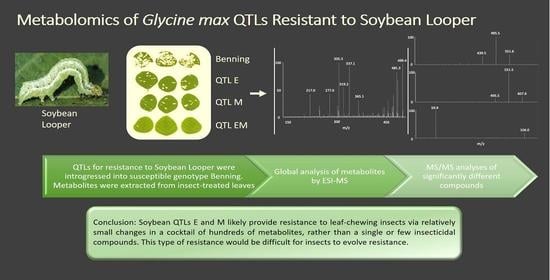
Metabolomics Differences of Glycine max QTLs Resistant to Soybean Looper
Abstract
:1. Introduction
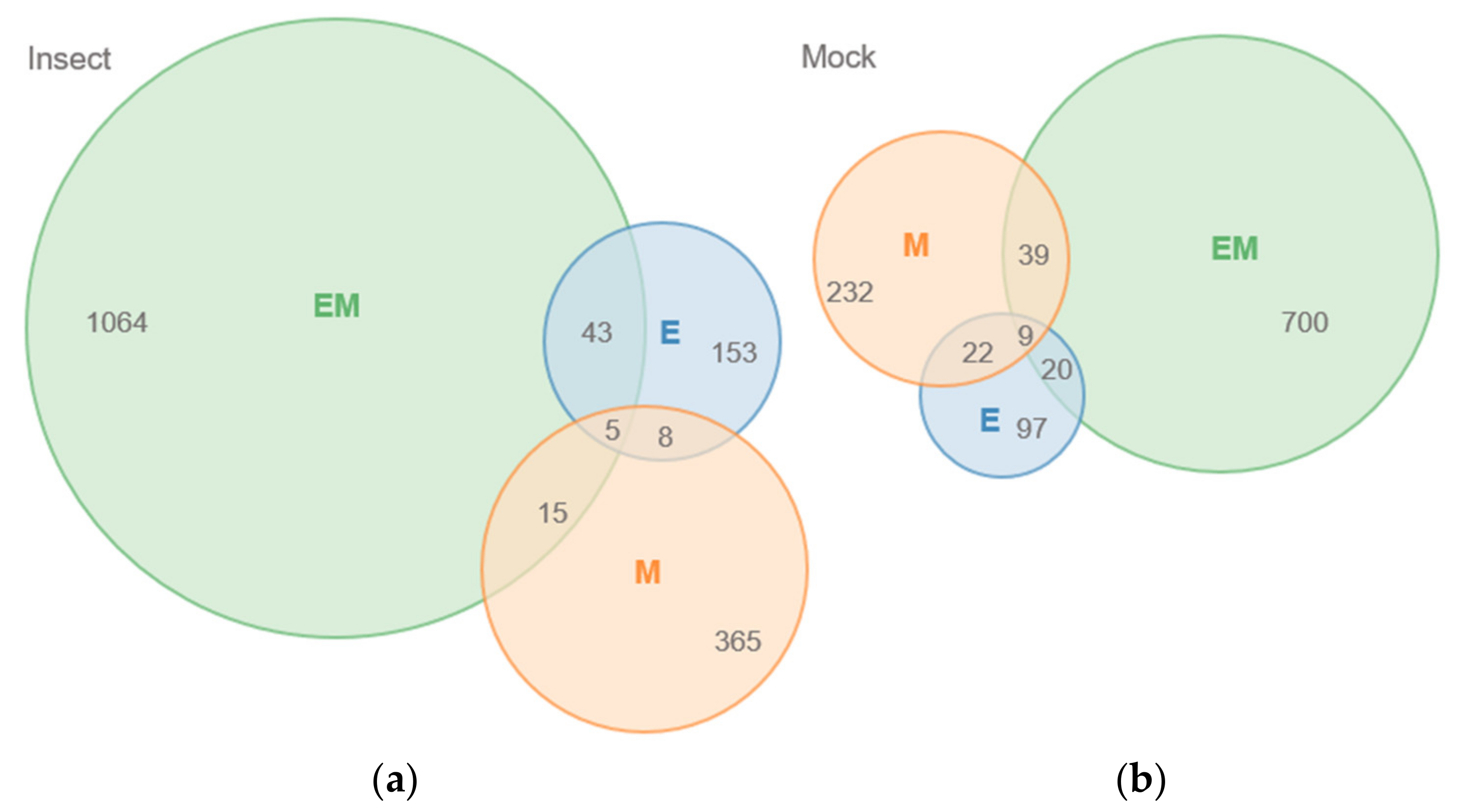
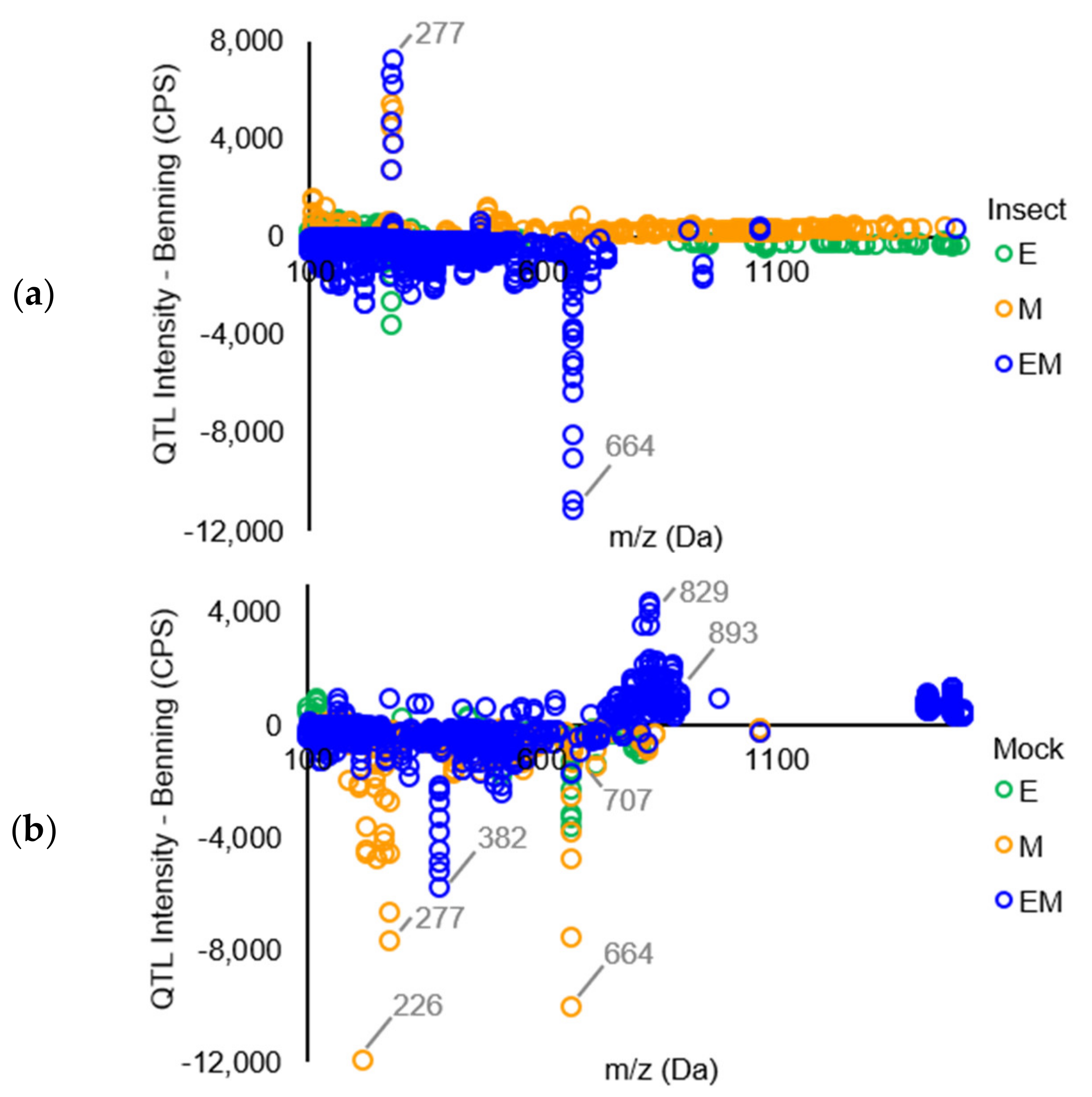
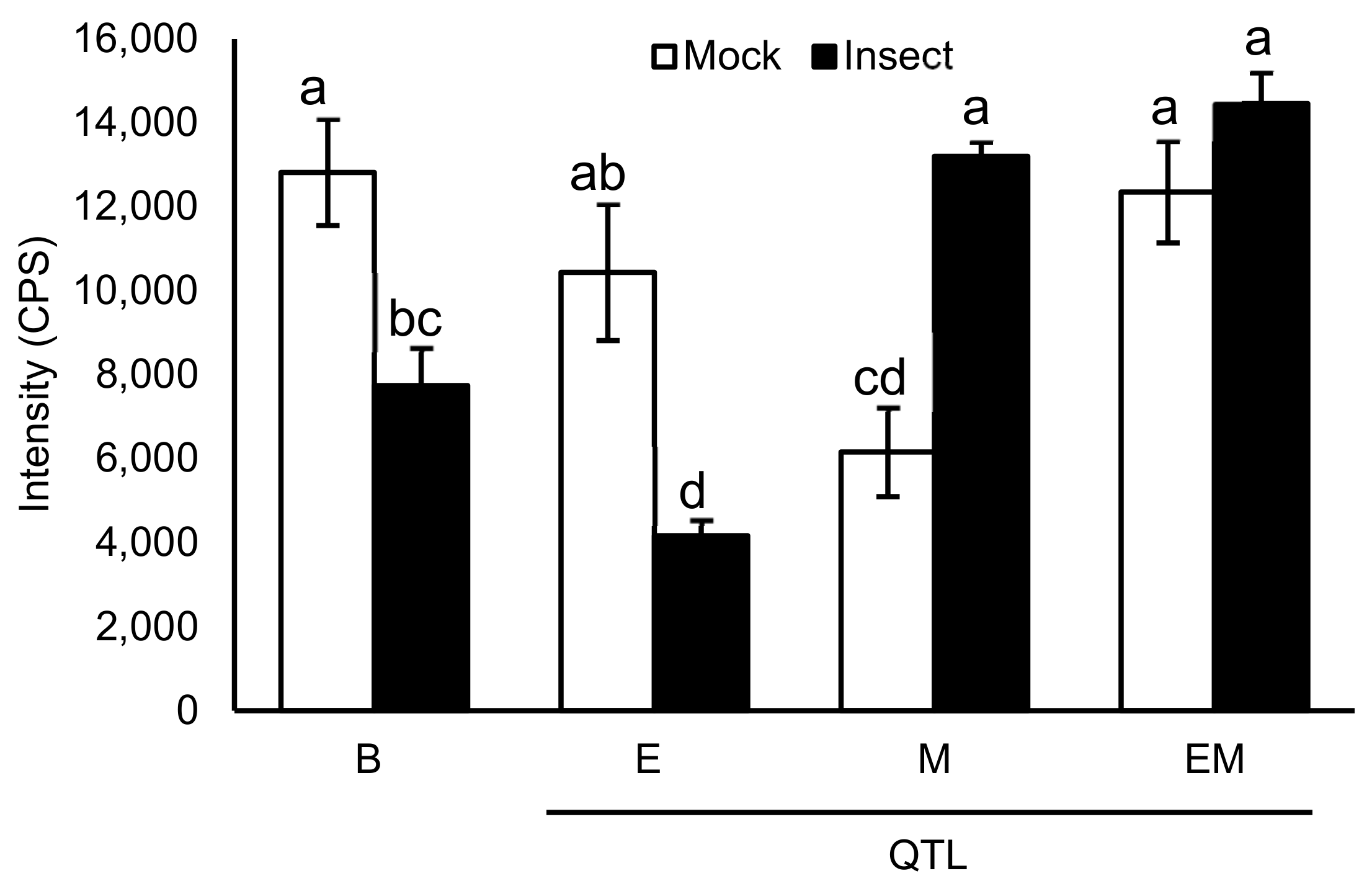
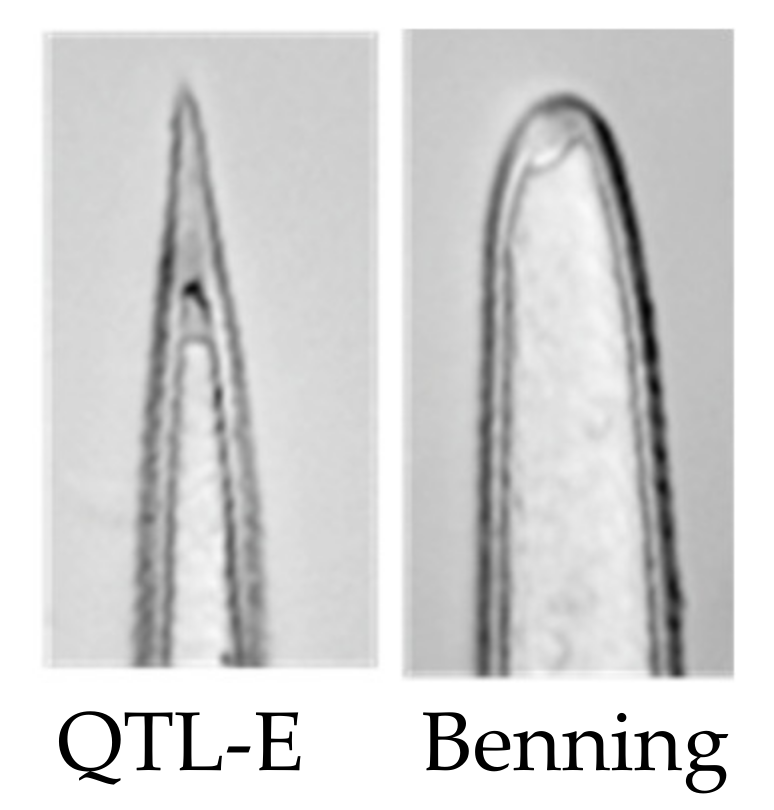
2. Results
Mass Spectrometric Comparision of Metabolites from Soybean QTLs Resistant to Soybean Looper
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Plant Growth Conditions
4.3. Insect Treatments
4.4. Extraction and Sample Preparations
4.5. Metabolite Analysis by ESI-MS
4.6. ESI-MS/MS and Database Search for Putative Compound Identification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Peshin, R.; Dhawan, A.K. (Eds.) Integrated Pest Management: Innovation-Development Process; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2009; pp. 83–87. [Google Scholar]
- Yang, Y.; Suh, S. Changes in environmental impacts of major crops in the US. Environ. Res. Lett. 2015, 10, 094016. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, C.; Brennan, R.M.; Graham, J.; Karley, A.J. Plant defense against herbivorous pests: Exploiting resistance and tolerance traits for sustainable crop protection. Front. Plant Sci. 2016, 7, 1132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, F.; Andow, D.A.; Buschman, L.L. Success of the high-dose/refuge resistance management strategy after 15 years of Bt crop use in North America. Entomol. Exp. Appl. 2011, 140, 1–16. [Google Scholar] [CrossRef]
- Storer, N.P.; Thompson, G.D.; Head, G.P. Application of pyramided traits against Lepidoptera in insect resistance management for Bt crops. GM Crop. Food 2012, 3, 154–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parrott, W.; Walker, D.; Zhu, S.; Boerma, H.R.; All, J. Genomics of insect-soybean interactions. In Genetics and Genomics of Soybean; Springer: Berlin/Heidelberg, Germany, 2008; pp. 269–291. [Google Scholar]
- Rector, B.; All, J.; Parrott, W.; Boerma, H. Identification of molecular markers linked to quantitative trait loci for soybean resistance to corn earworm. Theor. Appl. Genet. 1998, 96, 786–790. [Google Scholar] [CrossRef]
- Rector, B.; All, J.; Parrott, W.; Boerma, H. Quantitative trait loci for antibiosis resistance to corn earworm in soybean. Crop Sci. 2000, 40, 233–238. [Google Scholar] [CrossRef]
- Lourenção, A.L.; Miranda, M.A.; Pereira, J.C.; Ambrosano, G. Resistência de soja a insetos: X. Comportamento de cultivares e linhagens em relação a percevejos e desfolhadores. An. Soc. Entomol. Bras. 1997, 26, 543–550. [Google Scholar] [CrossRef] [Green Version]
- Narvel, J.M.; Walker, D.R.; Rector, B.G.; All, J.N.; Parrott, W.A.; Boerma, H.R. A retrospective DNA marker assessment of the development of insect resistant soybean. Crop Sci. 2001, 41, 1931–1939. [Google Scholar] [CrossRef]
- Komatsu, K.; Takahashi, M.; Nakazawa, Y. Antibiosis resistance of QTL introgressive soybean lines to common cutworm (Spodoptera litura Fabricius). Crop Sci. 2008, 48, 527–532. [Google Scholar] [CrossRef]
- Oki, N.; Komatsu, K.; Sayama, T.; Ishimoto, M.; Takahashi, M.; Takahashi, M. Genetic analysis of antixenosis resistance to the common cutworm (Spodoptera litura Fabricius) and its relationship with pubescence characteristics in soybean (Glycine max (L.) Merr.). Breed. Sci. 2012, 61, 608–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terry, L.; Chase, K.; Jarvik, T.; Orf, J.; Mansur, L.; Lark, K. Soybean quantitative trait loci for resistance to insects. Crop Sci. 2000, 40, 375–382. [Google Scholar] [CrossRef]
- Ortega, M.A.; Lail, L.A.; Wood, E.D.; All, J.N.; Li, Z.; Boerma, H.R.; Parrott, W.A. Registration of Two Soybean Germplasm Lines Containing Leaf-Chewing Insect Resistance QTLs from PI 229358 and PI 227687 Introgressed into ‘Benning’. J. Plant Regist. 2017, 11, 185–191. [Google Scholar] [CrossRef]
- Ortega, M.A.; All, J.N.; Boerma, H.R.; Parrott, W.A. Pyramids of QTLs enhance host–plant resistance and Bt-mediated resistance to leaf-chewing insects in soybean. Theor. Appl. Genet. 2016, 129, 703–715. [Google Scholar] [CrossRef] [Green Version]
- Smith, C.M.; Fischer, N. Chemical factors of an insect resistant soybean genotype affecting growth and survival of the soybean looper. Entomol. Exp. Appl. 1983, 33, 343–345. [Google Scholar] [CrossRef]
- Jahan, M.A.; Harris, B.; Lowery, M.; Infante, A.M.; Percifield, R.J.; Kovinich, N. Glyceollin transcription factor GmMYB29A2 regulates soybean resistance to Phytophthora sojae. Plant Physiol. 2020, 183, 530–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lygin, A.V.; Hill, C.B.; Zernova, O.V.; Crull, L.; Widholm, J.M.; Hartman, G.L.; Lozovaya, V.V. Response of soybean pathogens to glyceollin. Phytopathology 2010, 100, 897–903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veech, J.A. Phytoalexins and their role in the resistance of plants to nematodes. J. Nematol. 1982, 14, 2. [Google Scholar] [PubMed]
- Hart, S.V.; Kogan, M.; Paxton, J.D. Effect of soybean phytoalexins on the herbivorous insects Mexican bean beetle and soybean looper. J. Chem. Ecol. 1983, 9, 657–672. [Google Scholar] [CrossRef]
- Caballero, P.; Smith, C.M.; Fronczek, F.R.; Fischer, N.H. Isoflavones from an insect-resistant variety of soybean and the molecular structure of afrormosin. J. Nat. Prod. 1986, 49, 1126–1129. [Google Scholar] [CrossRef]
- Sharma, H.; Norris, D.M. Chemical basis of resistance in soya bean to cabbage looper, Trichoplusia ni. J. Sci. Food Agric. 1991, 55, 353–364. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Norris, D.M.; Hartwig, E.E.; Xu, M. Inducible phytoalexins in juvenile soybean genotypes predict soybean looper resistance in the fully developed plants. Plant Physiol. 1992, 100, 1479–1485. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Norris, D.M.; Xu, M. Insect resistance and glyceollin concentration in seedling soybeans support resistance ratings of fully developed plants. J. Econ. Entomol. 1993, 86, 401–406. [Google Scholar] [CrossRef]
- Piubelli, G.C.; Hoffmann-Campo, C.B.; Moscardi, F.; Miyakubo, S.H.; De Oliveira, M.C.N. Are chemical compounds important for soybean resistance to Anticarsia gemmatalis. J. Chem. Ecol. 2005, 31, 1509–1525. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann-Campo, C.B.; Ramos Neto, J.A.; Oliveira, M.C.N.D.; Oliveira, L.J. Detrimental effect of rutin on Anticarsia gemmatalis. Pesqui. Agropecu. Bras. 2006, 41, 1453–1459. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wang, H.; Fan, R.; Yang, Q.; Yu, D. Transcriptome analysis of soybean lines reveals transcript diversity and genes involved in the response to common cutworm (Spodoptera litura Fabricius) feeding. Plant Cell Environ. 2014, 37, 2086–2101. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, H.; Ma, Y.; Yang, W.; Yang, Q.; Yu, D. Identification of soybean herbivory-regulated genes and a transgenic investigation of their potential in insect resistance. Plant Cell Tissue Organ Cult. PCTOC 2015, 123, 321–340. [Google Scholar] [CrossRef]
- Zhao, G.; Jiang, Z.; Li, D.; Han, Y.; Hu, H.; Wu, L.; Wang, Y.; Gao, Y.; Teng, W.; Li, Y. Molecular loci associated with seed isoflavone content may underlie resistance to soybean pod borer (Leguminivora glycinivorella). Plant Breed. 2015, 134, 78–84. [Google Scholar] [CrossRef]
- Gómez, J.D.; Vital, C.E.; Oliveira, M.G.; Ramos, H.J. Broad range flavonoid profiling by LC/MS of soybean genotypes contrasting for resistance to Anticarsia gemmatalis (Lepidoptera: Noctuidae). PLoS ONE 2018, 13, e0205010. [Google Scholar] [CrossRef]
- Gómez, J.D.; Pinheiro, V.J.; Silva, J.C.; Romero, J.V.; Meriño-Cabrera, Y.; Coutinho, F.S.; Lourenção, A.L.; Serrão, J.E.; Vital, C.E.; Fontes, E.P. Leaf metabolic profiles of two soybean genotypes differentially affect the survival and the digestibility of Anticarsia gemmatalis caterpillars. Plant Physiol. Biochem. 2020, 155, 196–212. [Google Scholar] [CrossRef]
- Yesudas, C.; Sharma, H.; Lightfoot, D. Identification of QTL in soybean underlying resistance to herbivory by Japanese beetles (Popillia japonica, Newman). Theor. Appl. Genet. 2010, 121, 353–362. [Google Scholar] [CrossRef]
- O’neill, B.F.; Zangerl, A.R.; Dermody, O.; Bilgin, D.D.; Casteel, C.L.; Zavala, J.A.; DeLucia, E.H.; Berenbaum, M.R. Impact of elevated levels of atmospheric CO2 and herbivory on flavonoids of soybean (Glycine max Linnaeus). J. Chem. Ecol. 2010, 36, 35–45. [Google Scholar] [CrossRef]
- Hay, W.T.; Behle, R.W.; Berhow, M.A.; Miller, A.C.; Selling, G.W. Biopesticide synergy when combining plant flavonoids and entomopathogenic baculovirus. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hohenstein, J.D.; Studham, M.E.; Klein, A.; Kovinich, N.; Barry, K.; Lee, Y.-J.; MacIntosh, G.C. Transcriptional and chemical changes in soybean leaves in response to long-term aphid colonization. Front. Plant Sci. 2019, 10, 310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kikuta, S. The Cytotoxic Effect of Genistein, a Soybean Isoflavone, against Cultured Tribolium Cells. Insects 2020, 11, 241. [Google Scholar] [CrossRef] [PubMed]
- Sabljic, I.; Barneto, J.A.; Balestrasse, K.B.; Zavala, J.A.; Pagano, E.A. Role of reactive oxygen species and isoflavonoids in soybean resistance to the attack of the southern green stink bug. PeerJ 2020, 8, e9956. [Google Scholar] [CrossRef] [PubMed]
- Murakami, S.; Nakata, R.; Aboshi, T.; Yoshinaga, N.; Teraishi, M.; Okumoto, Y.; Ishihara, A.; Morisaka, H.; Huffaker, A.; Schmelz, E.A. Insect-induced daidzein, formononetin and their conjugates in soybean leaves. Metabolites 2014, 4, 532–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakata, R.; Kimura, Y.; Aoki, K.; Yoshinaga, N.; Teraishi, M.; Okumoto, Y.; Huffaker, A.; Schmelz, E.A.; Mori, N. Inducible de novo biosynthesis of isoflavonoids in soybean leaves by Spodoptera litura derived elicitors: Tracer techniques aided by high resolution LCMS. J. Chem. Ecol. 2016, 42, 1226–1236. [Google Scholar] [CrossRef] [Green Version]
- Hulburt, D.J.; Boerma, H.R.; All, J.N. Effect of pubescence tip on soybean resistance to lepidopteran insects. J. Econ. Entomol. 2004, 97, 621–627. [Google Scholar] [CrossRef]
- Hollowell, E.; Johnson, H. Correlation between rough-hairy pubescence in soybean and freedom from injury by Empoasca fabae. Phytopathology 1934, 24, 12. [Google Scholar]
- Kanno, H. Role of leaf pubescence in soybean resistance to the false melon beetle. Atrachya Menetriesi 1996, 31, 597–603. [Google Scholar]
- Hulburt, D. Identifying Additional Insect Resistance Quantitative Trait Loci in Soybean Using Simple Sequence Repeats. Master’s Thesis, University of Georgia, Athens, GA, USA, 2002. [Google Scholar]
- Ting, C. Genetic Studies on the Wild and Cultivated Soybeans 1. Agron. J. 1946, 38, 381–393. [Google Scholar] [CrossRef]
- Lambert, L.; Kilen, T. Insect Resistance Factor in Soybean PI’s 229358 and 227687 Demonstrated by Grafting 1. Crop Sci. 1984, 24, 163–165. [Google Scholar] [CrossRef]
- Fehr, W.; Caviness, C.; Burmood, D.; Pennington, J. Stage of development descriptions for soybeans, Glycine max (L.) Merrill. Crop Sci. 1971, 11, 929–931. [Google Scholar] [CrossRef]
- Allen, F.; Greiner, R.; Wishart, D. Competitive fragmentation modeling of ESI-MS/MS spectra for putative metabolite identification. Metabolomics 2015, 11, 98–110. [Google Scholar] [CrossRef] [Green Version]
- Smith, C.A.; O’Maille, G.; Want, E.J.; Qin, C.; Trauger, S.A.; Brandon, T.R.; Custodio, D.E.; Abagyan, R.; Siuzdak, G. METLIN: A metabolite mass spectral database. Ther. Drug Monit. 2005, 27, 747–751. [Google Scholar] [CrossRef] [PubMed]
- National Institute of Standards and Technology. EPA/NIH Mass Spectral Library; National Institute of Standards and Technology: Gaithersburg, MD, USA, 2014.
- Huang, F.-Q.; Dong, X.; Yin, X.; Fan, Y.; Fan, Y.; Mao, C.; Zhou, W. A mass spectrometry database for identification of saponins in plants. J. Chromatogr. A 2020, 1625, 461296. [Google Scholar] [CrossRef] [PubMed]
- Blaženović, I.; Kind, T.; Ji, J.; Fiehn, O. Software tools and approaches for compound identification of LC-MS/MS data in metabolomics. Metabolites 2018, 8, 31. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yousefi-Taemeh, M.; Lin, J.; Ifa, D.R.; Parrott, W.; Kovinich, N. Metabolomics Differences of Glycine max QTLs Resistant to Soybean Looper. Metabolites 2021, 11, 710. https://doi.org/10.3390/metabo11100710
Yousefi-Taemeh M, Lin J, Ifa DR, Parrott W, Kovinich N. Metabolomics Differences of Glycine max QTLs Resistant to Soybean Looper. Metabolites. 2021; 11(10):710. https://doi.org/10.3390/metabo11100710
Chicago/Turabian StyleYousefi-Taemeh, Maryam, Jie Lin, Demian R. Ifa, Wayne Parrott, and Nik Kovinich. 2021. "Metabolomics Differences of Glycine max QTLs Resistant to Soybean Looper" Metabolites 11, no. 10: 710. https://doi.org/10.3390/metabo11100710
APA StyleYousefi-Taemeh, M., Lin, J., Ifa, D. R., Parrott, W., & Kovinich, N. (2021). Metabolomics Differences of Glycine max QTLs Resistant to Soybean Looper. Metabolites, 11(10), 710. https://doi.org/10.3390/metabo11100710