analysis of Haematococcus pluvialis of multiple defensive systems against nitrogen starvation. Enzym. Microb. Technol. 2020, 134, 109487. [Google Scholar] [CrossRef]
- Delmer, D.P. Cellulose biosynthesis: Exciting Times for A Difficult Field of Study. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 245–276. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef]
- Aleamotu’a, M.; McCurdy, D.W.; Collings, D.A. Phi thickenings in roots: Novel secondary wall structures responsive to biotic and abiotic stresses. J. Exp. Bot. 2019, 70, 4631–4642. [Google Scholar] [CrossRef]
- Carroll, A.; Specht, C.D. Understanding Plant Cellulose Synthases through a Comprehensive Investigation of the Cellulose Synthase Family Sequences. Front. Plant Sci. 2011, 2, 5. [Google Scholar] [CrossRef] [Green Version]
- Desprez, T.; Juraniec, M.; Crowell, E.F.; Jouy, H.; Pochylova, Z.; Parcy, F.; Höfte, H.; Gonneau, M.; Vernhettes, S. Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2007, 104, 15572–15577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Persson, S.; Paredez, A.; Carroll, A.; Palsdottir, H.; Doblin, M.; Poindexter, P.; Khitrov, N.; Auer, M.; Somerville, C.R. Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 15566–15571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, R.; Ye, Z.H. Complexity of the transcriptional network controlling secondary wall biosynthesis. Plant Sci. 2014, 229, 193–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singhania, R.R.; Patel, A.K.; Sukumaran, R.K.; Larroche, C.; Pandey, A. Role and significance of beta-glucosidases in the hydrolysis of cellulose for bioethanol production. Bioresour. Technol. 2013, 127, 500–507. [Google Scholar] [CrossRef]
- Gloster, T.M.; Macdonald, J.M.; Tarling, C.A.; Stick, R.V.; Withers, S.G.; Davies, G.J. Structural, thermodynamic, and kinetic analyses of tetrahydrooxazine-derived inhibitors bound to beta-glucosidases. J. Biol. Chem. 2004, 279, 49236–49242. [Google Scholar] [CrossRef] [Green Version]
- Stoop, J.M.H.; Williamson, J.D.; Conkling, M.A.; MacKay, J.J.; Pharr, D.M. Characterization of NAD-dependent mannitol dehydrogenase from celery as affected by ions, chelators, reducing agents and metabolites. Plant Sci. 1998, 131, 43–51. [Google Scholar] [CrossRef]
- Christ, T.N.; Deweese, K.A.; Woodyer, R.D. Directed evolution toward improved production of L-ribose from ribitol. Comb. Chem. High Throughput Screen. 2010, 13, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Kaup, B.; Bringer-Meyer, S.; Sahm, H. Metabolic engineering of Escherichia coli: Construction of an efficient biocatalyst for D-mannitol formation in a whole-cell biotransformation. Appl. Microbiol. Biotechnol. 2004, 64, 333–339. [Google Scholar] [CrossRef]
- Hu, Q.; Huang, D.; Li, A.; Hu, Z.; Gao, Z.; Yang, Y.; Wang, C. Transcriptome-based analysis of the effects of salicylic acid and high light on lipid and astaxanthin accumulation in Haematococcus pluvialis. Biotechnol. Biofuels 2021, 14. [Google Scholar] [CrossRef] [PubMed]
- Béguin, P. Molecular biology of cellulose degradation. Annu. Rev. Microbiol. 1990, 44, 219–248. [Google Scholar] [CrossRef] [PubMed]
- Brummell, D.A.; Hall, B.D.; Bennett, A.B. Antisense suppression of tomato endo-1,4-beta-glucanase Cel2 mRNA accumulation increases the force required to break fruit abscission zones but does not affect fruit softening. Plant Mol. Biol. 1999, 40, 615–622. [Google Scholar] [CrossRef]
- Saxena, I.M.; Lin, F.C.; Brown, R.M., Jr. Identification of a new gene in an operon for cellulose biosynthesis in Acetobacter xylinum. Plant Mol. Biol. 1991, 16, 947–954. [Google Scholar] [CrossRef] [PubMed]
- Saxena, I.M.; Brown, R.M., Jr. Are the reversibly glycosylated polypeptides implicated in plant cell wall biosynthesis non-processive beta-glycosyltransferases? Trends Plant Sci. 1999, 4, 6–7. [Google Scholar] [CrossRef]
- Wong, H.C.; Fear, A.L.; Calhoon, R.D.; Eichinger, G.H.; Mayer, R.; Amikam, D.; Benziman, M.; Gelfand, D.H.; Meade, J.H.; Emerick, A.W.; et al. Genetic organization of the cellulose synthase operon in Acetobacter xylinum. Proc. Natl. Acad. Sci. USA 1990, 87, 8130–8134. [Google Scholar] [CrossRef] [Green Version]
- Low, C.P.; Emerty, P.J.; Louis, L.R.; Norman, S. L-Ascorbic Acid Metabolism in the Ascorbate-Deficient Arabidopsis Mutant vtc1. Plant Physiol. 1997, 115, 1277–1285. [Google Scholar]
- Imai, T.; Ban, Y.; Terakami, S.; Yamamoto, T.; Moriguchi, T. L-Ascorbate biosynthesis in peach: Cloning of six L-galactose pathway-related genes and their expression during peach fruit development. Physiol. Plant. 2009, 136, 139–149. [Google Scholar] [CrossRef]
- Zou, L.P.; Li, H.X.; Ouyang, B.; Zhang, J.H.; Ye, Z.B. Cloning, expression, and mapping of GDP-D-mannose pyrophosphorylase cDNA from tomato (Lycopersicon esculentum). Acta Genet. Sin. 2006, 33, 757–764. [Google Scholar] [CrossRef]
- Bayer, E.A.; Shoham, Y.; Lamed, R. Cellulose-Decomposing Prokaryotes and Their Enzyme Systems; Springer: Berlin/Heidelberg, Germany, 2000; pp. 578–617. [Google Scholar]
- He, C.; Zeng, S.; Teixeira da Silva, J.A.; Yu, Z.; Tan, J.; Duan, J. Molecular cloning and functional analysis of the phosphomannomutase (PMM) gene from Dendrobium officinale and evidence for the involvement of an abiotic stress response during germination. Protoplasma 2017, 254, 1693–1704. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Yu, Z.; Teixeira da Silva, J.A.; Zhang, J.; Liu, X.; Wang, X.; Zhang, X.; Zeng, S.; Wu, K.; Tan, J.; et al. DoGMP1 from Dendrobium officinale contributes to mannose content of water-soluble polysaccharides and plays a role in salt stress response. Sci. Rep. 2017, 7, 41010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barth, C.; Gouzd, Z.A.; Steele, H.P.; Imperio, R.M. A mutation in GDP-mannose pyrophosphorylase causes conditional hypersensitivity to ammonium, resulting in Arabidopsis root growth inhibition, altered ammonium metabolism, and hormone homeostasis. J. Exp. Bot. 2010, 61, 379–394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, C.S.; Kim, J.E.; Choi, J.G.; Oh, D.K. Characterization of a recombinant cellobiose 2-epimerase from Caldicellulosiruptor saccharolyticus and its application in the production of mannose from glucose. Appl. Microbiol. Biotechnol. 2011, 92, 1187–1196. [Google Scholar] [CrossRef]
- Park, C.S.; Yeom, S.J.; Lim, Y.R.; Kim, Y.S.; Oh, D.K. Characterization of a recombinant thermostable L-rhamnose isomerase from Thermotoga maritima ATCC 43589 and its application in the production of L-lyxose and L-mannose. Biotechnol. Lett. 2010, 32, 1947–1953. [Google Scholar] [CrossRef]
- Park, C.S.; Kwon, H.J.; Yeom, S.J.; Oh, D.K. Mannose production from fructose by free and immobilized D-lyxose isomerases from Providencia stuartii. Biotechnol. Lett. 2010, 32, 1305–1309. [Google Scholar] [CrossRef]
- Ma, R.; Thomas-Hall, S.R.; Chua, E.T.; Alsenani, F.; Eltanahy, E.; Netzel, M.E.; Netzel, G.; Lu, Y.; Schenk, P.M. Gene expression profiling of astaxanthin and fatty acid pathways in Haematococcus pluvialis in response to different LED lighting conditions. Bioresour. Technol. 2018, 250, 591–602. [Google Scholar] [CrossRef] [Green Version]
- Eom, H.; Lee, C.G.; Jin, E. Gene expression profile analysis in astaxanthin-induced Haematococcus pluvialis using a cDNA microarray. Planta 2006, 223, 1231–1242. [Google Scholar] [CrossRef]
- Lei, A.; Chen, H.; Shen, G.; Hu, Z.; Chen, L.; Wang, J. Expression of fatty acid synthesis genes and fatty acid accumulation in haematococcus pluvialis under different stressors. Biotechnol. Biofuels 2012, 5. [Google Scholar] [CrossRef] [Green Version]
- Ramakers, C.; Ruijter, J.M.; Deprez, R.H.; Moorman, A.F. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Nymark, M.; Valle, K.C.; Brembu, T.; Hancke, K.; Winge, P.; Andresen, K.; Johnsen, G.; Bones, A.M. An integrated analysis of molecular acclimation to high light in the marine diatom Phaeodactylum tricornutum. PLoS ONE 2009, 4, e7743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armin, K.J.L.; Beck, T.D.S. Analysis of relative gene expression data using real time quantitative PCR and the 2−ΔΔCT method. Methods Enzymol. 2001, 25, 402–408. [Google Scholar]
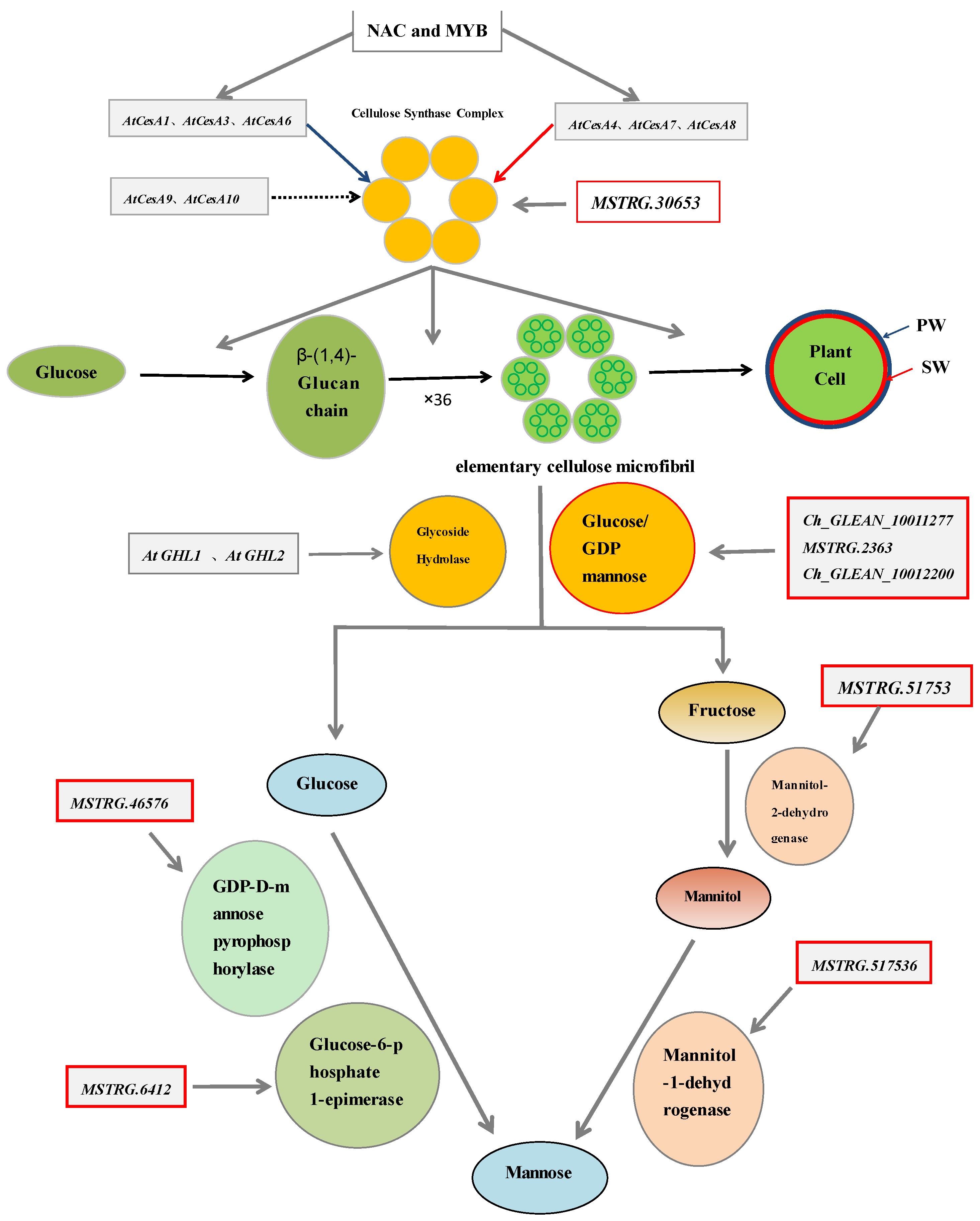


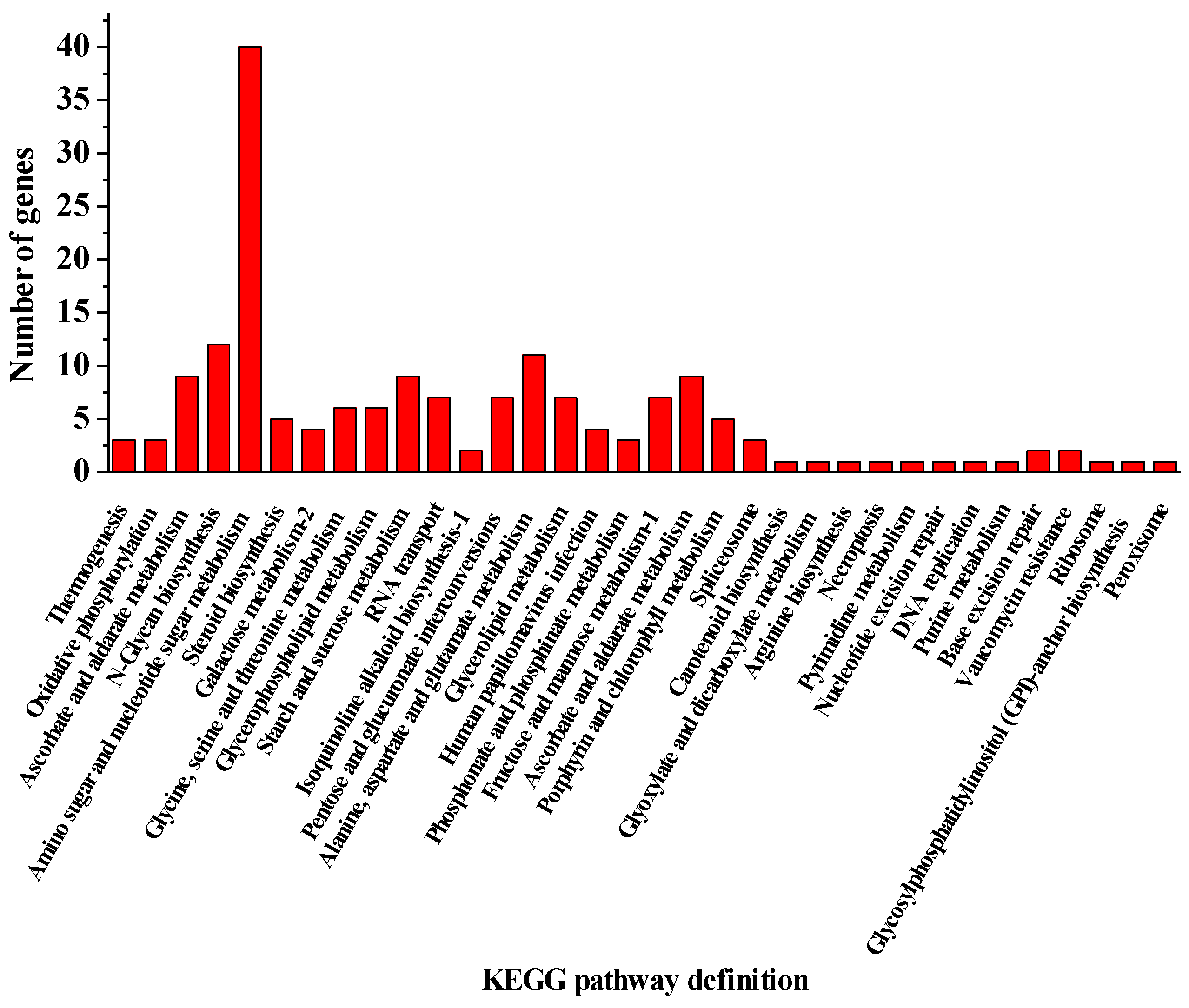
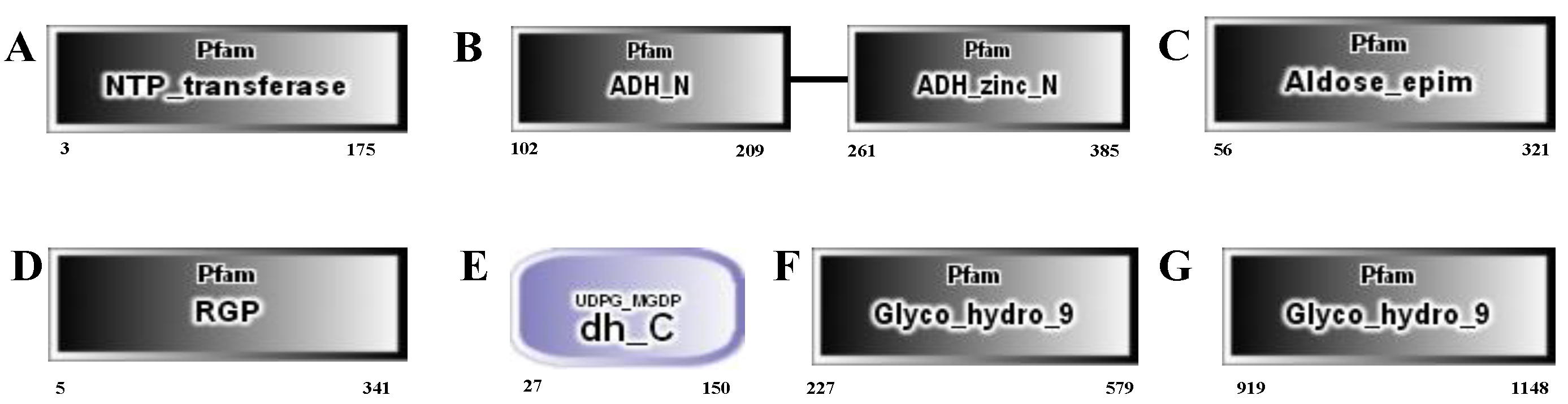
| Measurements | Control (0) | SAHL_1 | SAHL_6 | SAHL_12 | SAHL_24 | SAHL_48 |
|---|---|---|---|---|---|---|
| MC | 180.13± 2.12 d | 234.96± 1.92 c | 242.14 ± 1.66 c | 262.89 ± 2.88 b | 274.16 ± 3.46 b | 293.89 ± 1.58 a |
| CC | 300.16 ± 2.74 d | 352.99 ± 4.18 c | 366.51 ± 4.18 b | 372.01 ± 6.73 b | 369.47 ± 3.64 b | 388.49 ± 4.67 a |
| FC | 0.83 ± 0.01 d | 1.05 ± 0.01 c | 1.07 ± 0.02 c | 1.12 ± 0.01 c | 1.20 ± 0.01 b | 1.33 ± 0.01 a |
| GC | 0.76 ± 0.01 d | 0.99 ± 0.01 c | 1.01 ± 0.01 c | 1.06 ± 0.01 b | 1.10 ± 0.01 b | 1.16 ± 0.01 a |
| Gene_id | COG_Functional Categories | Pfam Functional Domain | Control | SAHL_1 | SAHL_6 | SAHL_12 | SAHL_24 | SAHL_48 |
|---|---|---|---|---|---|---|---|---|
| Ch_GLEAN_10012200 | Cell wall/membrane/envelope biogenesis | O-Glycosyl hydrolases | 1.15 | 16.96 | 2.41 | 24.55 | 13.52 | 12.32 |
| Ch_GLEAN_10011277 | Cell wall/membrane/envelope biogenesis | O-Glycosyl hydrolases | 3.11 | 3.69 | 3.32 | 3.74 | 2.31 | 3.16 |
| MSTRG.46576 | Cell wall/membrane/envelope biogenesis | GDP-D-mannose pyrophosphorylase | 3.46 | 6.09 | 5.31 | 11.02 | 25.98 | 149.19 |
| MSTRG.2363 | Cell wall/membrane/envelope biogenesis | Glucose/GDP mannose dehydrogenase | 7.52 | 11.54 | 7.79 | 11.96 | 8.85 | 7.84 |
| MSTRG.30653 | Cell wall/membrane/envelope biogenesis | UDP-arabinopyranose mutases | 2.78 | 4.06 | 4.35 | 4.81 | 7.22 | 6.24 |
| MSTRG.51753 | Carbohydrate transport and metabolism | Mannitol dehydrogenase | 2.7 | 2.8 | 3.66 | 3.62 | 4.16 | 4.28 |
| MSTRG.6412 | Carbohydrate transport and metabolism | Glucose-6-phosphate 1-epimerase | 4.13 | 3.91 | 5.06 | 4.82 | 4.95 | 7.05 |
| Genes | Ch_GLEAN_10012200 | Ch_GLEAN_10011277 | MSTRG.46576 | MSTRG.2363 | MSTRG.30653 | MSTRG.51753 | MSTRG.6412 |
|---|---|---|---|---|---|---|---|
| MC | 0.552 | −0.176 | 0.878 * | 0.155 | 0.885 * | 0.893 * | 0.891 * |
| CC | 0.862 * | −0.002 | 0.542 | 0.818 * | 0.881 * | 0.826 * | 0.679 |
| FC | 0.465 | −0.210 | 0.747 | 0.052 | 0.879 * | 0.890 * | 0.858 * |
| GC | 0.558 | −0.130 | 0.596 | 0.199 | 0.863 * | 0.860 * | 0.693 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, C.; Mei, R.; Anwar, M.; Zhao, D.; Lan, C.; Jiang, Y.; Zhuang, J.; Wang, C.; Hu, Z. The Functionally Characterization of Putative Genes Involved in the Formation of Mannose in the Aplanospore Cell Wall of Haematococcus pluvialis (Volvocales, Chlorophyta). Metabolites 2021, 11, 725. https://doi.org/10.3390/metabo11110725
Guo C, Mei R, Anwar M, Zhao D, Lan C, Jiang Y, Zhuang J, Wang C, Hu Z. The Functionally Characterization of Putative Genes Involved in the Formation of Mannose in the Aplanospore Cell Wall of Haematococcus pluvialis (Volvocales, Chlorophyta). Metabolites. 2021; 11(11):725. https://doi.org/10.3390/metabo11110725
Chicago/Turabian StyleGuo, Chunli, Rui Mei, Muhammad Anwar, Di Zhao, Chengxiang Lan, Yanan Jiang, Jieyi Zhuang, Chaogang Wang, and Zhangli Hu. 2021. "The Functionally Characterization of Putative Genes Involved in the Formation of Mannose in the Aplanospore Cell Wall of Haematococcus pluvialis (Volvocales, Chlorophyta)" Metabolites 11, no. 11: 725. https://doi.org/10.3390/metabo11110725
APA StyleGuo, C., Mei, R., Anwar, M., Zhao, D., Lan, C., Jiang, Y., Zhuang, J., Wang, C., & Hu, Z. (2021). The Functionally Characterization of Putative Genes Involved in the Formation of Mannose in the Aplanospore Cell Wall of Haematococcus pluvialis (Volvocales, Chlorophyta). Metabolites, 11(11), 725. https://doi.org/10.3390/metabo11110725








