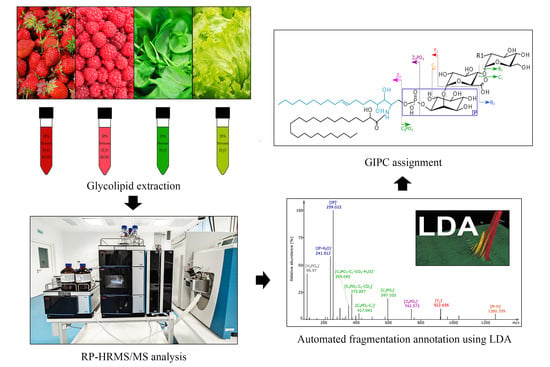
Chasing the Major Sphingolipids on Earth: Automated Annotation of Plant Glycosyl Inositol Phospho Ceramides by Glycolipidomics
Abstract
1. Introduction
2. Results
2.1. Method Development for Automated GIPC Assignment
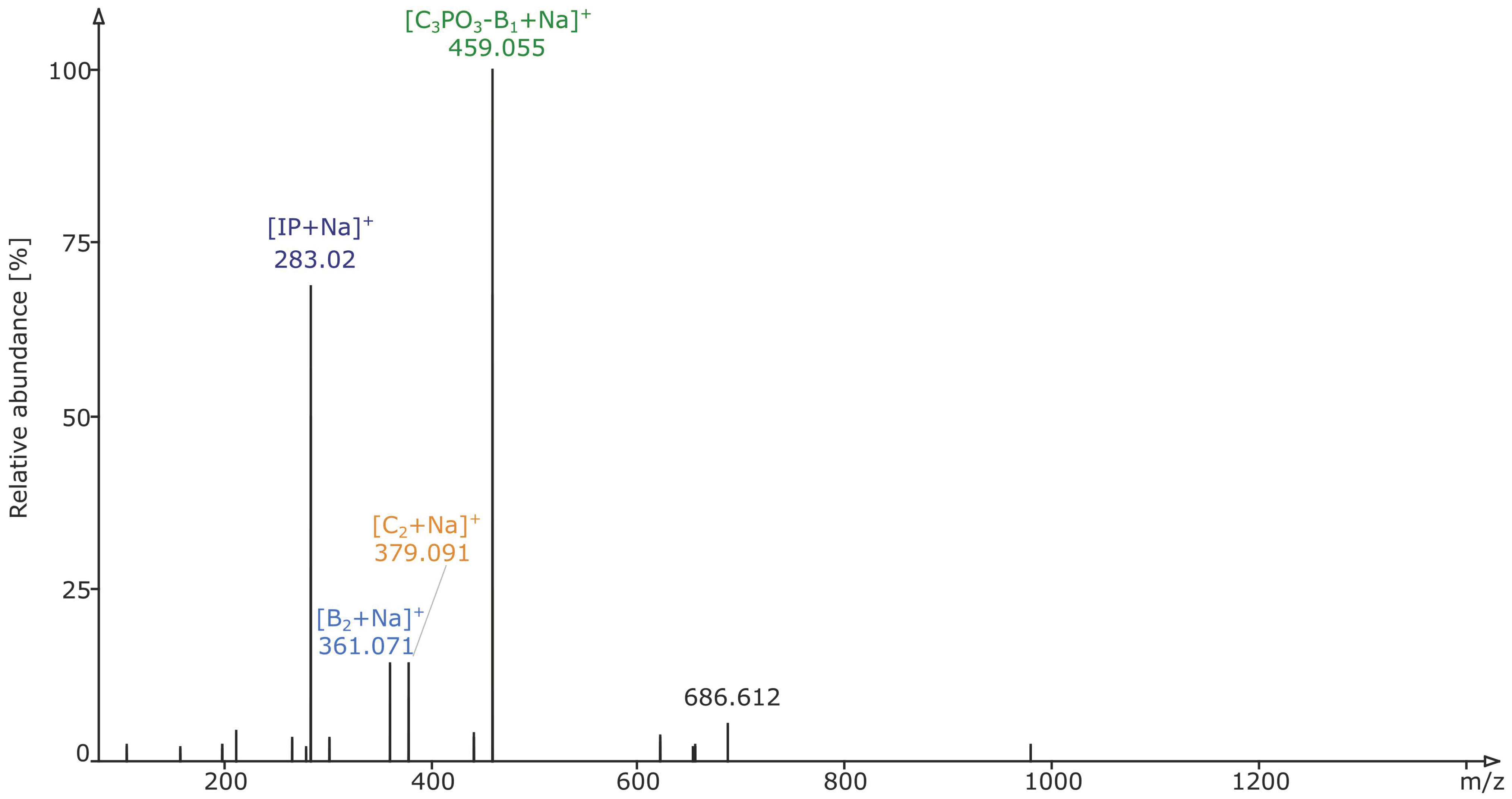
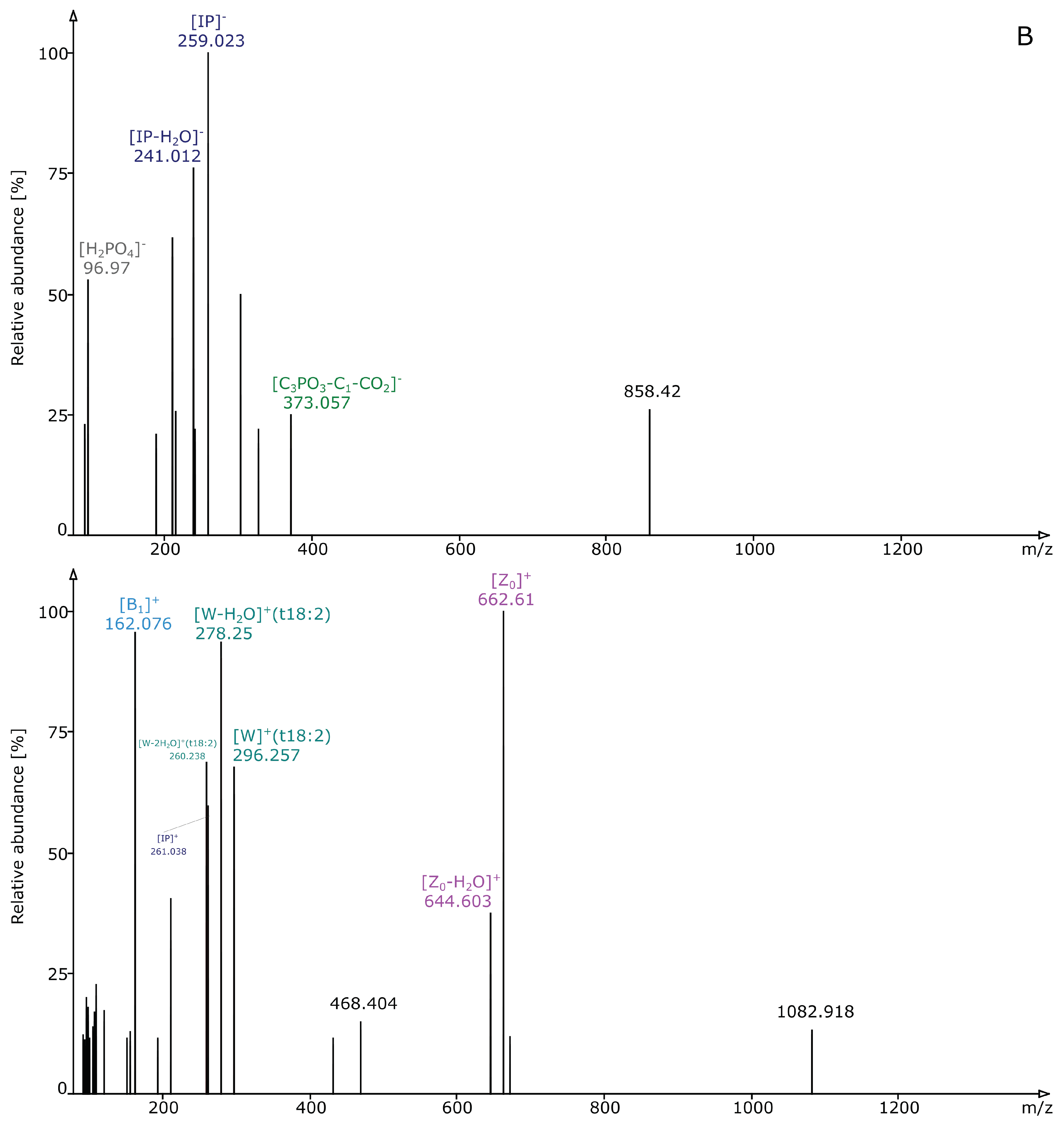
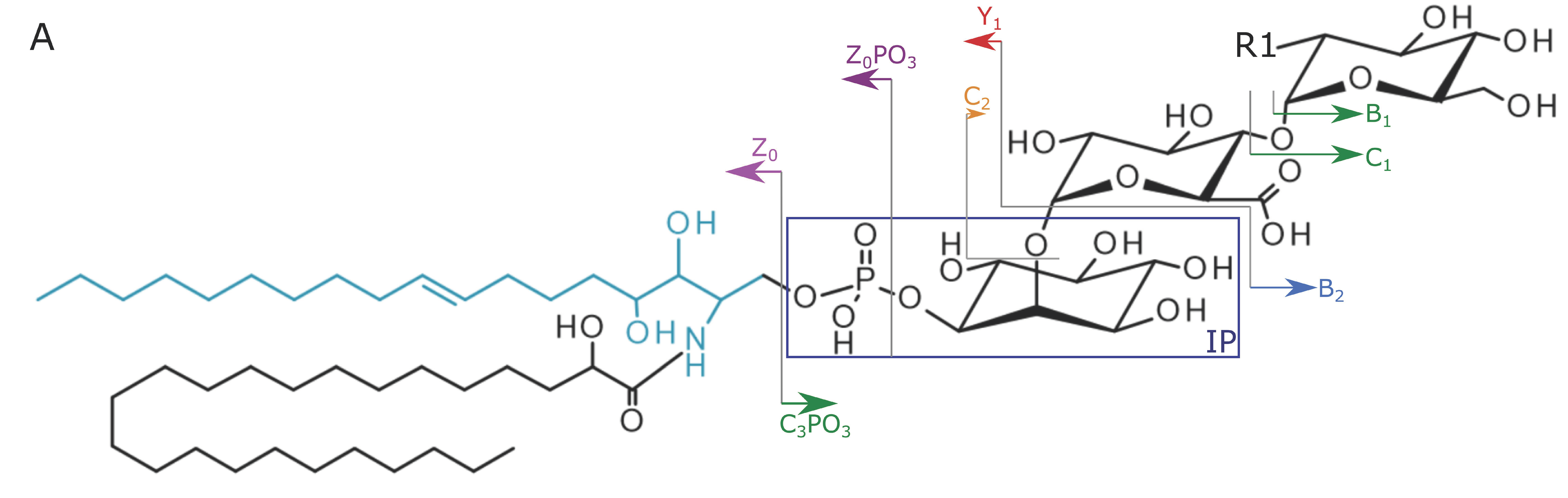
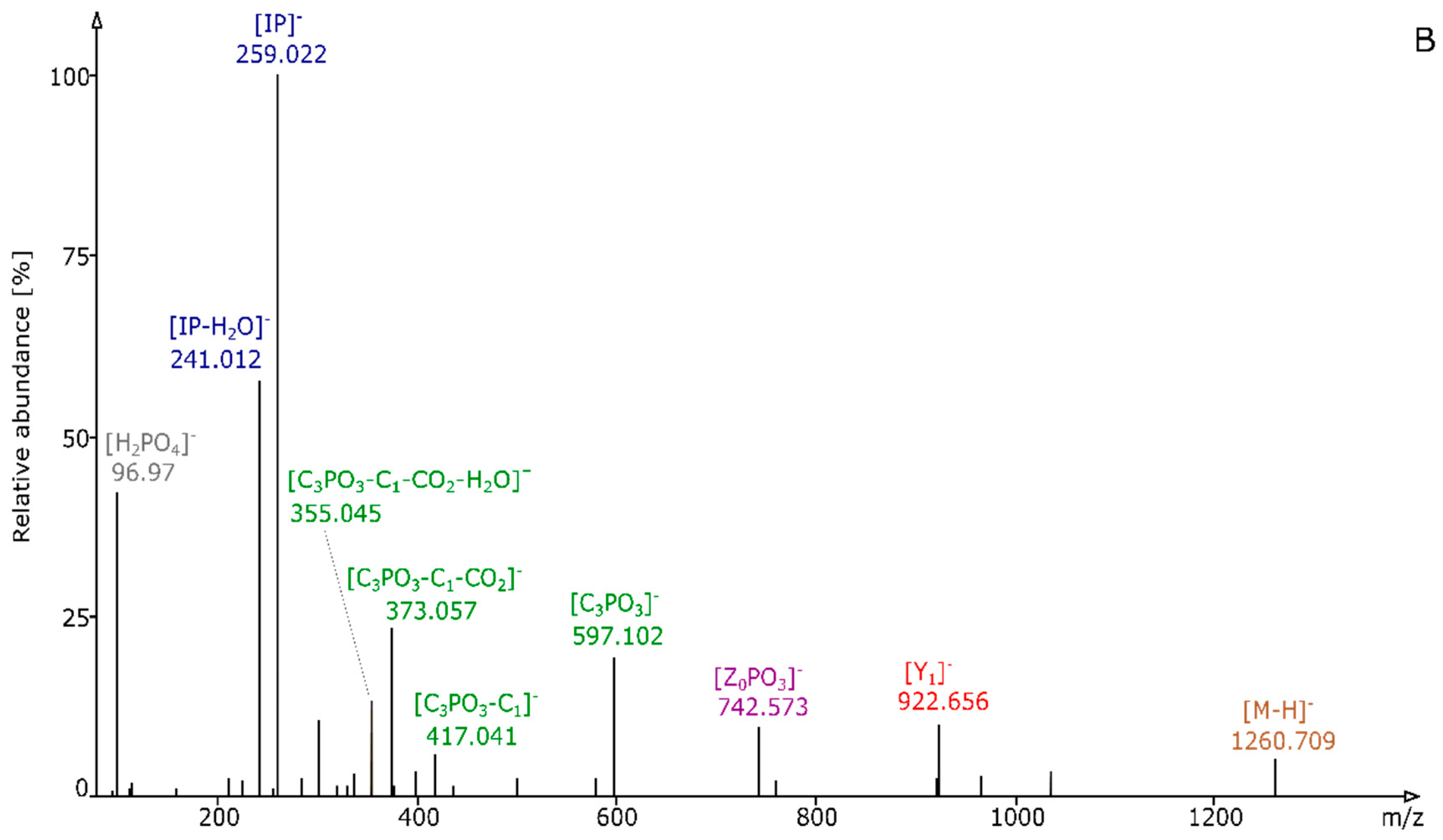
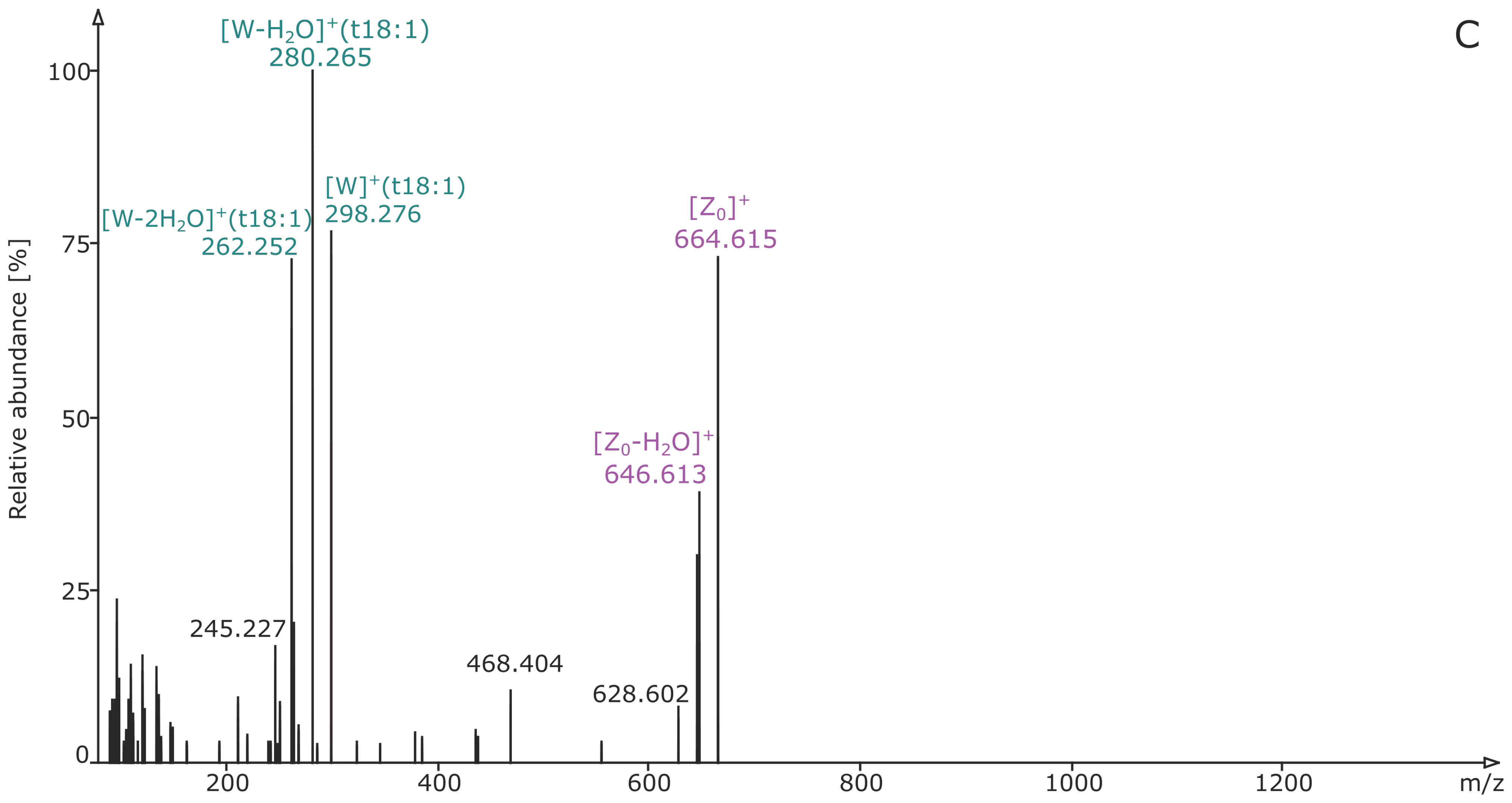
2.2. Structural Elucidation and GIPC Annotation Based on MS2 Information
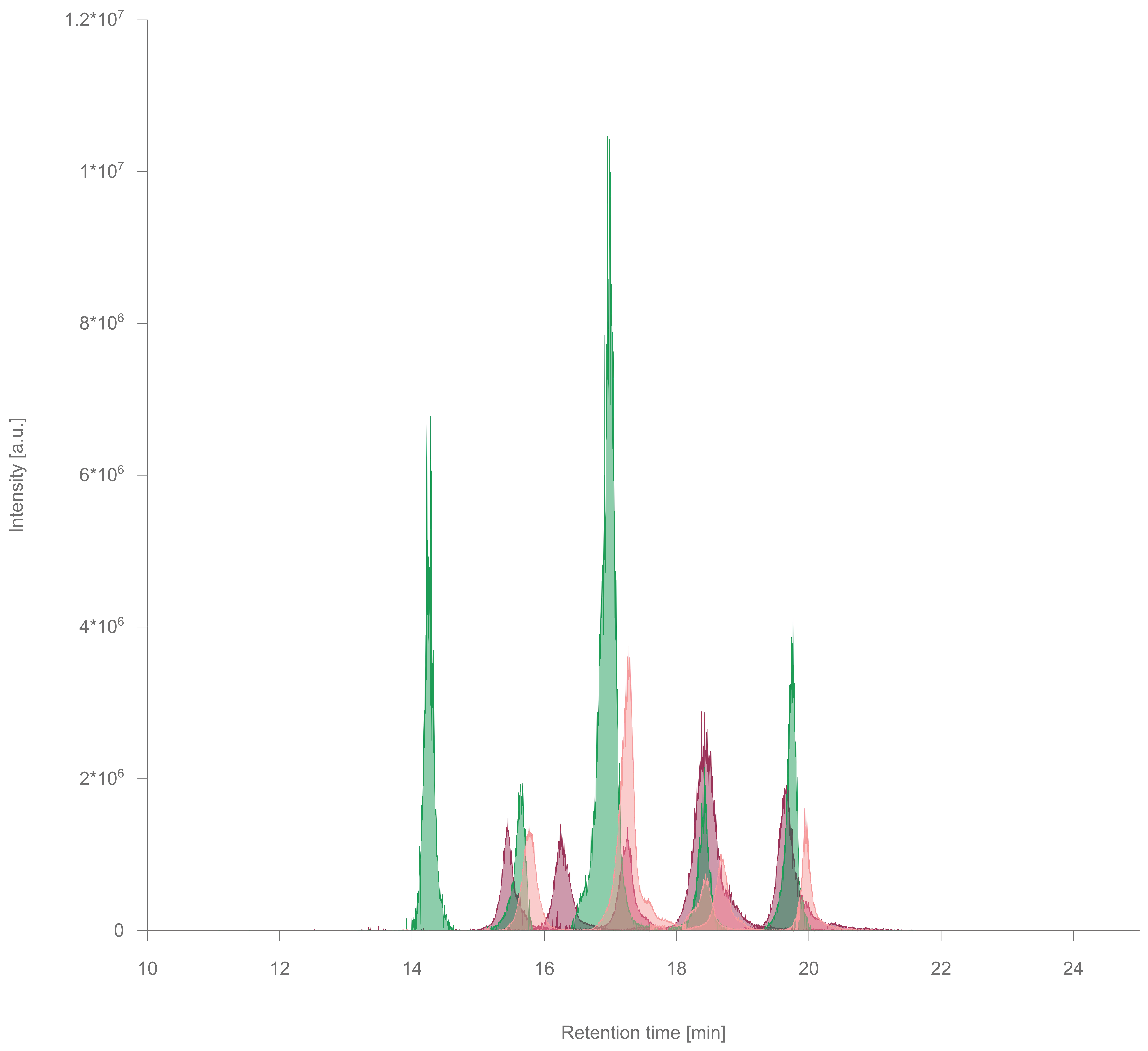
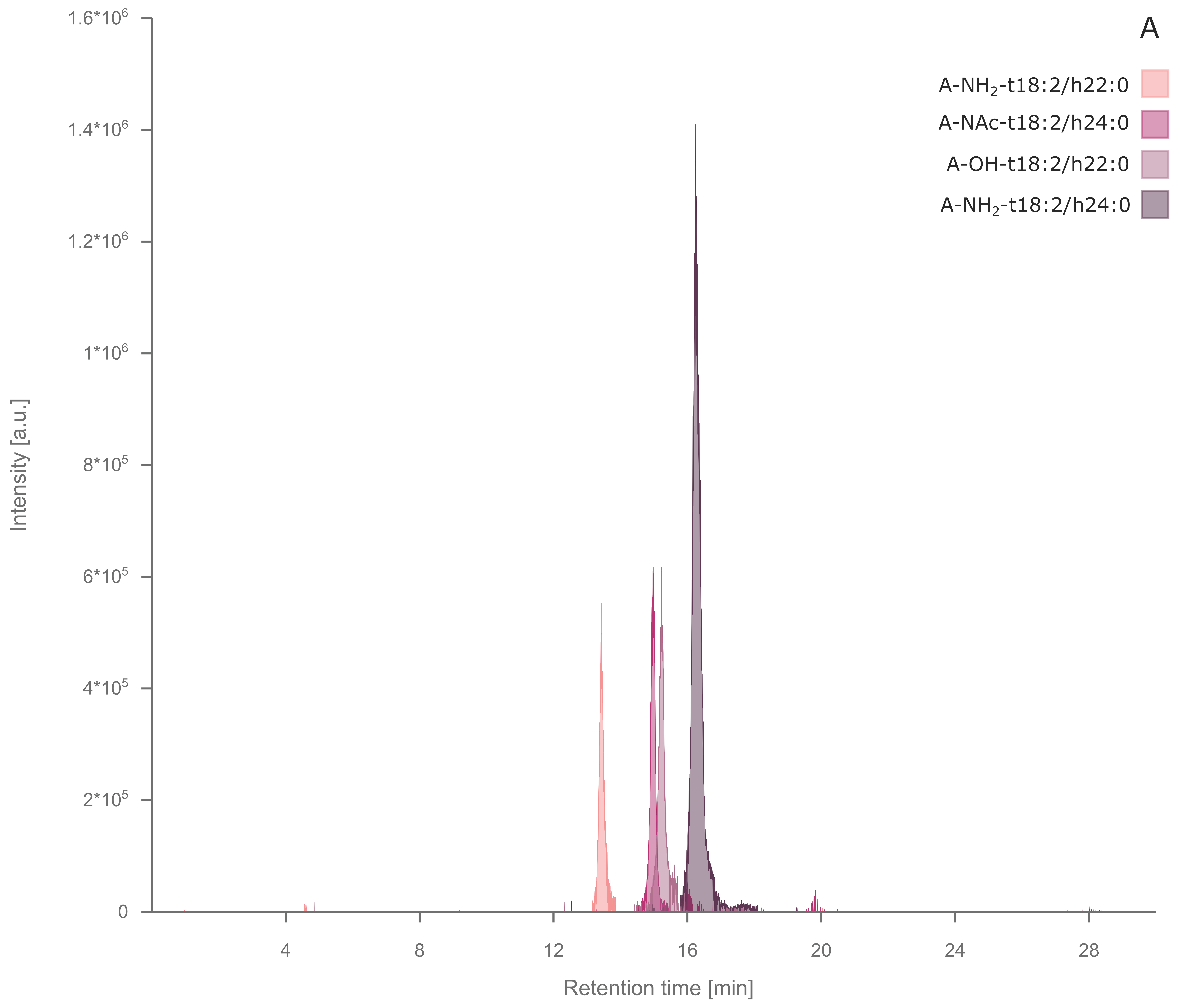
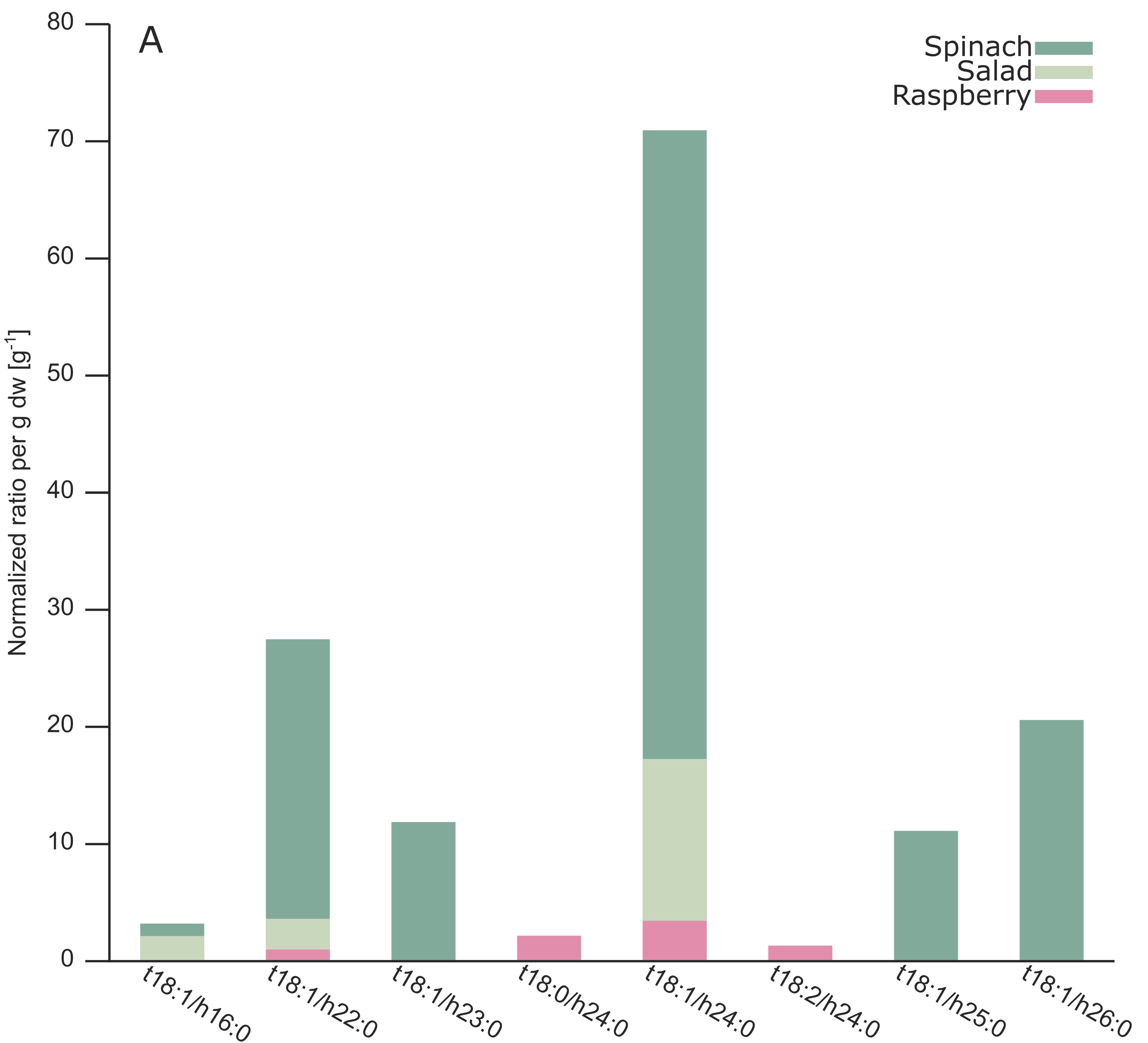
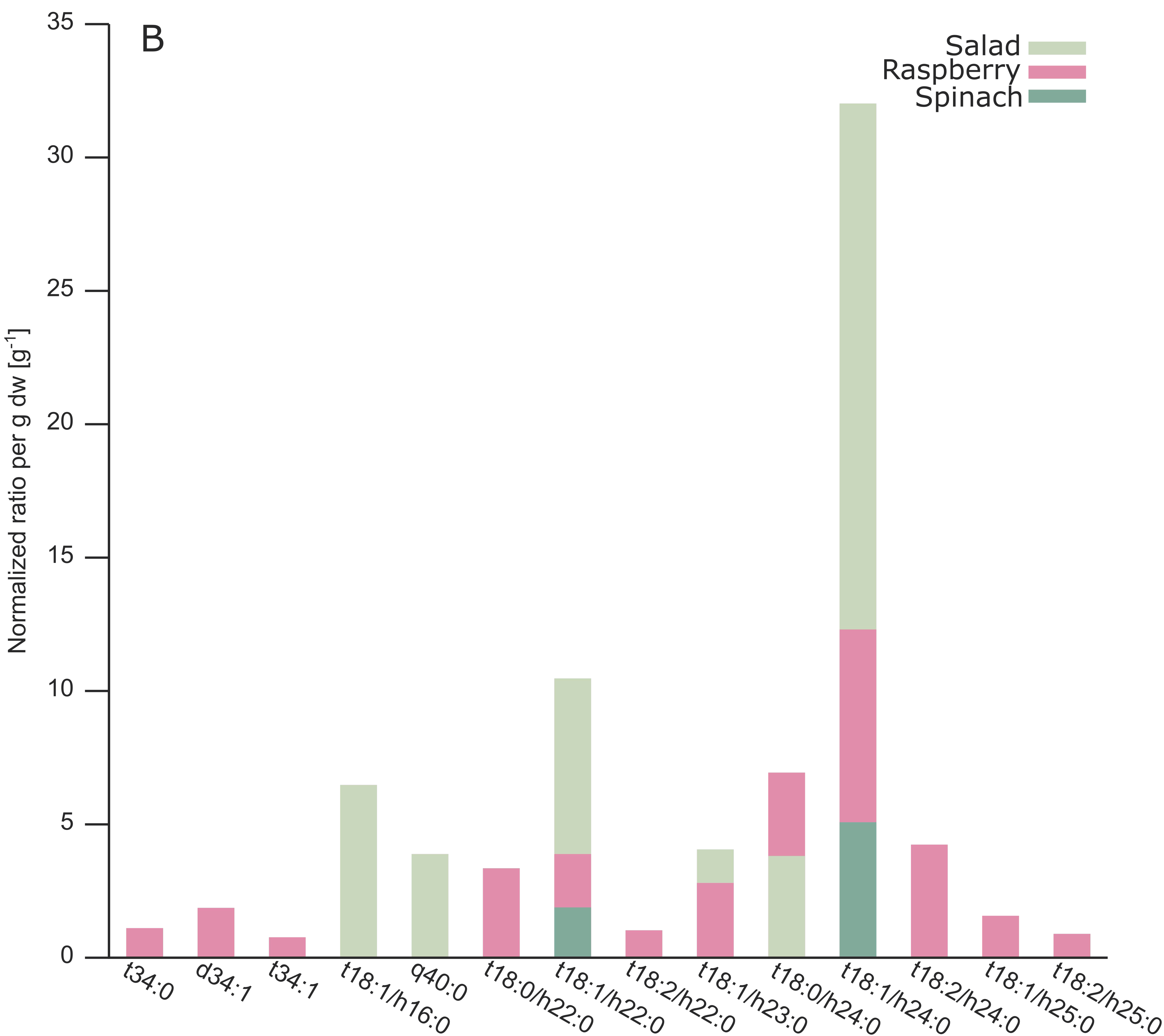
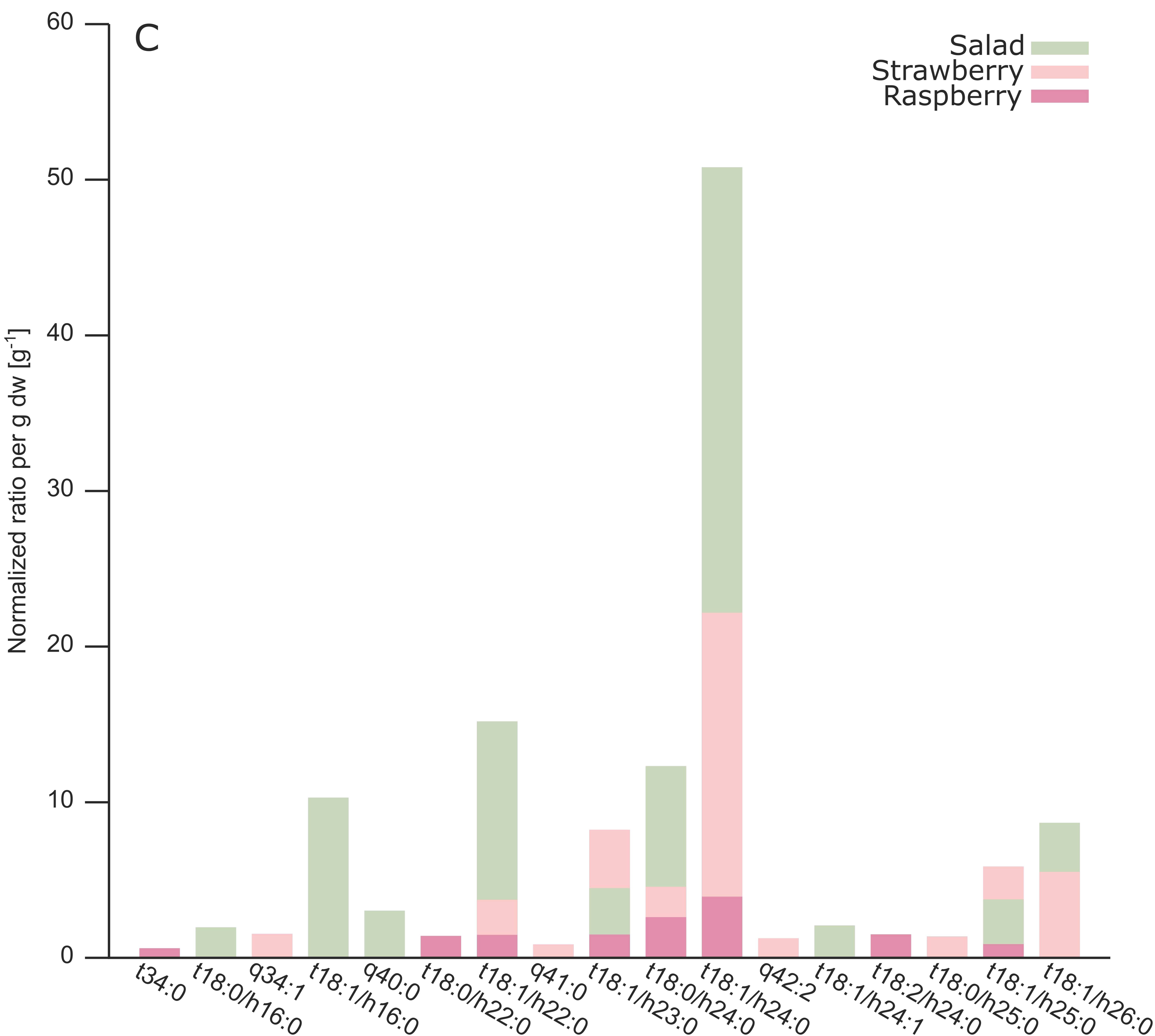
2.3. Analysis of Different Plant GIPCs by UHPLC-HRMS Suggesting t18:2 LCB
3. Discussion
4. Materials and Methods
4.1. Material
4.2. Sample Preparation
4.2.1. One-Phase Extraction
4.2.2. One-Phase Extraction Combined with Alkaline Hydrolysis
4.2.3. Centrifugation, Drying and Reconstitution
4.3. Reversed-Phase Chromatography
4.4. High-Resolution Mass Spectrometry
4.5. Data Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Composition | Plant | m/z [M − H]− | m/z [M + H]+ | Rt_neg [min] | Rt_pos [min] | Level | Normalized Ratio/g dw [g−1] | CV [%] |
|---|---|---|---|---|---|---|---|---|
| A-NAc-t18:1/h16:0 | Spinach | 1189.6246 | 1191.6388 | 8.00 | 8.00 | 2 | 3.15 | 25 |
| A-NAc-t18:1/h16:0 | Salad | 1189.6246 | 1191.6397 | 8.01 | 8.00 | 2 | 2.10 | 28 |
| A-NAc-t18:1/h22:0 | Spinach | 1273.7179 | 1275.7359 | 14.25 | 14.25 | 2 | 27.43 | 19 |
| A-NAc-t18:1/h22:0 | Salad | 1273.7193 | 1275.7344 | 14.19 | 14.26 | 2 | 3.57 | 44 |
| A-NAc-t18:1/h22:0 | Raspberry | 1273.7172 | 1275.7341 | 14.21 | 14.19 | 2 | 0.94 | 6 |
| A-NAc-t18:1/h23:0 | Spinach | 1287.7325 | 1289.7504 | 15.64 | 15.65 | 2 | 11.83 | 21 |
| A-NAc-t18:0/h24:0 | Raspberry | 1303.7648 | 1305.7813 | 17.94 | 18.17 | 2 | 2.13 | 4 |
| A-NAc-t18:1/h24:0 | Spinach | 1301.7502 | 1303.7657 | 16.98 | 16.98 | 2 | 70.89 | 21 |
| A-NAc-t18:1/h24:0 | Salad | 1301.7498 | 1303.7637 | 16.98 | 16.94 | 2 | 17.23 | 32 |
| A-NAc-t18:1/h24:0 | Raspberry | 1301.7491 | 1303.7649 | 16.96 | 16.94 | 2 | 3.40 | 5 |
| A-NAc-t18:2/h24:0 | Raspberry | 1299.7322 | 1301.749 | 14.98 | 14.97 | 2 | 1.27 | 3 |
| A-NAc-t18:1/h25:0 | Spinach | 1315.7652 | 1317.7811 | 18.41 | 18.41 | 2 | 11.08 | 20 |
| A-NAc-t18:1/h26:0 | Spinach | 1329.7811 | 1331.797 | 19.75 | 19.75 | 2 | 20.54 | 16 |
| A-NH2-t34:0 | Raspberry | 1133.6346 | 1135.6506 | 10.91 | 10.92 | 2 | 1.09 | 6 |
| A-NH2-d34:1 | Raspberry | 1115.6248 * | 1117.6415 | 10.89 | 10.90 | 3 | 1.85 | 7 |
| A-NH2-t34:1 | Raspberry | 1131.6196 * | 1133.6365 | 10.27 | 10.27 | 3 | 0.75 | 8 |
| A-NH2-t18:1/h16:0 | Salad | 1147.6148 | 1149.6302 | 8.64 | 8.52 | 2 | 6.46 | 5 |
| A-NH2-q40:0 | Salad | 1233.7225 * | 1235.7372 | 16.82 | 16.76 | 3 | 3.87 | 27 |
| A-NH2-t18:0/h22:0 | Raspberry | 1233.7242 | 1235.7389 | 16.64 | 16.55 | 2 | 3.33 | 7 |
| A-NH2-t18:1/h22:0 | Spinach | 1231.7058 * | 1233.7229 | 15.56 | 15.52 | 3 | 1.87 | 18 |
| A-NH2-t18:1/h22:0 | Salad | 1231.7083 | 1233.7226 | 15.51 | 15.49 | 2 | 10.45 | 11 |
| A-NH2-t18:1/h22:0 | Raspberry | 1231.7082 | 1233.7234 | 15.41 | 15.25 | 2 | 3.87 | 7 |
| A-NH2-t18:2/h22:0 | Raspberry | 1229.6945 * | 1231.7076 | 13.42 | 13.42 | 3 | 1.01 | 4 |
| A-NH2-t18:1/h23:0 | Salad | 1245.7242 * | 1247.7400 | 16.78 | 16.87 | 3 | 4.04 | 15 |
| A-NH2-t18:1/h23:0 | Raspberry | 1245.7248 | 1247.7388 | 16.89 | 16.85 | 2 | 2.79 | 7 |
| A-NH2-t18:0/h24:0 | Raspberry | 1261.7551 | 1263.7704 | 19.62 | 19.64 | 2 | 6.92 | 9 |
| A-NH2-t18:0/h24:0 | Salad | 1261.7531* | 1263.7701 | 19.82 | 19.53 | 3 | 3.80 | 27 |
| A-NH2-t18:1/h24:0 | Spinach | 1259.7391 | 1261.7565 | 18.32 | 18.30 | 2 | 5.06 | 18 |
| A-NH2-t18:1/h24:0 | Salad | 1259.7400 | 1261.7554 | 18.52 | 18.44 | 2 | 32.00 | 6 |
| A-NH2-t18:1/h24:0 | Raspberry | 1259.7400 | 1261.7543 | 18.46 | 18.42 | 2 | 12.28 | 8 |
| A-NH2-t18:2/h24:0 | Raspberry | 1257.7231 | 1259.7397 | 15.98 | 16.25 | 2 | 4.22 | 8 |
| A-NH2-t18:1/h25:0 | Raspberry | 1273.7539 * | 1275.7692 | 19.55 | 19.78 | 3 | 1.55 | 11 |
| A-NH2-q43:2 | Raspberry | 1271.7419 * | 1273.7571 | 17.71 | 17.73 | 3 | 0.87 | 9 |
| A-OH-t34:0 | Raspberry | 1134.6192 | 1136.6369 | 10.21 | 10.21 | 2 | 0.58 | 8 |
| A-OH-t18:0/h16:0 | Salad | 1150.6148 | 1152.6313 | 8.84 | 8.83 | 2 | 1.93 | 0 |
| A-OH-t18:1/h16:0 | Salad | 1148.5984 | 1150.6138 | 8.14 | 8.13 | 2 | 10.27 | 16 |
| A-OH-q34:1 | Strawberry | 1148.5982 | 1150.6144 | 8.11 | 8.13 | 2 | 1.51 | 6 |
| A-OH-q40:0 | Salad | 1234.7095 | 1236.7239 * | 15.66 | 15.60 | 3 | 3.00 | 2 |
| A-OH-t18:0/h22:0 | Raspberry | 1234.7084 | 1236.7247 | 15.36 | 15.63 | 2 | 1.37 | 6 |
| A-OH-t18:1/h22:0 | Salad | 1232.6921 | 1234.7071 | 14.47 | 14.44 | 2 | 15.16 | 19 |
| A-OH-t18:1/h22:0 | Strawberry | 1232.6921 | 1234.7083 | 14.25 | 14.43 | 2 | 3.69 | 2 |
| A-OH-t18:1/h22:0 | Raspberry | 1232.6924 | 1234.7055 | 14.43 | 14.43 | 2 | 1.44 | 6 |
| A-OH-q41:0 | Strawberry | 1248.7232 | 1250.7395 * | 17.01 | 16.98 | 3 | 0.83 | 2 |
| A-OH-t18:1/h23:0 | Raspberry | 1246.7076 | 1248.78221 | 15.82 | 15.77 | 2 | 1.46 | 7 |
| A-OH-t18:1/h23:0 | Salad | 1246.7079 | 1248.7209 | 15.85 | 15.84 | 2 | 4.45 | 18 |
| A-OH-t18:1/h23:0 | Strawberry | 1246.7076 | 1248.7238 | 15.53 | 15.76 | 2 | 8.20 | 1 |
| A-OH-t18:0/h24:0 | Salad | 1262.7397 | 1264.7564 | 18.55 | 18.50 | 2 | 12.30 | 4 |
| A-OH-t18:0/h24:0 | Raspberry | 1262.7385 | 1264.754 | 18.51 | 18.53 | 2 | 2.58 | 6 |
| A-OH-t18:0/h24:0 | Strawberry | 1262.7387 | 1264.7553 | 18.44 | 18.32 | 2 | 4.53 | 1 |
| A-OH-t18:1/h24:0 | Salad | 1260.7237 | 1262.7389 | 17.30 | 17.26 | 2 | 50.77 | 14 |
| A-OH-t18:1/h24:0 | Strawberry | 1260.7227 | 1262.7391 | 17.26 | 17.00 | 2 | 22.15 | 2 |
| A-OH-t18:1/h24:0 | Raspberry | 1260.7240 | 1262.7388 | 17.25 | 17.23 | 2 | 3.89 | 6 |
| A-OH-q42:2 | Strawberry | 1258.7073 | 1260.7238 * | 15.21 | 15.22 | 3 | 1.22 | 2 |
| A-OH-t18:1/h24:1 | Salad | 1258.7067 | 1260.7243 | 14.36 | 14.33 | 2 | 2.04 | 10 |
| A-OH-t18:2/h24:0 | Raspberry | 1258.7082 | 1260.7234 | 15.23 | 15.23 | 2 | 1.47 | 5 |
| A-OH-t18:0/h25:0 | Strawberry | 1276.7542 | 1278.7687 | 19.80 | 19.80 | 2 | 1.33 | 5 |
| A-OH-t18:1/h25:0 | Salad | 1274.7387 | 1276.7533 | 18.76 | 18.75 | 2 | 3.72 | 11 |
| A-OH-t18:1/h25:0 | Strawberry | 1274.7371 | 1276.7557 | 18.42 | 18.48 | 2 | 5.83 | 2 |
| A-OH-t18:1/h25:0 | Raspberry | 1274.7381 | 1276.7548 | 18.71 | 18.73 | 2 | 0.84 | 9 |
| A-OH-t18:1/h26:0 | Salad | 1288.7553 | 1290.7706 | 20.01 | 20.01 | 2 | 8.65 | 18 |
| A-OH-t18:1/h26:0 | Strawberry | 1288.7540 * | 1290.7697 | 19.76 | 19.94 | 3 | 5.49 | 2 |
| A-NH2-d18:2/16:0 | Raspberry | 1113.6100 * | 1115.6255 | 9.89 | 9.89 | 3 ** | 0.57 | 10 |
| A-NH2-q41:0 | Raspberry | 1247.7383 * | 1249.7544 | 18.16 | 18.17 | 3 ** | 2.03 | 7 |
| A-NH2-q41:2 | Raspberry | 1243.7076 * | 1245.7232 | 14.77 | 14.80 | 3 ** | 0.87 | 7 |

| Plant Species | Origin | Replicates | Fresh Weight [g] | Dry Weight [g] |
|---|---|---|---|---|
| Salad | Local supermarket | 4 | ~1 | ~0.04 |
| Spinach | Local supermarket | 4 | ~1.2 | ~0.08 |
| Strawberries | 47°58′ N, 16°6′ O | 5 | ~1.1 | ~0.10 |
| Raspberries | 47°58′ N, 16°6′ O | 5 | ~1.1 | ~0.16 |
| Parameter | Setting |
|---|---|
| Time before tol. | 1 min |
| Time after tol. | 1 min |
| Rel. Base-peak cutoff | 0.1‰ |
| Rt-shift | 0.0 min |
| Isotopic quantitation of _ isotopes where _ isotopic peak(s) have to match | 2, 1 |
| Find molecules where retention time is unknown | yes |
| LDA-version | 2.8.0 |
| machineName | OrbiTrap_exactive |
| neutronMass | 1.005 |
| coarseChromMzTolerance | 0.015 |
| MS2 | true |
| basePeakCutoff | 0.1 |
| massShift | 0.0 |
| threeDViewerDefaultTimeResolution | 2 |
| threeDViewerDefaultMZResolution | 0.005 |
| ms2PrecursorTolerance | 0.013 |
| ms2MzTolerance | 0.02 |
| ms2MinIntsForNoiseRemoval | 100 |
| ms2IsobarSCExclusionRatio | 0.01 |
| ms2IsobarSCFarExclusionRatio | 0.1 |
| ms2IsobaricOtherRtDifference | 2.0 |
| chainCutoffValue | 0.01 |
| ms2ChromMultiplicationFactorForInt | 10 |
| threeDViewerMs2DefaultTimeResolution | 1 |
| threeDViewerMs2DefaultMZResolution | 1 |
| maxFileSizeForChromTranslationAtOnce | 500 |
| chromMultiplicationFactorForInt | 1000 |
| chromLowestResolution | 1 |
| chromSmoothRange | 8.0 |
| chromSmoothRepeats | 4 |
| use3D | true |
| isotopeCorrection | false |
| removeFromOtherIsotopes | true |
| respectIsotopicDistribution | true |
| checkChainLabelCombinationFromSpeciesName | false |
| useNoiseCutoff | true |
| noiseCutoffDeviationValue | 2.0 |
| scanStep | 2 |
| profileMzRangeExtraction | 0.05 |
| profileTimeTolerance | 5.0 |
| profileIntThreshold | 5.0 |
| broaderProfileTimeTolerance | 3.0 |
| profileSmoothRange | 0.0025 |
| profileSmoothRepeats | 1 |
| profileMeanSmoothRepeats | 2 |
| profileMzMinRange | 0.002 |
| profileSteepnessChange1 | 1.5 |
| profileSteepnessChange2 | 1.8 |
| profileIntensityCutoff1 | 0.15 |
| profileIntensityCutoff2 | 0.2 |
| profileGeneralIntCutoff | 0.03 |
| profilePeakAcceptanceRange | 0.012 |
| profileSmoothingCorrection | 0.0 |
| profileMaxRange | 0.03 |
| smallChromMzRange | 0.004 |
| smallChromSmoothRepeats | 3 |
| smallChromMeanSmoothRepeats | 0 |
| smallChromSmoothRange | 2.0 |
| smallChromIntensityCutoff | 0.03 |
| broadChromSmoothRepeats | 5 |
| broadChromMeanSmoothRepeats | 0 |
| broadChromSmoothRange | 2 |
| broadChromIntensityCutoff | 0.0 |
| broadChromSteepnessChangeNoSmall | 1.33 |
| broadChromIntensityCutoffNoSmall | 0.05 |
| finalProbeTimeCompTolerance | 0.1 |
| finalProbeMzCompTolerance | 5.0E-4 |
| overlapDistanceDeviationFactor | 1.5 |
| overlapPossibleIntensityThreshold | 0.15 |
| overlapSureIntensityThreshold | 0.7 |
| overlapPeakDistanceDivisor | 3.0 |
| overlapFullDistanceDivisor | 6.0 |
| peakDiscardingAreaFactor | 1000 |
| isotopeInBetweenTime | 30 |
| isoInBetweenAreaFactor | 3.0 |
| isoNearNormalProbeTime | 30 |
| relativeAreaCutoff | 0.05 |
| relativeFarAreaCutoff | 0.05 |
| relativeFarAreaTimeSpace | 30 |
| relativeIsoInBetweenCutoff | 0.5 |
| isoInBetweenMaxTimeDistance | 300 |
| twinPeakMzTolerance | 0.01 |
| closePeakTimeTolerance | 10 |
| twinInBetweenCutoff | 0.95 |
| unionInBetweenCutoff | 0.8 |
| sparseData | false |
Appendix B
References
- Gronnier, J.; Germain, V.; Gouguet, P.; Cacas, J.-L.; Mongrand, S. GIPC: Glycosyl Inositol Phospho Ceramides, the major sphingolipids on earth. Plant Signal. Behav. 2016, 11, e1152438. [Google Scholar] [CrossRef] [PubMed]
- Carter, H.E.; Gigg, R.H.; Laws, J.H. Structure of Phytoglyolipide. J. Biol. Chem. 1958, 233, 1309–1314. [Google Scholar] [PubMed]
- Cacas, J.; Buré, C.; Grosjean, K.; Gerbeau-Pissot, P.; Lherminier, J.; Rombouts, Y.; Maes, E.; Bossard, C.; Gronnier, J.; Furt, F.; et al. Revisiting Plant Plasma Membrane Lipids in Tobacco: A Focus on Sphingolipids. Plant Physiol. 2016, 170, 367–384. [Google Scholar] [CrossRef] [PubMed]
- Carter, H.E.; Strobach, D.R.; Hawthorne, J.N. Biochemistry of the Sphingolipids. XVIII. Complete Structure of Tetrasaccharide Phytoglycolipid. Biochemistry 1969, 8, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Buré, C.; Cacas, J.; Mongrand, S.; Schmitter, J.-M. Characterization of glycosyl inositol phosphoryl ceramides from plants and fungi by mass spectrometry. Anal. Bioanal. Chem. 2014, 406, 995–1010. [Google Scholar] [CrossRef]
- LIPID MAPS® Lipidomics Gateway. Available online: https://lipidmaps.org/resources/lipidweb/index.php?page=lipids/sphingo/glyP_ino/index.htm (accessed on 4 August 2020).
- Buré, C.; Cacas, J.-L.; Wang, F.; Gaudin, K.; Domergue, F.; Mongrand, S.; Schmitter, J.-M. Fast screening of highly glycosylated plant sphingolipids by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2011, 25, 3131–3145. [Google Scholar] [CrossRef]
- Hastings, J.; Owen, G.; Dekker, A.; Ennis, M.; Kale, N.; Muthukrishnan, V.; Turner, S.; Swainston, N.; Mendes, P.; Steinbeck, C. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016, 44, D1214–D1219. [Google Scholar] [CrossRef]
- Sud, M.; Fahy, E.; Cotter, D.; Brown, A.; Dennis, E.A.; Glass, C.K.; Merrill, A.H.; Murphy, R.C.; Raetz, C.R.H.; Russell, D.W.; et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007, 35, 527–532. [Google Scholar] [CrossRef]
- Barrientos, R.C.; Zhang, Q. Recent advances in the mass spectrometric analysis of glycosphingolipidome—A review. Anal. Chim. Acta 2020. [Google Scholar] [CrossRef]
- Markham, J.E.; Jaworski, J.G. Rapid measurement of sphingolipids from Arabidopsis thaliana by reversed-phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2007, 21, 1304–1314. [Google Scholar] [CrossRef]
- Blaas, N.; Humpf, H.U. Structural profiling and quantitation of glycosyl inositol phosphoceramides in plants with fourier transform mass spectrometry. J. Agric. Food Chem. 2013, 61, 4257–4269. [Google Scholar] [CrossRef] [PubMed]
- Cacas, J.; Buré, C.; Furt, F.; Maalouf, J.; Badoc, A.; Cluzet, S.; Schmitter, J.; Antajan, E.; Mongrand, S. Biochemical survey of the polar head of plant glycosylinositolphosphoceramides unravels broad diversity. Phytochemistry 2013, 96, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Domon, B.; Costello, C.E. Structure Elucidation of Glycosphingolipids and Gangliosides Using High-Performance Tandem Mass Spectrometry. Biochemistry 1988, 27, 1534–1543. [Google Scholar] [CrossRef] [PubMed]
- Domon, B.; Costello, C.E. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj. J. 1988, 5, 397–409. [Google Scholar] [CrossRef]
- Hsu, F.; Turk, J. Glycosphingolipids as Lithiated Adducts by Dissociation on a Triple Stage Quadrupole Instrument. J. Am. Soc. Mass Spectrom. 2001, 12, 61–79. [Google Scholar] [CrossRef]
- Hartler, J.; Triebl, A.; Ziegl, A.; Trötzmüller, M.; Rechberger, G.N.; Zeleznik, O.A.; Zierler, K.A.; Torta, F.; Cazenave-Gassiot, A.; Wenk, M.R.; et al. Deciphering lipid structures based on platform-independent decision rules. Nat. Methods 2017, 14, 1171–1174. [Google Scholar] [CrossRef]
- Markham, J.E.; Li, J.; Cahoon, E.B.; Jaworski, J.G. Separation and Identification of Major Plant Sphingolipid Classes from Leaves. J. Biol. Chem. 2006, 281, 22684–22694. [Google Scholar] [CrossRef]
- Dugo, P.; Cacciola, F.; Kumm, T.; Dugo, G.; Mondello, L. Comprehensive multidimensional liquid chromatography: Theory and applications. J. Chromatogr. A 2008, 1184, 353–368. [Google Scholar] [CrossRef]
- Lísa, M.; Holčapek, M. Triacylglycerols profiling in plant oils important in food industry, dietetics and cosmetics using high-performance liquid chromatography–atmospheric pressure chemical ionization mass spectrometry. J. Chromatogr. A 2008, 1198–1199, 115–130. [Google Scholar] [CrossRef]
- Ovčačíková, M.; Lísa, M.; Cífková, E.; Holčapek, M. Retention behavior of lipids in reversed-phase ultrahigh-performance liquid chromatography–electrospray ionization mass spectrometry. J. Chromatogr. A 2016, 1450, 76–85. [Google Scholar] [CrossRef]
- Hartler, J.; Trötzmüller, M.; Chitraju, C.; Spener, F.; Köfeler, H.C.; Thallinger, G.G. Lipid Data Analyzer: Unattended identification and quantitation of lipids in LC-MS data. Bioinformatics 2011, 27, 572–577. [Google Scholar] [CrossRef] [PubMed]
- LIPID MAPS® Lipidomics Gateway. Available online: https://www.lipidmaps.org/data/LMSDRecord.php?LMID=LMSP03030005 (accessed on 15 March 2020).
- Salek, R.M.; Steinbeck, C.; Viant, M.R.; Goodacre, R.; Dunn, W.B. The role of reporting standards for metabolite annotation and identification in metabolomic studies. Gigascience 2013, 2, 2047-217X-2-13. [Google Scholar] [CrossRef] [PubMed]
- Adams, K.J.; Pratt, B.; Bose, N.; Dubois, L.G.; St. John-Williams, L.; Perrott, K.M.; Ky, K.; Kapahi, P.; Sharma, V.; Maccoss, M.J.; et al. Skyline for Small Molecules: A Unifying Software Package for Quantitative Metabolomics. J. Proteome Res. 2020, 19, 1447–1458. [Google Scholar] [CrossRef] [PubMed]
- Metabolomics Society. Available online: http://metabolomicssociety.org/ (accessed on 14 July 2020).
- Shiva, S.; Enninful, R.; Roth, M.R.; Tamura, P.; Jagadish, K.; Welti, R. An efficient modified method for plant leaf lipid extraction results in improved recovery of phosphatidic acid. Plant Methods 2018, 14, 1–8. [Google Scholar] [CrossRef]
- Peng, B.; Weintraub, S.T.; Coman, C.; Ponnaiyan, S.; Sharma, R.; Tews, B.; Winter, D.; Ahrends, R. A Comprehensive High-Resolution Targeted Workflow for the Deep Profiling of Sphingolipids. Anal. Chem. 2017, 89, 12480–12487. [Google Scholar] [CrossRef] [PubMed]
- Loos, M.; Gerber, C.; Corona, F.; Hollender, J.; Singer, H. Accelerated Isotope Fine Structure Calculation Using Pruned Transition Trees. Anal. Chem. 2015, 87, 5738–5744. [Google Scholar] [CrossRef] [PubMed]
- Koelmel, J.P.; Kroeger, N.M.; Gill, E.L.; Ulmer, C.Z.; Bowden, J.A.; Patterson, R.E.; Yost, R.A.; Garrett, T.J. Expanding Lipidome Coverage Using LC-MS/MS Data-Dependent Acquisition with Automated Exclusion List Generation. J. Am. Soc. Mass Spectrom. 2017, 28, 908–917. [Google Scholar] [CrossRef]
- LDA User Manual. Available online: http://genome.tugraz.at/lda2/2.6/LDA_2.6.pdf (accessed on 17 September 2020).

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panzenboeck, L.; Troppmair, N.; Schlachter, S.; Koellensperger, G.; Hartler, J.; Rampler, E. Chasing the Major Sphingolipids on Earth: Automated Annotation of Plant Glycosyl Inositol Phospho Ceramides by Glycolipidomics. Metabolites 2020, 10, 375. https://doi.org/10.3390/metabo10090375
Panzenboeck L, Troppmair N, Schlachter S, Koellensperger G, Hartler J, Rampler E. Chasing the Major Sphingolipids on Earth: Automated Annotation of Plant Glycosyl Inositol Phospho Ceramides by Glycolipidomics. Metabolites. 2020; 10(9):375. https://doi.org/10.3390/metabo10090375
Chicago/Turabian StylePanzenboeck, Lisa, Nina Troppmair, Sara Schlachter, Gunda Koellensperger, Jürgen Hartler, and Evelyn Rampler. 2020. "Chasing the Major Sphingolipids on Earth: Automated Annotation of Plant Glycosyl Inositol Phospho Ceramides by Glycolipidomics" Metabolites 10, no. 9: 375. https://doi.org/10.3390/metabo10090375
APA StylePanzenboeck, L., Troppmair, N., Schlachter, S., Koellensperger, G., Hartler, J., & Rampler, E. (2020). Chasing the Major Sphingolipids on Earth: Automated Annotation of Plant Glycosyl Inositol Phospho Ceramides by Glycolipidomics. Metabolites, 10(9), 375. https://doi.org/10.3390/metabo10090375