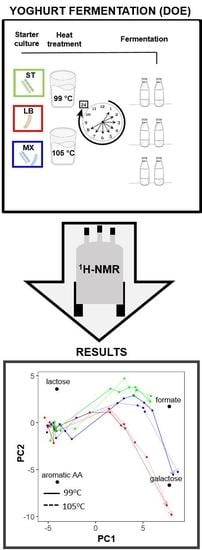
An NMR Metabolomics Approach to Investigate Factors Affecting the Yoghurt Fermentation Process and Quality
Abstract
1. Introduction
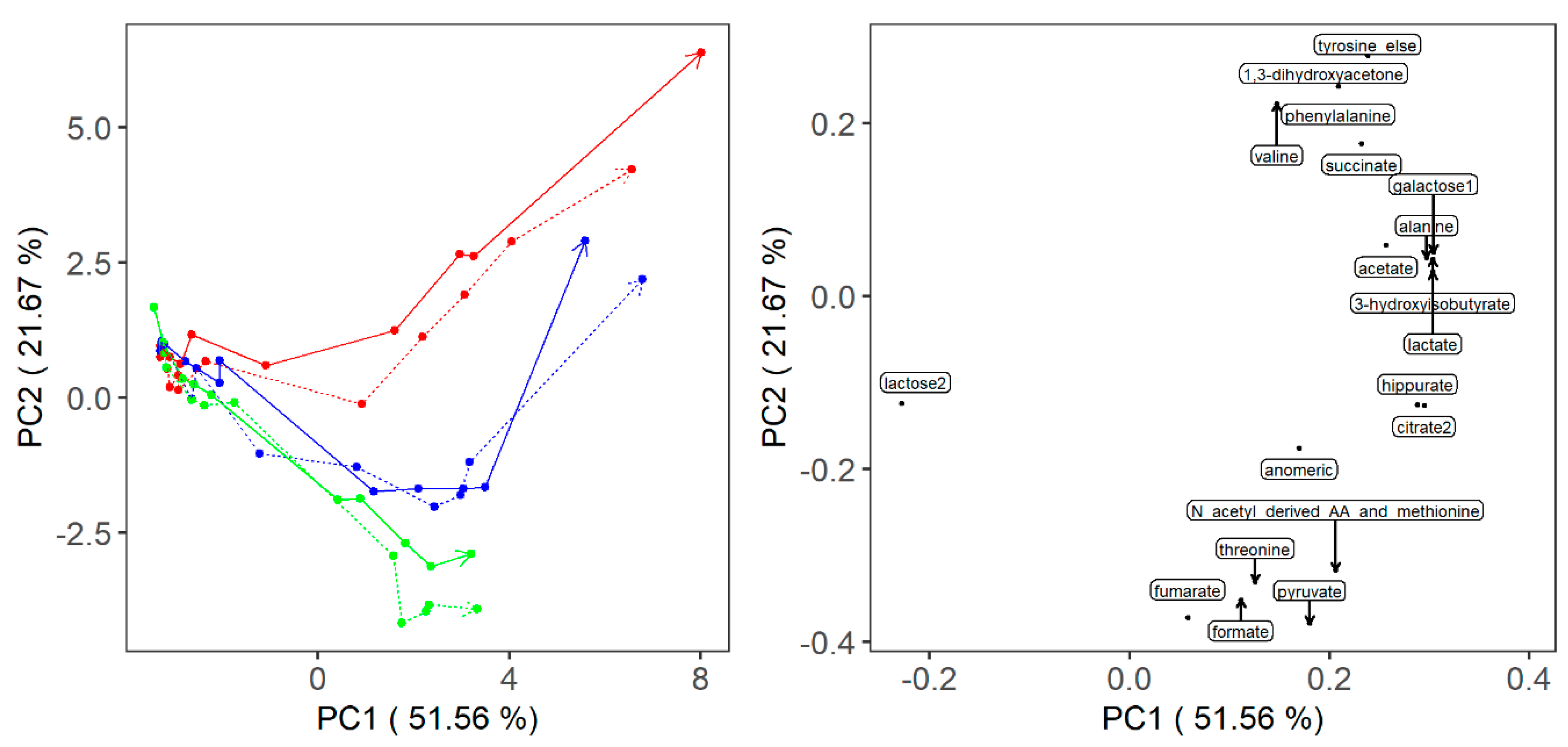
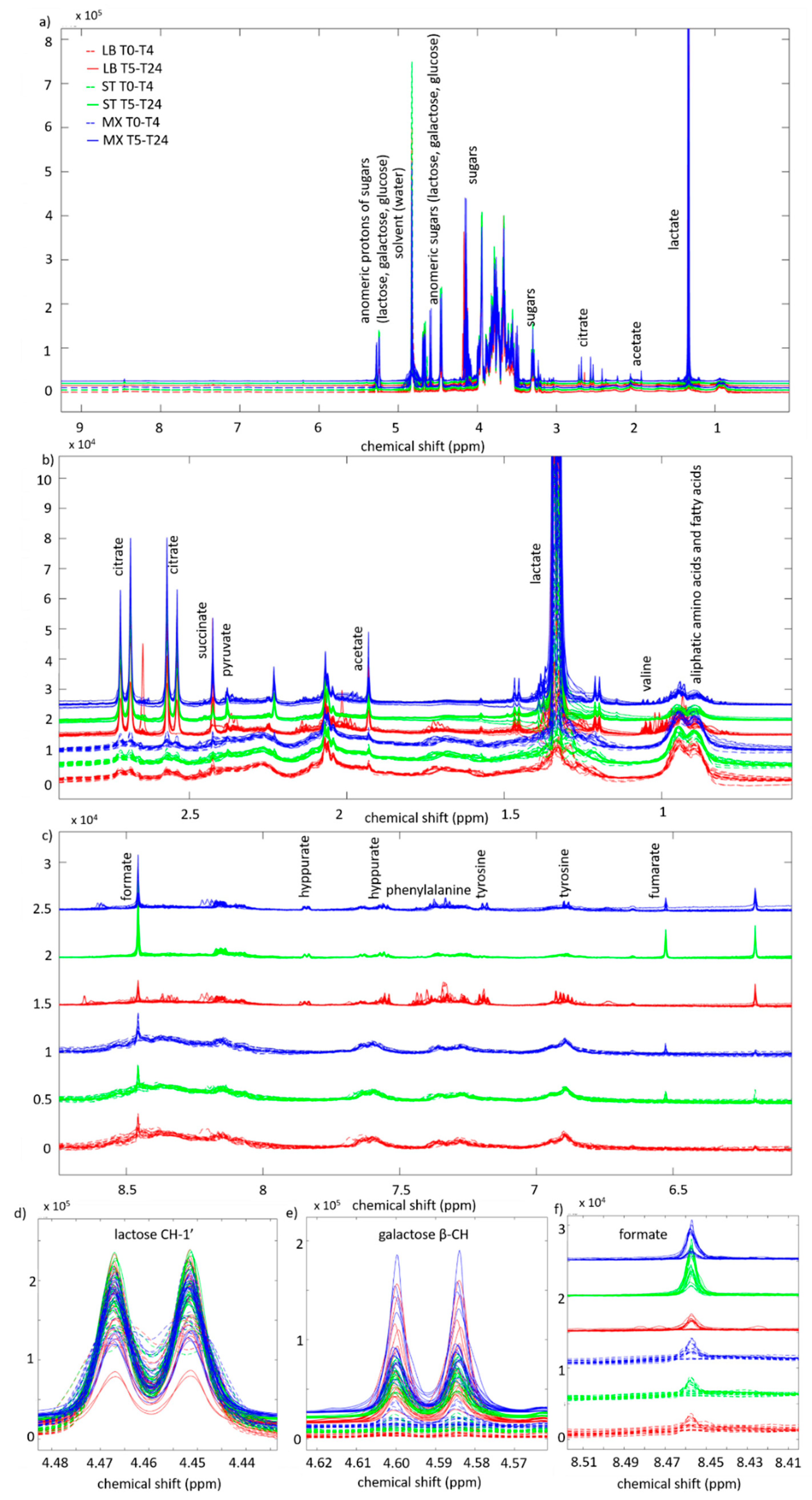
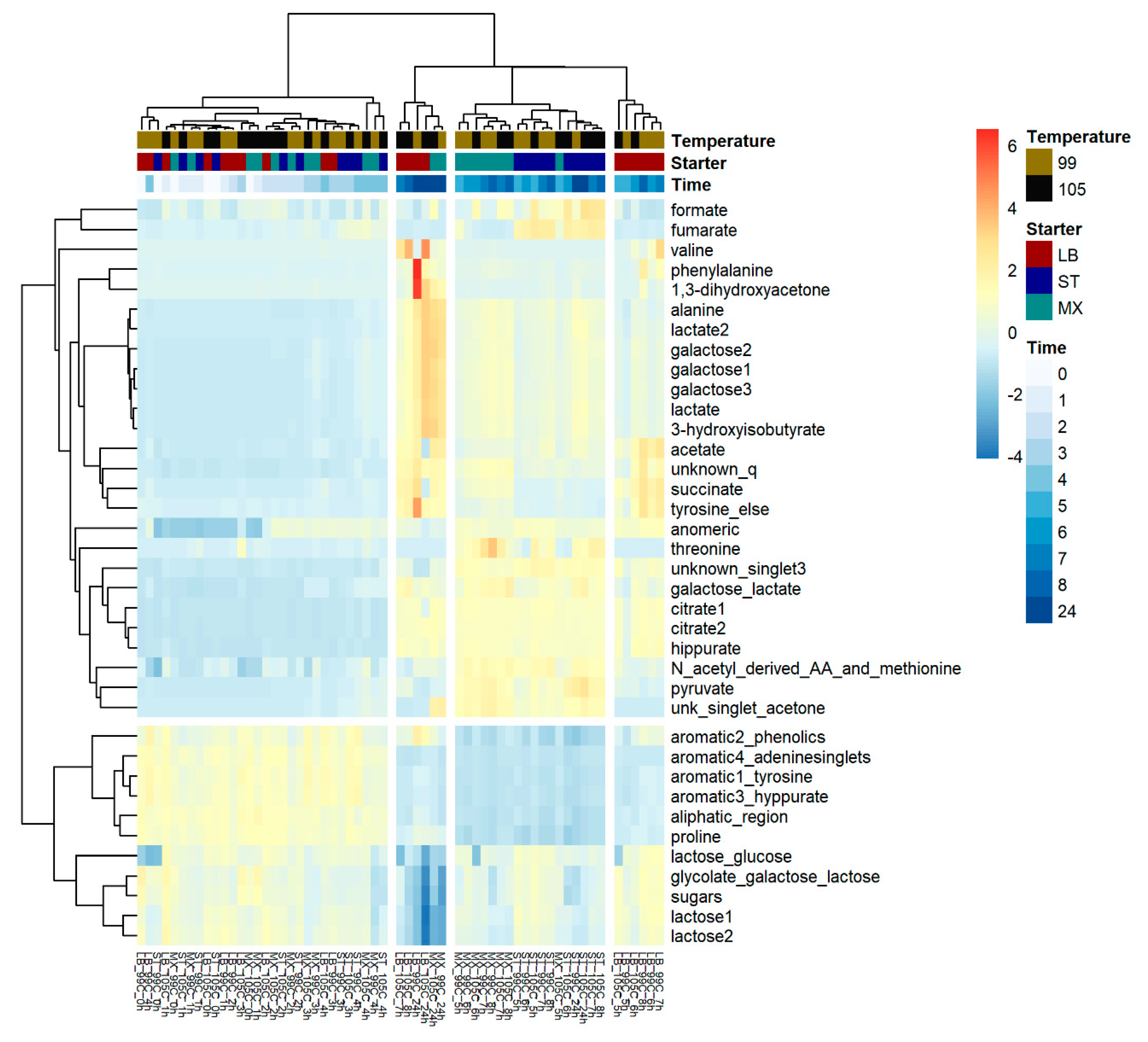
2. Results
3. Discussion
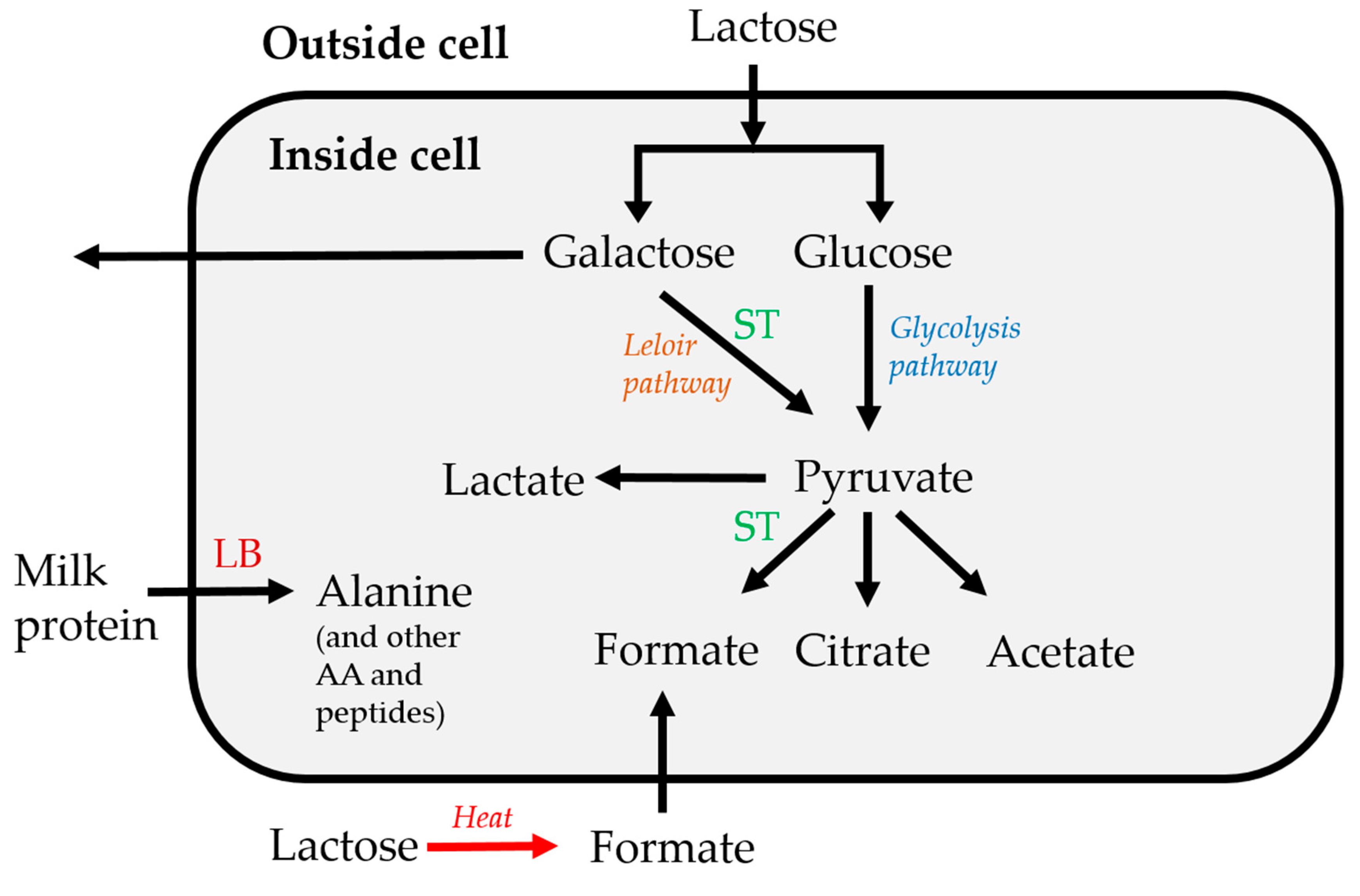
3.1. Fermentation Kinetics of Sugars
3.2. Fermentation Kinetics in Deprotonated Organic Acids and Other Molecules
3.3. The Investigation of Fermentation through Application of NMR and Chemometrics
4. Materials and Methods
4.1. Starter Cultures and Media
4.2. Preparation of Milk Samples for Fermentation
4.3. Chemicals
4.4. Preparation of NMR Samples
4.5. NMR Analysis
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Interval Name | End ppm | Initial ppm |
|---|---|---|
| Valine | 1.07 | 1.03 |
| 3-hydroxyisobutyrate | 1.22 | 1.18 |
| Lactate | 1.36 | 1.30 |
| Threonine | 1.40 | 1.36 |
| Alanine | 1.48 | 1.45 |
| Acetate | 1.95 | 1.90 |
| N_acetyl_derived_AA_and_methionine | 2.08 | 2.03 |
| Pyruvate | 2.39 | 2.38 |
| Succinate | 2.45 | 2.41 |
| Citrate1 | 2.60 | 2.51 |
| Citrate2 | 2.74 | 2.64 |
| Lactose | 3.34 | 3.27 |
| Galactose | 3.52 | 3.47 |
| Galactose | 4.10 | 4.07 |
| Lactate2 | 4.19 | 4.11 |
| 1,3-dihydroxyacetone | 4.43 | 4.42 |
| Lactose | 4.49 | 4.43 |
| Galactose | 4.62 | 4.56 |
| Lactose_glucose | 4.71 | 4.64 |
| Anomeric | 5.31 | 5.20 |
| Fumarate | 6.55 | 6.51 |
| Tyrosine_else | 7.21 | 7.17 |
| Phenylalanine | 7.46 | 7.41 |
| Hippurate | 7.87 | 7.81 |
| Formate | 8.48 | 8.44 |
| Unk_singlet_acetone * (SUS) | 2.24 | 2.22 |
| Unknown_singlet3 (SUS) | 6.22 | 6.18 |
| Unknown_q (SUS) | 9.71 | 9.64 |
| Aliphatic_region (BIN) | 1.00 | 0.83 |
| Proline (BIN) | 2.03 | 1.95 |
| Sugars (BIN) | 3.90 | 3.53 |
| Glycolate_galactose_lactose (BIN) | 4.02 | 3.90 |
| Galactose_lactate (BIN) | 4.11 | 4.10 |
| Aromatic1_tyrosine (BIN) | 7.04 | 6.78 |
| Aromatic2_phenolics (BIN) | 7.41 | 7.21 |
| Aromatic3_hyppurate (BIN) | 7.77 | 7.51 |
| Aromatic4_adeninesinglets (BIN) | 8.40 | 7.94 |
References
- Minervini, F.; De Angelis, M.; Di Cagno, R.; Gobbetti, M. Ecological parameters influencing microbial diversity and stability of traditional sourdough. Int. J. Food Microbiol. 2014, 171, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Vaningelgem, F.; Zamfir, M.; Adriany, T.; De Vuyst, L. Fermentation conditions affecting the bacterial growth and exopolysaccharide production by Streptococcus thermophilus ST 111 in milk-based medium. J. Appl. Microbiol. 2004, 97, 1257–1273. [Google Scholar] [CrossRef] [PubMed]
- Aichinger, P.-A.; Michel, M.; Servais, C.; Dillmann, M.-L.; Rouvet, M.; D’Amico, N.; Zink, R.; Klostermeyer, H.; Horne, D.S. Fermentation of a skim milk concentrate with Streptococcus thermophilus and chymosin: Structure, viscoelasticity and syneresis of gels. Colloids Surf. B Biointerfaces 2003, 31, 243–255. [Google Scholar] [CrossRef]
- Casarotti, S.N.; Monteiro, D.A.; Moretti, M.M.; Penna, A.L.B. Influence of the combination of probiotic cultures during fermentation and storage of fermented milk. Food Res. Int. 2014, 59, 67–75. [Google Scholar] [CrossRef]
- Leroy, F.; De Vuyst, L. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci. Technol. 2004, 15, 67–78. [Google Scholar] [CrossRef]
- Rhee, S.J.; Lee, J.-E.; Lee, C.-H. Importance of lactic acid bacteria in Asian fermented foods. Microbl. Cell Fact. 2011, 10, S5. [Google Scholar] [CrossRef]
- Papamanoli, E.; Tzanetakis, N.; Litopoulou-Tzanetaki, E.; Kotzekidou, P. Characterization of lactic acid bacteria isolated from a Greek dry-fermented sausage in respect of their technological and probiotic properties. Meat Sci. 2003, 65, 859–867. [Google Scholar] [CrossRef]
- Son, H.-S.; Hwang, G.-S.; Park, W.-M.; Hong, Y.-S.; Lee, C.-H. Metabolomic characterization of malolactic fermentation and fermentative behaviors of wine yeasts in grape wine. J. Agric. Food Chem. 2009, 57, 4801–4809. [Google Scholar] [CrossRef]
- Lee, J.-E.; Hwang, G.-S.; Lee, C.-H.; Hong, Y.-S. Metabolomics reveals alterations in both primary and secondary metabolites by wine bacteria. J. Agric. Food Chem. 2009, 57, 10772–10783. [Google Scholar] [CrossRef]
- Piras, C.; Marincola, F.C.; Savorani, F.; Engelsen, S.B.; Cosentino, S.; Viale, S.; Pisano, M.B. A NMR metabolomics study of the ripening process of the Fiore Sardo cheese produced with autochthonous adjunct cultures. Food Chem. 2013, 141, 2137–2147. [Google Scholar] [CrossRef]
- Ferri, M.; Serrazanetti, D.I.; Tassoni, A.; Baldissarri, M.; Gianotti, A. Improving the functional and sensorial profile of cereal-based fermented foods by selecting Lactobacillus plantarum strains via a metabolomics approach. Food Res. Int. 2016, 89, 1095–1105. [Google Scholar] [CrossRef]
- Sundekilde, U.K.; Larsen, L.B.; Bertram, H.C. NMR-based milk metabolomics. Metabolites 2013, 3, 204–222. [Google Scholar] [CrossRef] [PubMed]
- Savorani, F.; Khakimov, B.; Viereck, N.; Engelsen, S.B. NMR Foodomics. In NMR-Based Metabolomics; Royal Society of Chemistry: London, UK, 2018; pp. 183–245. [Google Scholar]
- Ebrahimi, P.; Larsen, F.H.; Jensen, H.M.; Vogensen, F.K.; Engelsen, S.B. Real-time metabolomic analysis of lactic acid bacteria as monitored by in vitro NMR and chemometrics. Metabolomics 2016, 12, 77. [Google Scholar] [CrossRef]
- Walstra, P.; Wouters, J.T.; Geurts, T.J. Dairy Science and Technology; CRC press: Boca Raton, FL, USA, 2005. [Google Scholar]
- Lamanna, R.; Braca, A.; Di Paolo, E.; Imparato, G. Identification of milk mixtures by 1H NMR profiling. Magn. Reson. Chem. 2011, 49, S22–S26. [Google Scholar] [CrossRef]
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. An Introduction to Statistical Learning; Springer Science+Business Media: New York, NY, USA, 2013; Volume 112. [Google Scholar]
- Tibshirani, R.; Walther, G.; Hastie, T. Estimating the number of clusters in a data set via the gap statistic. J. R. Stat. Soc. Ser. B (Stat. Methodol.) 2001, 63, 411–423. [Google Scholar] [CrossRef]
- Sørensen, K.I.; Curic-Bawden, M.; Junge, M.P.; Janzen, T.; Johansen, E. Enhancing the sweetness of yoghurt through metabolic remodeling of carbohydrate metabolism in Streptococcus thermophilus and Lactobacillus delbrueckii subsp. bulgaricus. Appl. Environ. Microbiol. 2016, 82, 3683–3692. [Google Scholar] [CrossRef]
- Angelov, M.; Kostov, G.; Simova, E.; Beshkova, D.; Koprinkova-Hristova, P. Proto-cooperation factors in yogurt starter cultures. Revue de Génie Industriel. 2009, 3, 4–12. [Google Scholar]
- Sieuwerts, S.; De Bok, F.A.; Hugenholtz, J.; van Hylckama Vlieg, J.E. Unraveling microbial interactions in food fermentations: From classical to genomics approaches. Appl. Environ. Microbiol. 2008, 74, 4997–5007. [Google Scholar] [CrossRef]
- Lu, Y.; Ishikawa, H.; Kwon, Y.; Hu, F.; Miyakawa, T.; Tanokura, M. Real-time monitoring of chemical changes in three kinds of fermented milk products during fermentation using quantitative difference nuclear magnetic resonance spectroscopy. J. Agric. Food Chem. 2018, 66, 1479–1487. [Google Scholar] [CrossRef]
- Liu, S.-Q. Practical implications of lactate and pyruvate metabolism by lactic acid bacteria in food and beverage fermentations. Int. J. Food Microbiol. 2003, 83, 115–131. [Google Scholar] [CrossRef]
- Özer, B. Strategies for yogurt manufacturing. In Development and Manufacture of Yogurt and Other Functional Dairy Products; CRC Press: Boca Raton, FL, USA, 2010; Volume 47. [Google Scholar]
- Settachaimongkon, S.; Nout, M.R.; Fernandes, E.C.A.; Hettinga, K.A.; Vervoort, J.M.; van Hooijdonk, T.C.; Zwietering, M.H.; Smid, E.J.; van Valenberg, H.J. Influence of different proteolytic strains of Streptococcus thermophilus in co-culture with Lactobacillus delbrueckii subsp. bulgaricus on the metabolite profile of set-yoghurt. Int. J. Food Microbiol. 2014, 177, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, J.; Makino, S.; Kimura, K.; Isogai, E.; Saito, T. Influence of Different Sterilization Conditions on the Growth and Exopolysaccharide of Streptococcus thermophilus and Co-Cultivation with Lactobacillus delbrueckii subsp. bulgaricus OLL1073R-1. Adv. Microbiol. 2015, 5, 760. [Google Scholar] [CrossRef][Green Version]
- Bouteille, R.; Gaudet, M.; Lecanu, B.; This, H. Monitoring lactic acid production during milk fermentation by in situ quantitative proton nuclear magnetic resonance spectroscopy. J. Dairy Sci. 2013, 96, 2071–2080. [Google Scholar] [CrossRef]
- Hickey, M.W.; Hillier, A.J.; Jago, G.R. Transport and metabolism of lactose, glucose, and galactose in homofermentative lactobacilli. Appl. Environ. Microbiol. 1986, 51, 825–831. [Google Scholar] [CrossRef] [PubMed]
- Beshkova, D.M.; Simova, E.D.; Frengova, G.I.; Simov, Z.I.; Adilov, E.F. Production of amino acids by yogurt bacteria. Biotechnol. Prog. 1998, 14, 963–965. [Google Scholar] [CrossRef]
- Radke-Mitchell, L.; Sandine, W. Associative growth and differential enumeration of Streptococcus thermophilus and Lactobacillus bulgaricus: A review. J. Food Prot. 1984, 47, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Fox, P.; Uniacke-Lowe, T.; McSweeney, P.; O’Mahony, J. Heat-induced changes in milk. In Dairy Chemistry and Biochemistry; Springer International Publishing: Cham, Switzerland, 2015; pp. 345–375. [Google Scholar]
- Homoud, A.M. Characterisation of Proteins in Camel Milk, the Effect of Heat Treatment on Physicochemical and Functional Properties Related to Yogurt. Ph.D. Thesis, Heriot-Watt University, Edinburgh, UK, 2015. [Google Scholar]
- Gentès, M.-C.; St-Gelais, D.; Turgeon, S.L. Exopolysaccharide–milk protein interactions in a dairy model system simulating yoghurt conditions. Dairy Sci. Technol. 2013, 93, 255–271. [Google Scholar] [CrossRef]
- Gonnella, N.C. LC-NMR: Expanding the Limits of Structure Elucidation; CRC Press: Boca Raton, FL, USA, 2020. [Google Scholar]
- Khakimov, B.; Tseng, L.H.; Godejohann, M.; Bak, S.; Engelsen, S.B. Screening for triterpenoid saponins in plants using hyphenated analytical platforms. Molecules 2016, 21, 1614. [Google Scholar] [CrossRef]
- Khakimov, B.; Mobaraki, N.; Trimigno, A.; Aru, V.; Engelsen, S.B. Signature Mapping (SigMa): An efficient approach for processing of complex human urine 1H-NMR metabolomics data. Anal. Chim. Acta 2020, 1108, 142–151. [Google Scholar] [CrossRef]
- Savorani, F.; Tomasi, G.; Engelsen, S.B. icoshift: A versatile tool for the rapid alignment of 1D NMR spectra. J. Magn. Reson. 2010, 202, 190–202. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Evironment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria. Available online: https://www.R-project.org/ (accessed on 16 July 2020).
- Kassambara, A.; Mundt, F. Package ‘factoextra’:Extract and Visualize the Results of Multivariate Data Analyses. 2017. R Packages. Available online: https://rpkgs.datanovia.com/factoextra/index.html (accessed on 16 July 2020).
- Kolde, R. Pheatmap: Pretty Heatmaps (Version 1.0 12). 2019. CRAN. Available online: https://cran.r-project.org/web/packages/pheatmap/index.html (accessed on 16 July 2020).
- Wickham, H. ggplot2. Wiley Interdiscip Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]


| Strain | Temperature (°C) | Inoculation % | 1st Dilution (g) | 2nd Dilution (g) | Analysis Time (h) |
|---|---|---|---|---|---|
| Streptococcus thermophilus | 37 | 0.00172 | 4.00 | 0.94 | 24 |
| Lactobacillus delbrueckii ssp. bulgaricus | 43 | 0.00050 | 2.00 | 0.54 | 24 |
| Streptococcus thermophilus and Lactobacillus delbrueckii ssp. bulgaricus | 37 | 0.00340 | 4.00 | 1.86 | 24 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trimigno, A.; Bøge Lyndgaard, C.; Atladóttir, G.A.; Aru, V.; Balling Engelsen, S.; Harder Clemmensen, L.K. An NMR Metabolomics Approach to Investigate Factors Affecting the Yoghurt Fermentation Process and Quality. Metabolites 2020, 10, 293. https://doi.org/10.3390/metabo10070293
Trimigno A, Bøge Lyndgaard C, Atladóttir GA, Aru V, Balling Engelsen S, Harder Clemmensen LK. An NMR Metabolomics Approach to Investigate Factors Affecting the Yoghurt Fermentation Process and Quality. Metabolites. 2020; 10(7):293. https://doi.org/10.3390/metabo10070293
Chicago/Turabian StyleTrimigno, Alessia, Christian Bøge Lyndgaard, Guðrún Anna Atladóttir, Violetta Aru, Søren Balling Engelsen, and Line Katrine Harder Clemmensen. 2020. "An NMR Metabolomics Approach to Investigate Factors Affecting the Yoghurt Fermentation Process and Quality" Metabolites 10, no. 7: 293. https://doi.org/10.3390/metabo10070293
APA StyleTrimigno, A., Bøge Lyndgaard, C., Atladóttir, G. A., Aru, V., Balling Engelsen, S., & Harder Clemmensen, L. K. (2020). An NMR Metabolomics Approach to Investigate Factors Affecting the Yoghurt Fermentation Process and Quality. Metabolites, 10(7), 293. https://doi.org/10.3390/metabo10070293