Tumor Tissue-Specific Biomarkers of Colorectal Cancer by Anatomic Location and Stage
Abstract
1. Introduction
2. Results
2.1. Metabolic Differences between Colon Cancer and Normal Controls
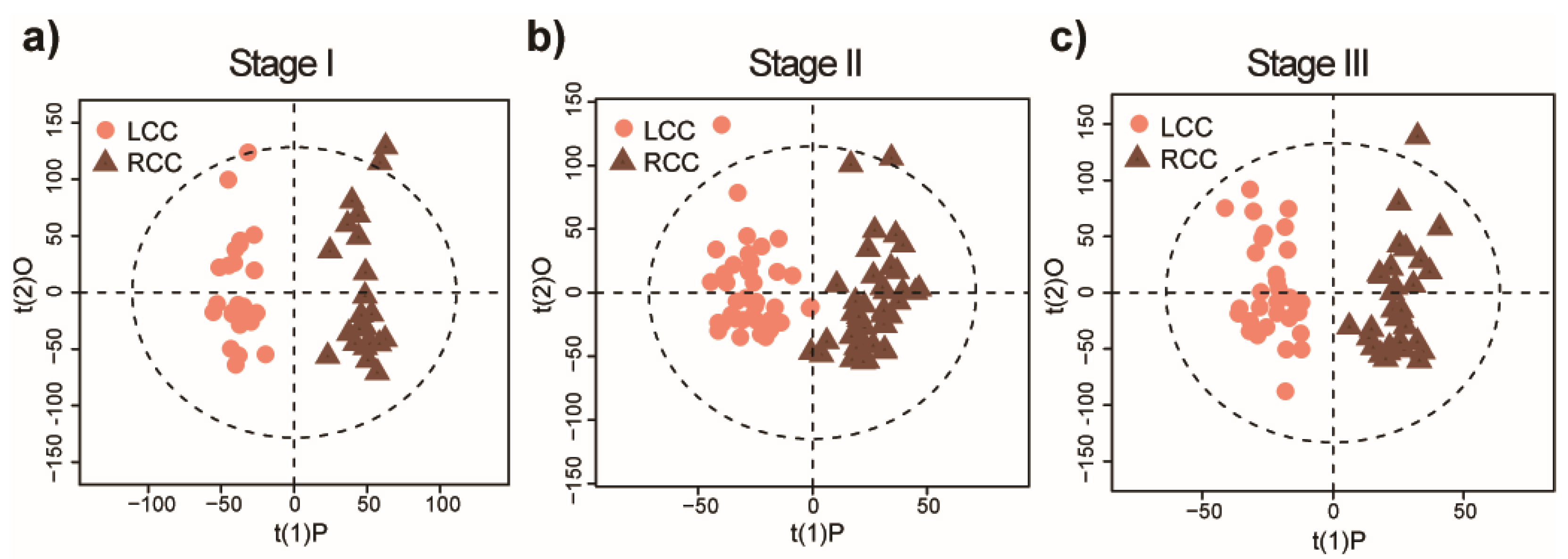
2.2. Distinct Metabolomic Signatures Associated with Anatomic Location
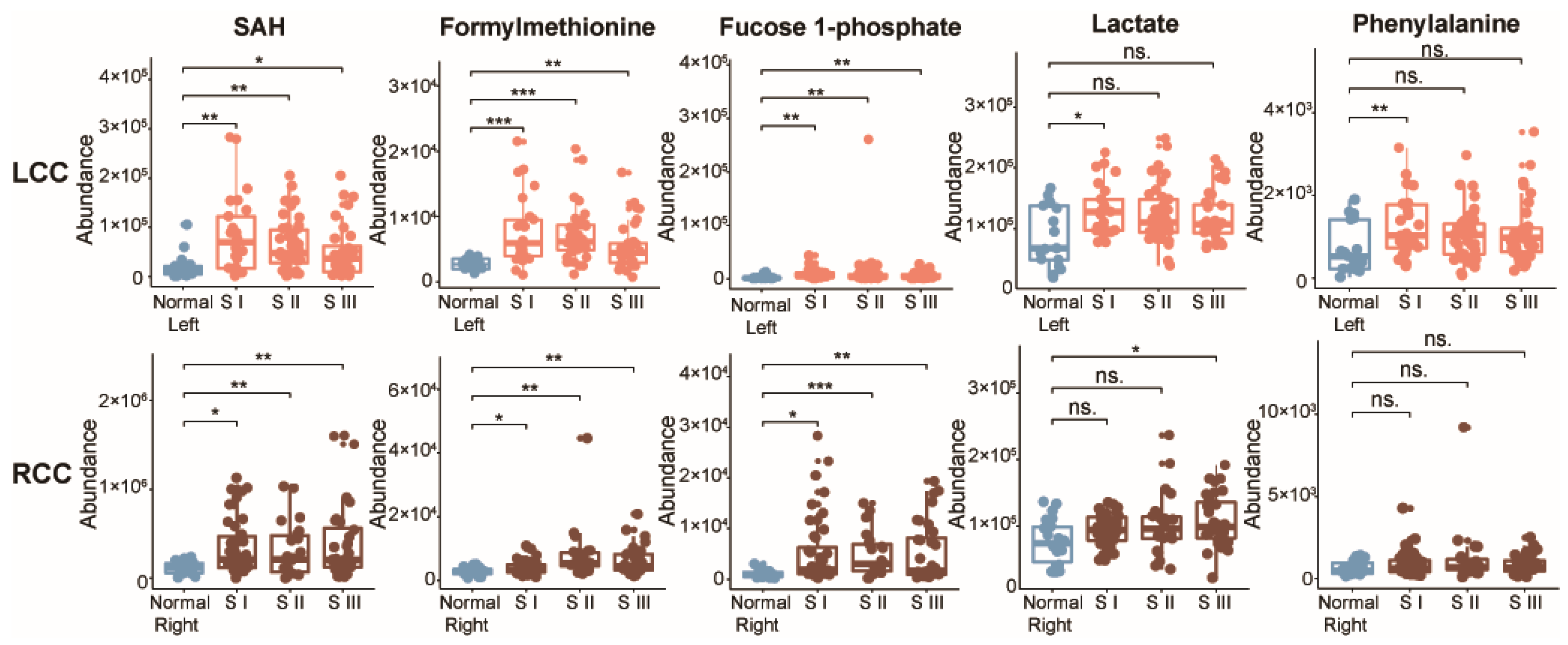
2.3. Potential Metabolic Biomarkers for Early-Stage LCC and RCC
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Sample Collection
4.3. Tissue Metabolite Extraction
4.4. UPLC-MS Analysis
4.5. UPLC-MS Data Processing
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Crooke, H.; Kobayashi, M.; Mitchell, B.; Nwokeji, E.; Laurie, M.; Kamble, S.; McKenna, M.; Masood, A.; Korytowsky, B. Estimating 1- and 5-year relative survival trends in colorectal cancer (CRC) in the United States: 2004 to 2014. J. Clin. Oncol. 2018, 36, 587. [Google Scholar] [CrossRef]
- Wolf, A.M.D.; Fontham, E.T.H.; Church, T.R.; Flowers, C.R.; Guerra, C.E.; LaMonte, S.J.; Etzioni, R.; McKenna, M.T.; Oeffinger, K.C.; Shih, Y.T.; et al. Colorectal cancer screening for average-risk adults: 2018 guideline update from the American Cancer Society. CA Cancer J. Clin. 2018, 68, 250–281. [Google Scholar] [CrossRef] [PubMed]
- Muthukrishnan, M.; Arnold, L.D.; James, A.S. Patients’ self-reported barriers to colon cancer screening in federally qualified health center settings. Prev. Med. Rep. 2019, 15, 100896. [Google Scholar] [CrossRef]
- Zhang, F.; Zhang, Y.Y.; Zhao, W.W.; Deng, K.; Wang, Z.Z.; Yang, C.Y.; Ma, L.B.; Openkova, M.S.; Hou, Y.; Li, K. Metabolomics for biomarker discovery in the diagnosis, prognosis, survival and recurrence of colorectal cancer: A systematic review. Oncotarget 2017, 8, 35460–35472. [Google Scholar] [CrossRef]
- Erben, V.; Bhardwaj, M.; Schrotz-King, P.; Brenner, H. Metabolomics Biomarkers for Detection of Colorectal Neoplasms: A Systematic Review. Cancers 2018, 10, 246. [Google Scholar] [CrossRef]
- Cheng, Y.; Xie, G.; Chen, T.; Qiu, Y.; Zou, X.; Zheng, M.; Tan, B.; Feng, B.; Dong, T.; He, P.; et al. Distinct urinary metabolic profile of human colorectal cancer. J. Proteome Res. 2012, 11, 1354–1363. [Google Scholar] [CrossRef]
- Zhu, J.J.; Djukovic, D.; Deng, L.L.; Gu, H.W.; Himmati, F.; Chiorean, E.G.; Raftery, D. Colorectal Cancer Detection Using Targeted Serum Metabolic Profiling. J. Proteome Res. 2014, 13, 4120–4130. [Google Scholar] [CrossRef]
- Qiu, Y.; Cai, G.; Zhou, B.; Li, D.; Zhao, A.; Xie, G.; Li, H.; Cai, S.; Xie, D.; Huang, C.; et al. A distinct metabolic signature of human colorectal cancer with prognostic potential. Clin. Cancer Res. 2014, 20, 2136–2146. [Google Scholar] [CrossRef]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef]
- Tang, W.H.; Wang, Z.; Levison, B.S.; Koeth, R.A.; Britt, E.B.; Fu, X.; Wu, Y.; Hazen, S.L. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 2013, 368, 1575–1584. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Mironova, V.; Chen, Y.; Lundh, E.P.F.; Zhang, Q.; Cai, Y.; Vasiliou, V.; Zhang, Y.; Garcia-Milian, R.; Khan, S.A.; et al. Molecular Pathway Analysis Indicates a Distinct Metabolic Phenotype in Women With Right-Sided Colon Cancer. Transl. Oncol. 2020, 13, 42–56. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Rattray, N.J.W.; Zhang, Q.; Mironova, V.; Santos-Neto, A.; Hsu, K.S.; Rattray, Z.; Cross, J.R.; Zhang, Y.; Paty, P.B.; et al. Sex Differences in Colon Cancer Metabolism Reveal A Novel Subphenotype. Sci. Rep. 2020, 10, 4905. [Google Scholar] [CrossRef]
- Bufill, J.A. Colorectal-cancer-evidence for distinct genetic categories based on proximal or distal tumor location. Ann. Intern. Med. 1990, 113, 779–788. [Google Scholar] [CrossRef] [PubMed]
- Loupakis, F.; Yang, D.Y.; Yau, L.; Feng, S.B.; Cremolini, C.; Zhang, W.; Maus, M.K.H.; Antoniotti, C.; Langer, C.; Scherer, S.J.; et al. Primary tumor location as a prognostic factor in metastatic colorectal cancer. JNCI J. Nat. Cancer Inst. 2015, 107. [Google Scholar] [CrossRef] [PubMed]
- Gervaz, P.; Usel, M.; Rapiti, E.; Chappuis, P.; Neyroud-Kaspar, I.; Bouchardy, C. Right colon cancer: Left behind. Eur. J. Surg. Oncol. 2016, 42, 1343–1349. [Google Scholar] [CrossRef] [PubMed]
- Petrelli, F.; Tomasello, G.; Borgonovo, K.; Ghidini, M.; Turati, L.; Dallera, P.; Passalacqua, R.; Sgroi, G.; Barni, S. Prognostic survival associated with left-sided vs right-sided colon cancer a systematic review and meta-analysis. JAMA Oncol. 2017, 3, 211–219. [Google Scholar] [CrossRef]
- Schroll, M.M.; Ludwig, K.R.; Bauer, K.M.; Hummon, A.B. Calcitriol supplementation causes decreases in tumorigenic proteins and different proteomic and metabolomic signatures in right versus left-sided colon cancer. Metabolites 2018, 8, 5. [Google Scholar] [CrossRef]
- Gu, J.; Xiao, Y.; Shu, D.; Liang, X.; Hu, X.; Xie, Y.; Lin, D.; Li, H. Metabolomics analysis in serum from patients with colorectal polyp and colorectal cancer by (1)H-NMR spectrometry. Dis. Mark. 2019, 2019, 3491852. [Google Scholar] [CrossRef]
- Zha, H.; Cai, Y.; Yin, Y.; Wang, Z.; Li, K.; Zhu, Z.J. SWATHtoMRM: Development of high-coverage targeted metabolomics method using SWATH technology for biomarker discovery. Anal. Chem. 2018, 90, 4062–4070. [Google Scholar] [CrossRef]
- Ikeda, A.; Nishiumi, S.; Shinohara, M.; Yoshie, T.; Hatano, N.; Okuno, T.; Bamba, T.; Fukusaki, E.; Takenawa, T.; Azuma, T.; et al. Serum metabolomics as a novel diagnostic approach for gastrointestinal cancer. Biomed. Chromatogr. 2012, 26, 548–558. [Google Scholar] [CrossRef]
- Farshidfar, F.; Weljie, A.M.; Kopciuk, K.A.; Hilsden, R.; McGregor, S.E.; Buie, W.D.; MacLean, A.; Vogel, H.J.; Bathe, O.F. A validated metabolomic signature for colorectal cancer: Exploration of the clinical value of metabolomics. Br. J. Cancer 2016, 115, 848–857. [Google Scholar] [CrossRef] [PubMed]
- Efron, B.; Tibshirani, R. Improvements on cross-validation: The.632+ bootstrap method. J. Am. Stat. Assoc. 1997, 92, 548–560. [Google Scholar] [CrossRef]
- Peng, D.X.; Lai, F.J. Using partial least squares in operations management research: A practical guideline and summary of past research. J. Oper. Manag. 2012, 30, 467–480. [Google Scholar] [CrossRef]
- Cross, A.J.; Moore, S.C.; Boca, S.; Huang, W.Y.; Xiong, X.; Stolzenberg-Solomon, R.; Sinha, R.; Sampson, J.N. A prospective study of serum metabolites and colorectal cancer risk. Cancer 2014, 120, 3049–3057. [Google Scholar] [CrossRef]
- Liesenfeld, D.B.; Grapov, D.; Fahrmann, J.F.; Salou, M.; Scherer, D.; Toth, R.; Habermann, N.; Bohm, J.; Schrotz-King, P.; Gigic, B.; et al. Metabolomics and transcriptomics identify pathway differences between visceral and subcutaneous adipose tissue in colorectal cancer patients: The ColoCare study. Am. J. Clin. Nutr. 2015, 102, 433–443. [Google Scholar] [CrossRef]
- Rattray, N.J.W.; Charkoftaki, G.; Rattray, Z.; Hansen, J.E.; Vasiliou, V.; Johnson, C.H. Environmental influences in the etiology of colorectal cancer: The premise of metabolomics. Curr. Pharmacol. Rep. 2017, 3, 114–125. [Google Scholar] [CrossRef]
- Chan, E.C.Y.; Koh, P.K.; Mal, M.; Cheah, P.Y.; Eu, K.W.; Backshall, A.; Cavill, R.; Nicholson, J.K.; Keun, H.C. Metabolic profiling of human colorectal cancer using high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy and gas chromatography mass spectrometry (GC/MS). J. Proteome Res. 2009, 8, 352–361. [Google Scholar] [CrossRef]
- Jimenez, B.; Mirnezami, R.; Kinross, J.; Cloarec, O.; Keun, H.C.; Holmes, E.; Goldin, R.D.; Ziprin, P.; Darzi, A.; Nicholson, J.K. 1H HR-MAS NMR spectroscopy of tumor-induced local metabolic “field-effects” enables colorectal cancer staging and prognostication. J. Proteome Res. 2013, 12, 959–968. [Google Scholar] [CrossRef]
- Mirnezami, R.; Jimenez, B.; Li, J.V.; Kinross, J.M.; Veselkov, K.; Goldin, R.D.; Holmes, E.; Nicholson, J.K.; Darzi, A. Rapid diagnosis and staging of colorectal cancer via high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy of intact tissue biopsies. Ann. Surg. 2014, 259, 1138–1149. [Google Scholar] [CrossRef]
- Qiu, Y.; Cai, G.; Su, M.; Chen, T.; Liu, Y.; Xu, Y.; Ni, Y.; Zhao, A.; Cai, S.; Xu, L.X.; et al. Urinary metabonomic study on colorectal cancer. J. Proteome Res. 2010, 9, 1627–1634. [Google Scholar] [CrossRef] [PubMed]


| Metabolite | m/z | RT | FC a | VIP b | pc | pd | pe | pf |
|---|---|---|---|---|---|---|---|---|
| SAH | 385.1282 | 363.2 | 0.29 | 1.8 | 0.028 | 0.001 | 0.026 | N.S. |
| Formylmethionine | 176.0393 | 188.9 | 0.64 | 1.9 | 0.010 | <0.001 | 0.033 | N.S. |
| Fucose 1-phosphate | 243.0285 | 438.0 | 0.38 | 1.3 | 0.026 | 0.001 | 0.010 | N.S. |
| Lactate | 89.0233 | 218.2 | 0.75 | 2.1 | 0.008 | 0.046 | N.S. | N.S. |
| Phenylalanine | 207.1130 | 252.0 | 0.53 | 2.3 | 0.003 | 0.019 | N.S. | N.S. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, Y.; Rattray, N.J.W.; Zhang, Q.; Mironova, V.; Santos-Neto, A.; Muca, E.; Vollmar, A.K.R.; Hsu, K.-S.; Rattray, Z.; Cross, J.R.; et al. Tumor Tissue-Specific Biomarkers of Colorectal Cancer by Anatomic Location and Stage. Metabolites 2020, 10, 257. https://doi.org/10.3390/metabo10060257
Cai Y, Rattray NJW, Zhang Q, Mironova V, Santos-Neto A, Muca E, Vollmar AKR, Hsu K-S, Rattray Z, Cross JR, et al. Tumor Tissue-Specific Biomarkers of Colorectal Cancer by Anatomic Location and Stage. Metabolites. 2020; 10(6):257. https://doi.org/10.3390/metabo10060257
Chicago/Turabian StyleCai, Yuping, Nicholas J. W. Rattray, Qian Zhang, Varvara Mironova, Alvaro Santos-Neto, Engjel Muca, Ana K. Rosen Vollmar, Kuo-Shun Hsu, Zahra Rattray, Justin R. Cross, and et al. 2020. "Tumor Tissue-Specific Biomarkers of Colorectal Cancer by Anatomic Location and Stage" Metabolites 10, no. 6: 257. https://doi.org/10.3390/metabo10060257
APA StyleCai, Y., Rattray, N. J. W., Zhang, Q., Mironova, V., Santos-Neto, A., Muca, E., Vollmar, A. K. R., Hsu, K.-S., Rattray, Z., Cross, J. R., Zhang, Y., Paty, P. B., Khan, S. A., & Johnson, C. H. (2020). Tumor Tissue-Specific Biomarkers of Colorectal Cancer by Anatomic Location and Stage. Metabolites, 10(6), 257. https://doi.org/10.3390/metabo10060257








