Metabolomics and Inflammatory Mediator Profiling for the Differentiation of Life-Threatening and Non-Severe Appendicitis in the Pediatric Population
Abstract
:1. Introduction
2. Results
2.1. Sample Description
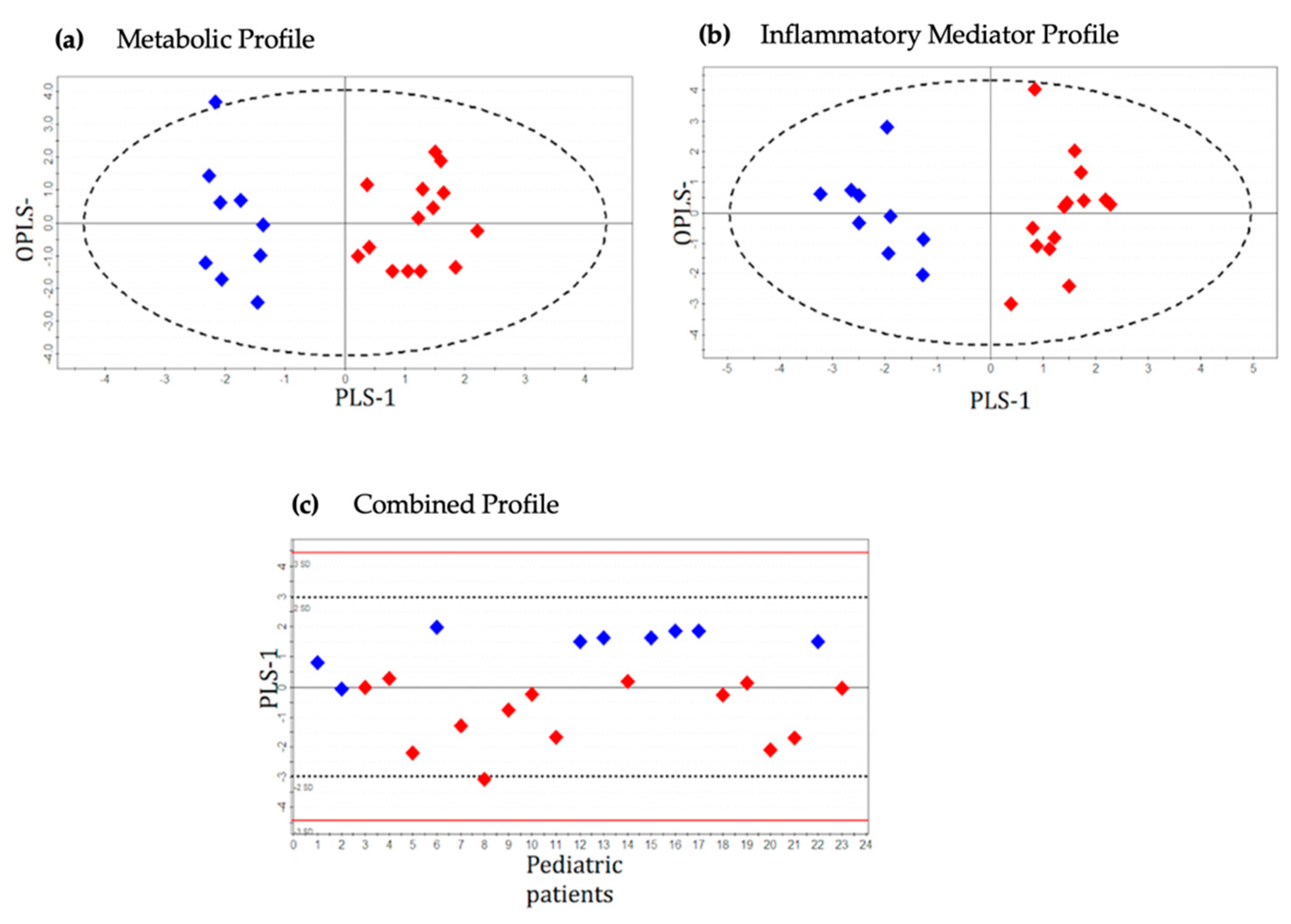
2.2. Metabolic Profile
2.3. Inflammatory Protein Mediator Profile
2.4. Integrated Metabolic and Protein Mediator Profile
3. Discussion
4. Materials and Methods
4.1. Study Design, Setting and Population
4.2. Sample Collection
4.3. Proton Nuclear Magnetic Resonance (1H NMR) Spectroscopy
4.4. Gas Chromatography-Mass Spectrometry (GC-MS)
4.5. Inflammatory Mediator Profiling
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Williams, N.; Kapila, L. Acute appendicitis in the under-5 year old. J. R. Cool Surg. Edinb. 1994, 39, 168–170. [Google Scholar]
- Guthery, S.L.; Hutchings, C.; Dean, J.M.; Hoff, C. National estimates of hospital utilization by children with gastrointestinal disorders: Analysis of the 1997 kids’ inpatient database. J. Pediatr. 2004, 144, 589–594. [Google Scholar] [CrossRef]
- Selbst, S.M.; Friedman, M.J.; Singh, S.B. Epidemiology and etiology of malpractice lawsuits involving children in US emergency departments and urgent care centers. Pediatr. Emerg. Care 2005, 21, 165–169. [Google Scholar]
- Andersson, R.E. Short and long-term mortality after appendectomy in Sweden 1987 to 2006. Influence of appendectomy diagnosis, sex, age, co-morbidity, surgical method, hospital volume, and time period. A national population-based cohort study. World J. Surg. 2013, 37, 974–981. [Google Scholar] [CrossRef]
- Blomqvist, P.G.; Andersson, R.E.; Granath, F.; Lambe, M.P.; Ekbom, A.R. Mortality after appendectomy in Sweden, 1987–1996. Ann. Surg. 2001, 233, 455–460. [Google Scholar] [CrossRef]
- Andersson, M.N.; Andersson, R.E. Causes of short-term mortality after appendectomy: A population-based case-controlled study. Ann. Surg. 2011, 254, 103–107. [Google Scholar] [CrossRef]
- Bundy, D.G.; Byerley, J.S.; Liles, E.A.; Perrin, E.M.; Katznelson, J.; Rice, H.E. Does this child have appendicitis? JAMA 2007, 298, 438–451. [Google Scholar] [CrossRef] [Green Version]
- Hennelly, K.E.; Bachur, R. Appendicitis update. Curr. Opin. Pediatr. 2011, 23, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Rothrock, S.G.; Pagane, J. Acute appendicitis in children: Emergency department diagnosis and management. Ann. Emerg. Med. 2000, 36, 39–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vissers, R.J.; Lennarz, W.B. Pitfalls in appendicitis. Emerg. Med. Clin. North. Am. 2010, 28, 103–118. [Google Scholar] [CrossRef] [PubMed]
- Zouari, M.; Jallouli, M.; Louati, H.; Kchaou, R.; Chtourou, R.; Kotti, A.; Dhaou, M.B.; Zitouni, H.; Mhiri, R. Predictive value of C-reactive protein, ultrasound and Alvarado score in acute appendicitis: A prospective pediatric cohort. Am. J. Emerg. Med. 2016, 34, 189–192. [Google Scholar] [CrossRef]
- Abbas, M.H.; Choudhry, M.N.; Hamza, N.; Ali, B.; Amin, A.A.; Ammori, B.J. Admission levels of serum amyloid a and procalcitonin are more predictive of the diagnosis of acute appendicitis compared with C-reactive protein. Surg. Laparosc. Endosc. Percutaneous Tech. 2014, 24, 488–494. [Google Scholar] [CrossRef]
- Soomro, A.G.; Siddiqui, F.G.; Abro, A.H.; Abro, S.; Shaikh, N.A.; Memon, A.S. Diagnostic accuracy of Alvarado scoring system in acute appendicitis. J. Liaquat. Univ. Med. Health Sci. 2008, 7, 93–96. [Google Scholar]
- Yegane, R.P.; Hajinasrollah, E.; Ahmadei, M. Evaluation of modified Alvarado score in acute appendicitis among Iranian patients. Acta Med. Iran. 2008, 46, 501–506. [Google Scholar]
- Curran, T.J.; Muenchow, S.K. The treatment of complicated appendicitis in children using peritoneal drainage: Results from a public hospital. J. Pediatr. Surg. 1993, 28, 204–208. [Google Scholar] [CrossRef]
- Acosta, R.; Crain, E.F.; Goldman, H.S. CT can reduce hospitalization for observation in children with suspected appendicitis. Pediatr. Radiol. 2005, 35, 495–500. [Google Scholar] [CrossRef]
- Andersson, R.E. Meta-analysis of the clinical and laboratory diagnosis of appendicitis. Br. J. Surg. 2004, 91, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Nance, M.L.; Adamson, W.T.; Hedrick, H.L. Appendicitis in the young child: A continuing diagnostic challenge. Pediatr. Emerg. Care 2000, 16, 160–162. [Google Scholar] [CrossRef] [PubMed]
- Rothrock, S.G.; Skeoch, G.; Rush, J.J.; Johnson, N.E. Clinical features of misdiagnosed appendicitis in children. Ann. Emerg. Med. 1991, 20, 45–50. [Google Scholar] [CrossRef]
- Mickiewicz, B.; Duggan, G.E.; Winston, B.W.; Doig, C.; Kubes, P.; Vogel, H.J. Metabolic profiling of serum samples by 1H nuclear magnetic resonance spectroscopy as a potential diagnostic approach for septic shock. Crit. Care Med. 2014, 42, 1140–1149. [Google Scholar] [CrossRef] [PubMed]
- Mickiewicz, B.; Kelly, J.J.; Ludwig, T.E.; Weljie, A.M.; Wiley, J.P.; Schmidt, T.A.; Vogel, H.J. Metabolic analysis of knee synovial fluid as a potential diagnostic approach for osteoarthritis. J. Orthop. Res. 2015, 33, 1631–1638. [Google Scholar] [CrossRef] [PubMed]
- Roberts, D.J.; Jenne, C.N.; Léger, C.; Kramer, A.H.; Gallagher, C.N.; Todd, S.; Parney, I.F.; Doig, C.J.; Yong, V.W.; Kubes, P.; et al. A prospective evaluation of the temporal matrix metalloproteinase response after severe traumatic brain injury in humans. J. Neurotrauma 2013, 30, 1717–1726. [Google Scholar] [CrossRef] [PubMed]
- Roberts, D.J.; Jenne, C.N.; Léger, C.; Kramer, A.H.; Gallagher, C.N.; Todd, S.; Parney, I.F.; Doig, C.J.; Yong, V.W.; Kubes, P.; et al. Association between the cerebral inflammatory and matrix metalloproteinase responses after severe traumatic brain injury in humans. J. Neurotrauma 2013, 30, 1727–1736. [Google Scholar] [CrossRef]
- Soininen, P.; Kangas, A.J.; Würtz, P.; Tukiainen, T.; Tynkkynen, T.; Laatikainen, R.; Järvelin, M.R.; Kähönen, M.; Lehtimäki, T.; Viikari, J.; et al. High-throughput serum NMR metabonomics for cost-effective holistic studies on systemic metabolism. Analyst 2009, 134, 1781–1785. [Google Scholar] [CrossRef]
- Shommu, N.S.; Jenne, C.N.; Blackwood, J.; Joffe, A.R.; Martin, D.A.; Thompson, G.C.; Vogel, H.J. Metabolomic and inflammatory mediator based biomarker profiling as a potential novel method to aid pediatric appendicitis identification. PLoS ONE 2018, 13, e0193563. [Google Scholar] [CrossRef]
- Shommu, N.S.; Jenne, C.N.; Blackwood, J.; Martin, D.A.; Joffe, A.R.; Eccles, R.; Brindle, M.; Khanafer, I.; Vogel, H.J.; Thompson, G.C. The use of metabolomic and inflammatory mediator profiling provides a novel approach to identifying pediatric appendicitis in the emergency department. Sci. Rep. 2018, 8, 4083. [Google Scholar] [CrossRef]
- Naqvi, S.A.; Thompson, G.C.; Joffe, A.R.; Blackwood, J.; Martin, D.A.; Brindle, M.; Barkema, H.W.; Jenne, C.N. Cytokines and chemokines in pediatric appendicitis: A multiplex analysis of inflammatory protein mediators. Mediat. Inflamm. 2019, 2019. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014, 42, D199–D205. [Google Scholar] [CrossRef] [Green Version]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Eisner, R.; Young, N.; Gautam, B.; Hau, D.D.; Psychogios, N.; Dong, E.; Bouatra, S.; et al. HMDB: A knowledgebase for the human metabolome. Nucleic Acids Res. 2009, 37, D603–D610. [Google Scholar] [CrossRef]
- Arlt, A.; Bharti, R.; Ilves, I.; Häsler, R.; Miettinen, P.; Paajanen, H.; Brunke, G.; Ellrichmann, M.; Rehman, A.; Hauser, C.; et al. Characteristic changes in microbial community composition and expression of innate immune genes in acute appendicitis. Innate Immun. 2015, 21, 30–41. [Google Scholar] [CrossRef]
- Ong, D.S.T.; Wang, L.; Zhu, Y.; Ho, B.; Ding, J.L. The response of ferritin to LPS and acute phase of Pseudomonas infection. J. Endotoxin Res. 2005, 11, 267–280. [Google Scholar] [CrossRef] [Green Version]
- Orino, K.; Watanabe, K. Molecular, physiological and clinical aspects of the iron storage protein ferritin. Vet. J. 2008, 178, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Hachim, M.Y.; Ahmed, A.H. The role of the cytokines and cell-adhesion molecules on the immunopathology of acute appendicitis. Saudi Med. J. 2006, 27, 1815–1821. [Google Scholar] [PubMed]
- Yildirim, O.; Solak, C.; Koçer, B.; Ünal, B.; Karabeyoğlu, M.; Bozkurt, B.; Aksaray, S.; Cengiz, Ö. The role of serum inflammatory markers in acute appendicitis and their success in preventing negative laparotomy. J. Investig. Surg. 2006, 19, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Ruber, M.; Berg, A.; Ekerfelt, C.; Olaison, G.; Andersson, R.E. Different cytokine profiles in patients with a history of gangrenous or phlegmonous appendicitis. Clin. Exp. Immunol. 2006, 143, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Chavez, F.A.; Wheeler, H.; Lindberg, G.; Munford, R.S.; O’Keefe, G.E. Regional and systemic cytokine responses to acute inflammation of the vermiform appendix. Ann. Surg. 2003, 237, 408–416. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira Machado, S.L.; Bagatini, M.D.; da Costa, P.; Baldissarelli, J.; Reichert, K.P.; de Oliveira, L.S.; Lemos, J.G.; Duarte, T.; Chitolina Schetinger, M.R.; Morsch, V.M. Evaluation of mediators of oxidative stress and inflammation in patients with acute appendicitis. Biomarkers 2016, 21, 530–537. [Google Scholar] [CrossRef]
- Mickiewicz, B.; Holinski, P.; Vogel, H.J.; Winston, B.W.; Wong, J.; Thompson, G.; Blackwood, J.; Joffe, A.R. Metabolomic profiling of serum samples using NMR spectroscopy to diagnose pediatric sepsis in the emergency department and intensive care unit. Pediatric Crit. Care Med. 2014, 15, 146. [Google Scholar] [CrossRef]
- Mickiewicz, B.; Thompson, G.C.; Blackwood, J.; Jenne, C.N.; Winston, B.W.; Vogel, H.J.; Joffe, A.R. Biomarker Phenotype for Early Diagnosis and Triage of Sepsis to the Pediatric Intensive Care Unit. Sci. Rep. 2018, 8, 16606. [Google Scholar] [CrossRef]
- Eriksson, L.; Byrne, T.; Johansson, E.; Trygg, J.; Vikström, C. Multi- and megavariate data analysis. In Part I: Basic Principles and Applications; Umetrics Academy: Umea, Sweden, 2006; Volume 425. [Google Scholar]
- Eriksson, L. Multi- and megavariate data analysis. In Part II: Advanced Applications and Method Extensions; Umetrics Academy: Umea, Sweden, 2006; Volume 307. [Google Scholar]
- Mickiewicz, B.; Thompson, G.C.; Blackwood, J.; Jenne, C.N.; Winston, B.W.; Vogel, H.J.; Joffe, A.R. Development of metabolic and inflammatory mediator biomarker phenotyping for early diagnosis and triage of pediatric sepsis. Crit. Care 2015, 19, 320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mickiewicz, B.; Tam, P.; Jenne, C.N.; Leger, C.; Wong, J.; Winston, B.W.; Doig, C.; Kubes, P.; Vogel, H.J. Integration of metabolic and inflammatory mediator profiles as a potential prognostic approach for septic shock in the intensive care unit. Crit. Care 2015, 19, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicholson, J.K.; Foxall, P.J.; Spraul, M.; Farrant, R.D.; Lindon, J.C. 750 MHz 1H and 1H-13C NMR spectroscopy of human blood plasma. Anal. Chem. 1995, 67, 793–811. [Google Scholar] [CrossRef]
- Weljie, A.M.; Newton, J.; Mercier, P.; Carlson, E.; Slupsky, C.M. Targeted profiling: Quantitative analysis of 1H NMR metabolomics data. Anal. Chem. 2006, 78, 4430–4442. [Google Scholar] [CrossRef]
- Hummel, J.S.; Walther, D.; Kopka, J. The Golm Metabolome Database: A database for GC-MS based metabolite profiling. In Metabolomics; Nielsen, J., Jewett, M.C., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 18, pp. 75–95. [Google Scholar]
- Madsen, R.; Lundstedt, T.; Trygg, J. Chemometrics in metabolomics--a review in human disease diagnosis. Anal. Chim. Acta 2010, 659, 23–33. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. Metabonomics: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef]
- Trygg, J.; Holmes, E.; Lundstedt, T. Chemometrics in metabonomics. J. Proteome Res. 2007, 6, 469–479. [Google Scholar] [CrossRef]
- van den Berg, R.A.; Hoefsloot, H.C.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef] [Green Version]
- Metz, C.E. Basic principles of ROC analysis. Semin. Nucl. Med. 1978, 8, 283–298. [Google Scholar] [CrossRef]



| Patient Demographic | Severe Appendicitis | Moderate Appendicitis | Mild Appendicitis |
|---|---|---|---|
| (n = 15) | (n = 9) | (n = 29) | |
| Age, median (IQR) | 10 (6–14) | 9 (8–14) | 11 (9–13) |
| Sex—Female; n (%) | 6 (40.0) | 6 (40.0) | 13 (44.8) |
| Documented RLQ Pain, n (%) | 12 (80.0) | 7 (77.8) | 23 (79.3) |
| Documented Nausea, n (%) | 6 (40.0) | 5 (55.6) | 19 (65.5) |
| Documented Vomiting, n (%) | 10 (66.7) | 5 (55.6) | 19 (65.5) |
| Documented Anorexia, n (%) | 12 (80.0) | 6 (66.7) | 17 (58.6) |
| Documented Fever in the ED, n (%) | 11 (73.3) | 5 (55.6) | 16 (55.2) |
| WBC in the ED; mean (SD) | 15.9 (2.1) | 19.8 (1.5) | 17.1 (1.2) |
| Neutrophils in the ED, mean (SD) | 13.0 (2.0) | 17.0 (1.5) | 14.2 (1.1) |
| US completed in the ED, n (%) | 10 (66.7) | 8 (88.9) | 29 (100.0) |
| CT completed in the ED, n (%) | 2 (13.3) | 1 (11.1) | 1 (3.5) |
| ED Disposition, n (%) | |||
| Surgery Ward | 0 (0.0) | 0 (0.0) | 5 (17.2) |
| OR | 10 (66.7) | 9 (100.0) | 24 (82.8) |
| PICU | 5 (33.3) | 0 (0.0) | 0 (0.0) |
| PRISM 3, median (IQR) | 3 (0–4) | - | - |
| PELOD (Day 1), median (IQR) | 2 (1–11) | - | - |
| Ventilator Days, median (IQR) | 0 (0–2) | - | - |
| Inotrope Days, median (IQR) | 1 (0–1) | - | - |
| PICU Length of Stay (days), median (IQR) | 3 (3–4) | - | - |
| Analysis | Sn:Sp | AUROC | CV-ANOVA p-Value | R2Y:Q2 | |
|---|---|---|---|---|---|
| Severe vs. moderate appendicitis | Metabolic profile | 1.00:0.86 | 1.00 ± 0.00 | 1.3 × 10−4 | 0.90:0.70 |
| Inflammatory mediator profile | 0.92:0.80 | 0.92 ± 0.08 | 9.9 × 10−4 | 0.66:0.56 | |
| Combined profile | 1.00:0.88 | 1.00 ± 0.00 | 2.4 × 10−4 | 0.78:0.65 | |
| Severe vs. mild appendicitis | Metabolic profile | 0.71:0.89 | 0.93 ± 0.04 | 4.8 × 10−6 | 0.60:0.48 |
| Inflammatory mediator profile | 0.92:0.90 | 0.96 ± 0.03 | 9.3 × 10−6 | 0.63:0.55 | |
| Combined profile | 0.91:0.90 | 0.97 ± 0.03 | 5.1 × 10−6 | 0.65:0.57 |
| Analysis | R2Y:Q2 Outliers Excluded | R2Y:Q2 Outliers included | |
|---|---|---|---|
| Metabolic profile | 0.90:0.70 | 0.83:0.70 | |
| Severe vs. moderate appendicitis | Inflammatory mediator profile | 0.66:0.56 | 0.61:0.53 |
| Combined profile | 0.78:0.65 | 0.73:0.59 | |
| Severe vs. mild appendicitis | Metabolic profile Inflammatory mediator profile Combined profile | 0.60:0.48 | 0.63:0.40 |
| 0.63:0.55 | 0.62:0.55 | ||
| 0.65:0.57 | 0.63:0.56 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shommu, N.S.; Blackwood, J.; Jenne, C.N.; Joffe, A.R.; Martin, D.-A.; Mickiewicz, B.; Brindle, M.; Eccles, R.; Vogel, H.J.; Thompson, G.C.; et al. Metabolomics and Inflammatory Mediator Profiling for the Differentiation of Life-Threatening and Non-Severe Appendicitis in the Pediatric Population. Metabolites 2021, 11, 664. https://doi.org/10.3390/metabo11100664
Shommu NS, Blackwood J, Jenne CN, Joffe AR, Martin D-A, Mickiewicz B, Brindle M, Eccles R, Vogel HJ, Thompson GC, et al. Metabolomics and Inflammatory Mediator Profiling for the Differentiation of Life-Threatening and Non-Severe Appendicitis in the Pediatric Population. Metabolites. 2021; 11(10):664. https://doi.org/10.3390/metabo11100664
Chicago/Turabian StyleShommu, Nusrat S., Jaime Blackwood, Craig N. Jenne, Ari R. Joffe, Dori-Ann Martin, Beata Mickiewicz, Mary Brindle, Robin Eccles, Hans J. Vogel, Graham C. Thompson, and et al. 2021. "Metabolomics and Inflammatory Mediator Profiling for the Differentiation of Life-Threatening and Non-Severe Appendicitis in the Pediatric Population" Metabolites 11, no. 10: 664. https://doi.org/10.3390/metabo11100664
APA StyleShommu, N. S., Blackwood, J., Jenne, C. N., Joffe, A. R., Martin, D.-A., Mickiewicz, B., Brindle, M., Eccles, R., Vogel, H. J., Thompson, G. C., & on behalf of the Alberta Sepsis Network. (2021). Metabolomics and Inflammatory Mediator Profiling for the Differentiation of Life-Threatening and Non-Severe Appendicitis in the Pediatric Population. Metabolites, 11(10), 664. https://doi.org/10.3390/metabo11100664







