Effects of Genetic Variation on Urinary Small Molecule Signatures of Mice after Exposure to Ionizing Radiation: A Study of p53 Deficiency
Abstract
1. Introduction
2. Material and Methods
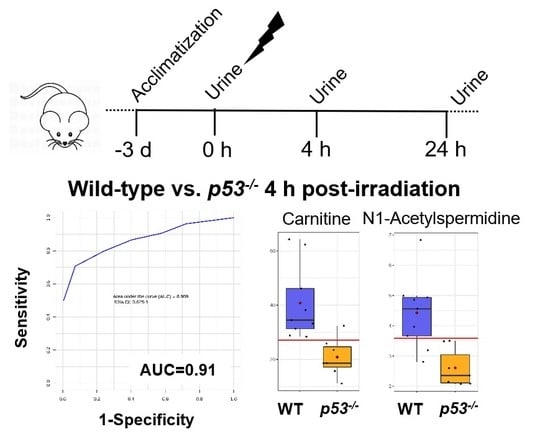
2.1. Animal Models and Radiation Exposure
2.2. Metabolite Extraction and LC-MS Analysis
2.3. Data Processing, Statistical Analysis, and Marker Validation
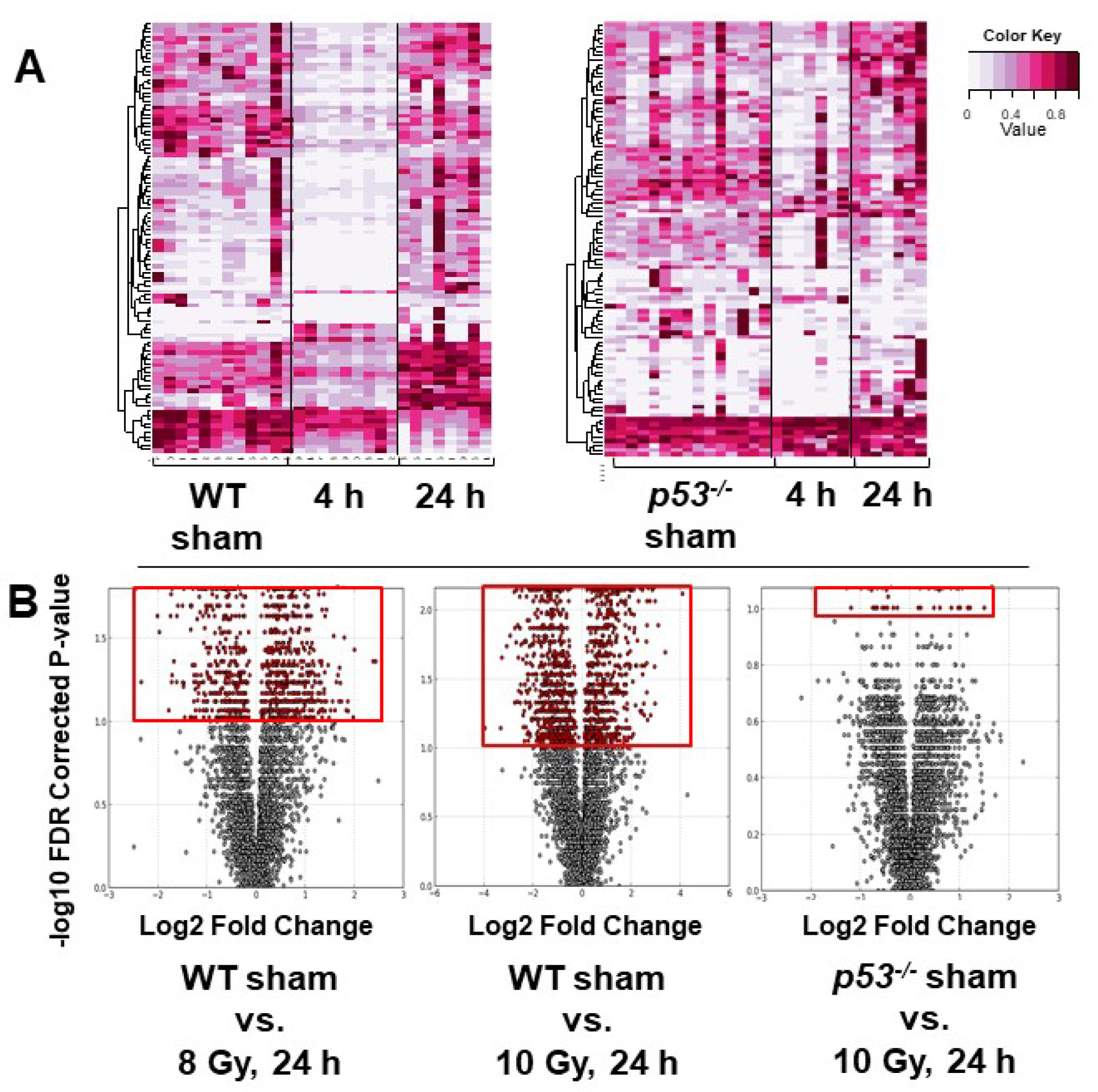
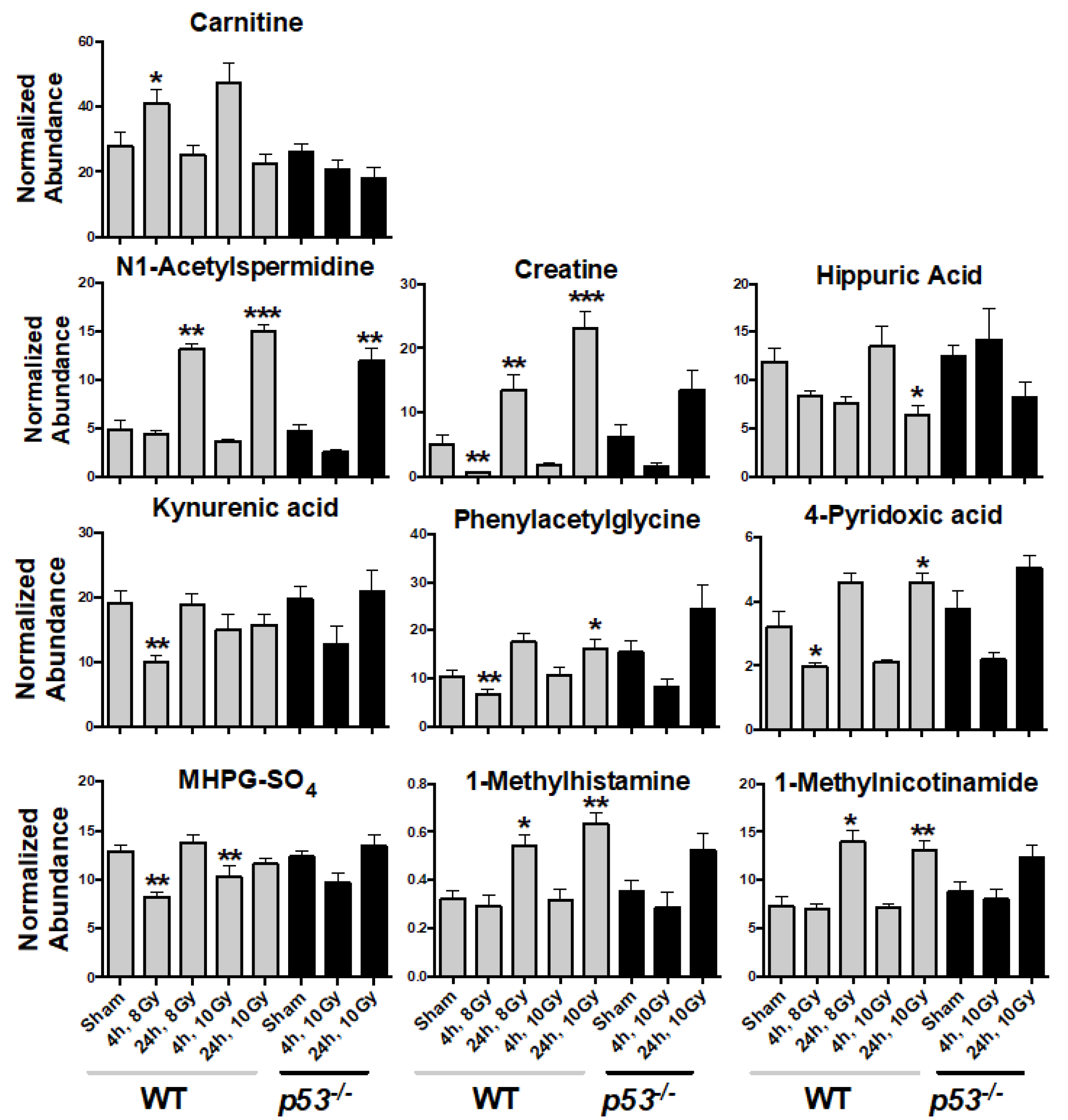
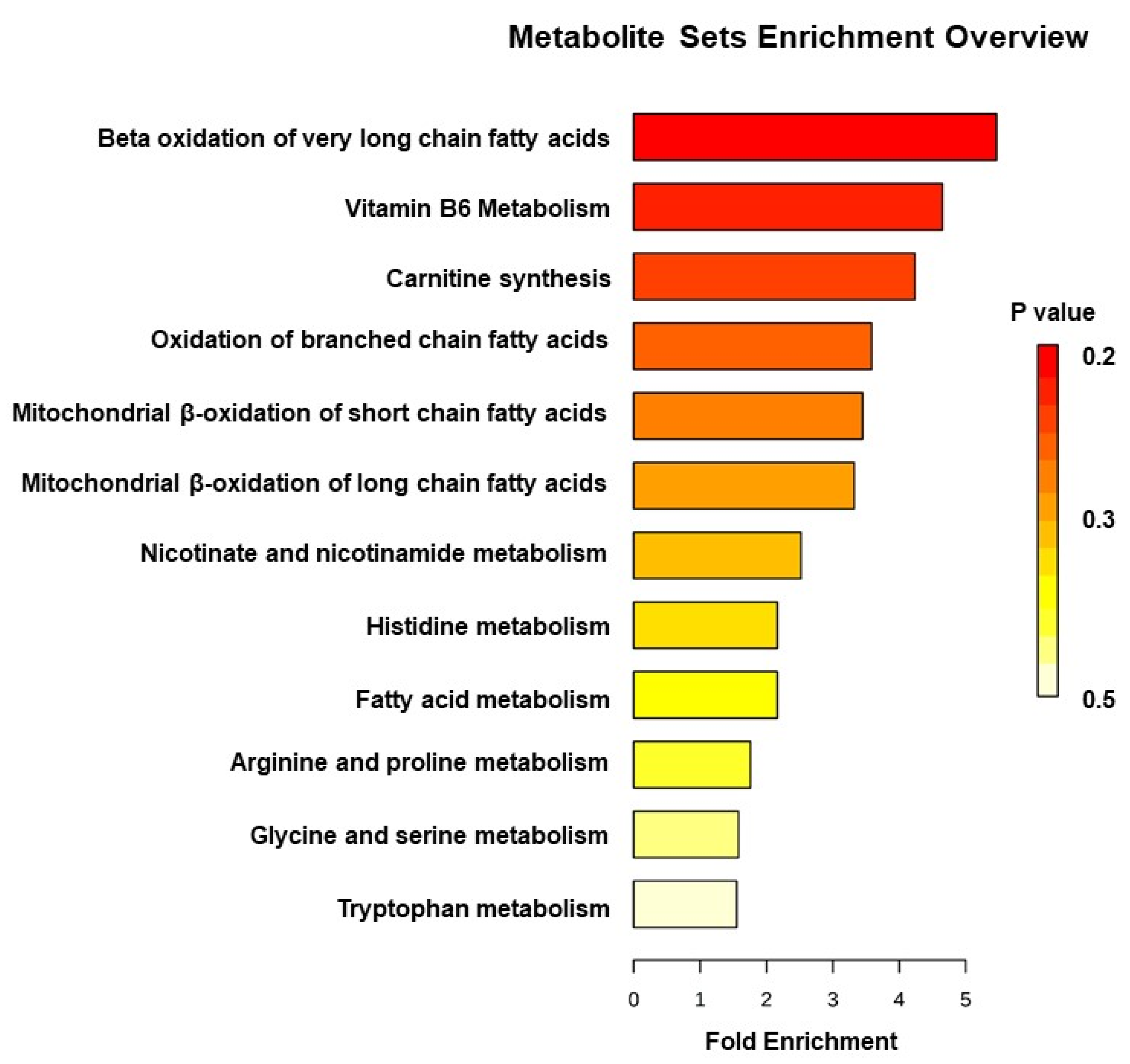
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pannkuk, E.L.; Fornace, A.J.; Laiakis, E.C. Metabolomic applications in radiation biodosimetry: Exploring radiation effects through small molecules. Int. J. Radiat. Boil. 2017, 93, 1151–1176. [Google Scholar] [CrossRef]
- Flood, A.B.; Ali, A.N.; Boyle, H.K.; Du, G.; Satinsky, V.A.; Swarts, S.G.; Williams, B.B.; Demidenko, E.; Schreiber, W.; Swartz, H.M. Evaluating the Special Needs of The Military for Radiation Biodosimetry for Tactical Warfare Against Deployed Troops: Comparing Military to Civilian Needs for Biodosimetry Methods. Health Phys. 2016, 111, 169–182. [Google Scholar] [CrossRef]
- Rajaraman, P.; Hauptmann, M.; Bouffler, S.; Wojcik, A. Human individual radiation sensitivity and prospects for prediction. Ann. ICRP 2018, 47, 126–141. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.; The Human Serum Metabolome (HUSERMET) Consortium; Broadhurst, D.; Begley, P.; Zelená, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.; Halsall, A.; et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef] [PubMed]
- Beckonert, O.; Keun, H.C.; Ebbels, T.M.D.; Bundy, J.G.; Holmes, E.; Lindon, J.C.; Nicholson, J. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2007, 2, 2692–2703. [Google Scholar] [CrossRef] [PubMed]
- Pannkuk, E.L.; Laiakis, E.C.; Gill, K.; Jain, S.K.; Mehta, K.Y.; Nishita, D.; Bujold, K.; Bakke, J.; Gahagen, J.; Authier, S.; et al. Liquid Chromatography–Mass Spectrometry-Based Metabolomics of Nonhuman Primates after 4 Gy Total Body Radiation Exposure: Global Effects and Targeted Panels. J. Proteome Res. 2019, 18, 2260–2269. [Google Scholar] [CrossRef]
- Pannkuk, E.L.; Laiakis, E.C.; Girgis, M.; Dowd, S.E.; Dhungana, S.; Nishita, D.; Bujold, K.; Bakke, J.; Gahagen, J.; Authier, S.; et al. Temporal Effects on Radiation Responses in Nonhuman Primates: Identification of Biofluid Small Molecule Signatures by Gas Chromatography⁻Mass Spectrometry Metabolomics. Metabolites 2019, 9, 98. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Coy, S.L.; Pannkuk, E.L.; Laiakis, E.C.; Fornace, A.J.; Vouros, P. Differential Mobility Spectrometry-Mass Spectrometry (DMS-MS) in Radiation Biodosimetry: Rapid and High-Throughput Quantitation of Multiple Radiation Biomarkers in Nonhuman Primate Urine. J. Am. Soc. Mass Spectrom. 2018, 29, 1650–1664. [Google Scholar] [CrossRef]
- Chaudhuri, A.R.; Nussenzweig, A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Boil. 2017, 18, 610–621. [Google Scholar] [CrossRef]
- Laiakis, E.C.; Pannkuk, E.L.; Diaz-Rubio, M.E.; Wang, Y.-W.; Mak, T.D.; Simbulan-Rosenthal, P.C.M.; Brenner, D.J.; Fornace, A.J. Implications of genotypic differences in the generation of a urinary metabolomics radiation signature. Mutat. Res. Mol. Mech. Mutagen. 2016, 788, 41–49. [Google Scholar] [CrossRef]
- Canman, C.E.; Lim, D.-S.; Cimprich, K.A.; Taya, Y.; Tamai, K.; Sakaguchi, K.; Appella, E.; Kastan, M.B.; Siliciano, J.D. Activation of the ATM Kinase by Ionizing Radiation and Phosphorylation of p53. Science 1998, 281, 1677–1679. [Google Scholar] [CrossRef] [PubMed]
- Laiakis, E.C.; Mak, T.D.; Strawn, S.J.; Wang, Y.W.; Moon, B.H.; Ake, P.; Fornace, A.J. Global metabolomic responses in urine from atm deficient mice in response to LD50/30 gamma irradiation doses. Environ. Mol. Mutagen 2018, 59, 576–585. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-L.; Blum, J.M.; Kirsch, D.G. Role of p53 in regulating tissue response to radiation by mechanisms independent of apoptosis. Transl. Cancer Res. 2013, 2, 412–421. [Google Scholar] [PubMed]
- Gudkov, A.V.; Komarova, E.A. The role of p53 in determining sensitivity to radiotherapy. Nat. Rev. Cancer 2003, 3, 117–129. [Google Scholar] [CrossRef]
- Cui, Y.F.; Zhou, P.K.; Woolford, L.B.; Lord, B.; Hendry, J.H.; Wang, D.W. Apoptosis in bone marrow cells of mice with different p53 genotypes after gamma-rays irradiation in vitro. J. Environ. Pathol. Toxicol. Oncol. 1995, 14, 159–163. [Google Scholar]
- Komarov, P.G.; Komarova, E.A.; Kondratov, R.V.; Christov-Tselkov, K.; Coon, J.S.; Chernov, M.V.; Gudkov, A.V. A Chemical Inhibitor of p53 That Protects Mice from the Side Effects of Cancer Therapy. Science 1999, 285, 1733–1737. [Google Scholar] [CrossRef]
- A Komarova, E.; Kondratov, R.V.; Wang, K.; Christov, K.; Golovkina, T.V.; Goldblum, J.R.; Gudkov, A.V. Dual effect of p53 on radiation sensitivity in vivo: p53 promotes hematopoietic injury, but protects from gastro-intestinal syndrome in mice. Oncogene 2004, 23, 3265–3271. [Google Scholar] [CrossRef]
- Soussi, T.; Beroud, C. Assessing TP53 status in human tumours to evaluate clinical outcome. Nat. Rev. Cancer 2001, 1, 233–239. [Google Scholar] [CrossRef]
- Jacks, T.; Remington, L.; Williams, B.O.; Schmitt, E.M.; Halachmi, S.; Bronson, R.T.; Weinberg, R.A. Tumor spectrum analysis in p53-mutant mice. Curr. Boil. 1994, 4, 1–7. [Google Scholar] [CrossRef]
- Pannkuk, E.L.; Laiakis, E.C.; Authier, S.; Wong, K.; Fornace, A.J., Jr. Global metabolomic identification of longer-term dose dependent urinary biomarkers in nonhuman primates exposed to ionizing radiation. Radiat. Res. 2015, 184, 121–131. [Google Scholar] [CrossRef]
- Pannkuk, E.L.; Laiakis, E.C.; Mak, T.D.; Astarita, G.; Authier, S.; Wong, K.; Fornace, A.J. A lipidomic and metabolomic serum signature from nonhuman primates exposed to ionizing radiation. Metabolomics 2016, 12, 80. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [PubMed]
- Mak, T.D.; Laiakis, E.C.; Goudarzi, M.; Fornace, A.J. MetaboLyzer: A Novel Statistical Workflow for Analyzing Postprocessed LC–MS Metabolomics Data. Anal. Chem. 2013, 86, 506–513. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Eisner, R.; Young, N.; Gautam, B.; Hau, D.D.; Psychogios, N.; Dong, E.; Bouatra, S.; et al. HMDB: A knowledgebase for the human metabolome. Nucleic Acids Res. 2008, 37, D603–D610. [Google Scholar] [CrossRef]
- Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Degtyarenko, K.; De Matos, P.; Ennis, M.; Hastings, J.; Zbinden, M.; McNaught, A.; Alcántara, R.; Darsow, M.; Guedj, M.; Ashburner, M. ChEBI: A database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2007, 36, D344–D350. [Google Scholar] [CrossRef]
- Guijas, C.; Montenegro-Burke, R.; Domingo-Almenara, X.; Palermo, A.; Warth, B.; Hermann, G.; Koellensperger, G.; Huan, T.; Uritboonthai, W.; Aisporna, A.E.; et al. METLIN: A Technology Platform for Identifying Knowns and Unknowns. Anal. Chem. 2018, 90, 3156–3164. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; A Barrett, D.; Beale, M.H.; Beger, R.D.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Huang, J.; Long, Z.; Lin, W.; Liao, X.; Xie, Y.; Liu, L.; Ma, W. Integrative omics analysis of p53-dependent regulation of metabolism. FEBS Lett. 2018, 592, 380–393. [Google Scholar] [CrossRef]
- Huang, R.; Liu, X.; Li, H.; Zhou, Y.; Zhou, P.-K. Integrated analysis of transcriptomic and metabolomic profiling reveal the p53 associated pathways underlying the response to ionizing radiation in HBE cells. Cell Biosci. 2020, 10, 56. [Google Scholar] [CrossRef] [PubMed]
- Aubrey, B.J.; Kelly, G.L.; Janic, A.; Herold, M.J.; Strasser, A. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression. Cell Death Differ. 2018, 25, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.A.S.; Laiakis, E.C.; Ghandhi, S.A.; Morton, S.R.; Fornace, J.A.J.; Amundson, S.A.; Fornace, A.J. Gene Expression in Parp1 Deficient Mice Exposed to a Median Lethal Dose of Gamma Rays. Radiat. Res. 2018, 190, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Rudqvist, N.-P.; Laiakis, E.C.; Ghandhi, S.A.; Kumar, S.; Knotts, J.D.; Chowdhury, M.; Fornace, A.J.; Amundson, S.A. Global Gene Expression Response in Mouse Models of DNA Repair Deficiency after Gamma Irradiation. Radiat. Res. 2018, 189, 337–344. [Google Scholar] [CrossRef]
- Goudarzi, M.; Mak, T.D.; Chen, C.; Smilenov, L.B.; Brenner, D.J.; Fornace, A.J. The effect of low dose rate on metabolomic response to radiation in mice. Radiat. Environ. Biophys. 2014, 53, 645–657. [Google Scholar] [CrossRef][Green Version]
- Platten, M.; Nollen, E.A.A.; Röhrig, U.F.; Fallarino, F.; Opitz, C. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat. Rev. Drug Discov. 2019, 18, 379–401. [Google Scholar] [CrossRef]
- Ohtsu, H. Histamine Synthesis and Lessons Learned from Histidine Decarboxylase Deficient Mice. Results Probl. Cell Differ. 2010, 709, 21–31. [Google Scholar] [CrossRef]
- Obeid, R.; Geisel, J.; A Nix, W. 4-Pyridoxic Acid/Pyridoxine Ratio in Patients with Type 2 Diabetes is Related to Global Cardiovascular Risk Scores. Diagnostic 2019, 9, 28. [Google Scholar] [CrossRef]
- Pallister, T.; Jackson, M.; Martin, T.C.; Zierer, J.; Jennings, A.; Mohney, R.P.; MacGregor, A.J.; Steves, C.J.; Cassidy, A.; Spector, T.D.; et al. Hippurate as a metabolomic marker of gut microbiome diversity: Modulation by diet and relationship to metabolic syndrome. Sci. Rep. 2017, 7, 13670. [Google Scholar] [CrossRef]
- Ferreira, M.R.; Andreyev, H.J.N.; Mohammed, K.; Truelove, L.; Gowan, S.M.; Li, J.; Gulliford, S.L.; Marchesi, J.R.; Dearnaley, D.P.; Andreyev, J. Microbiota- and Radiotherapy-Induced Gastrointestinal Side-Effects (MARS) Study: A Large Pilot Study of the Microbiome in Acute and Late-Radiation Enteropathy. Clin. Cancer Res. 2019, 25, 6487–6500. [Google Scholar] [CrossRef]
- Badenhorst, C.P.S.; Van Der Sluis, R.; Erasmus, E.; Van Dijk, A.A. Glycine conjugation: Importance in metabolism, the role of glycine N-acyltransferase, and factors that influence interindividual variation. Expert Opin. Drug Metab. Toxicol. 2013, 9, 1139–1153. [Google Scholar] [CrossRef]
- Iizuka, D.; Yoshioka, S.; Kawai, H.; Izumi, S.; Suzuki, F.; Kamiya, K. Metabolomic screening using ESI-FT MS identifies potential radiation-responsive molecules in mouse urine. J. Radiat. Res. 2016, 58, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Ellinger, F. The histamine hypothesis of biological radiation effects. Schweiz. Med. Wochenschr. 1951, 81, 61–65. [Google Scholar] [PubMed]
- Martinel Lamas, D.J.; Nicoud, M.B.; Sterle, H.A.; Cremaschi, G.A.; Medina, V.A. Histamine: A potential cytoprotective agent to improve cancer therapy. Cell Death Dis. 2015, 6, e2029. [Google Scholar] [CrossRef] [PubMed]
- A Medina, V.; Croci, M.; Mohamad, N.A.; Massari, N.; Garbarino, G.; Cricco, G.P.; Núñez, M.A.; Martín, G.A.; Crescenti, E.J.V.; Bergoc, R.; et al. Mechanisms underlying the radioprotective effect of histamine on small intestine. Int. J. Radiat. Boil. 2007, 83, 653–663. [Google Scholar] [CrossRef] [PubMed]
- E Lewis, J.; Singh, N.; Holmila, R.J.; Sumer, B.D.; Williams, N.S.; Furdui, C.M.; Kemp, M.L.; Boothman, D.A. Targeting NAD+ Metabolism to Enhance Radiation Therapy Responses. Semin. Radiat. Oncol. 2019, 29, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Vaziri, H.; Dessain, S.K.; Eaton, E.N.; Imai, S.-I.; Frye, R.A.; Pandita, T.K.; Guarente, L.; Weinberg, R.A. hSIR2SIRT1 Functions as an NAD-Dependent p53 Deacetylase. Cell 2001, 107, 149–159. [Google Scholar] [CrossRef]
- Goodall, M.C. Effect of Neutron and Gamma Radiation on Adrenaline and Noradrenaline Release in the Human. Heal. Phys. 1968, 14, 199–203. [Google Scholar] [CrossRef]
- Sanchez-Macedo, N.; Feng, J.; Faubert, B.; Chang, N.; Elia, A.; Rushing, E.J.; Tsuchihara, K.; Bungard, D.; Berger, S.L.; Jones, R.G.; et al. Depletion of the novel p53-target gene carnitine palmitoyltransferase 1C delays tumor growth in the neurofibromatosis type I tumor model. Cell Death Differ. 2013, 20, 659–668. [Google Scholar] [CrossRef]
- Yahagi, N.; Shimano, H.; Matsuzaka, T.; Najima, Y.; Sekiya, M.; Nakagawa, Y.; Ide, T.; Tomita, S.; Okazaki, H.; Tamura, Y.; et al. p53 Activation in Adipocytes of Obese Mice. J. Boil. Chem. 2003, 278, 25395–25400. [Google Scholar] [CrossRef]
- Napoli, M.; Flores, E.R. The p53 family orchestrates the regulation of metabolism: Physiological regulation and implications for cancer therapy. Br. J. Cancer 2016, 116, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Manna, S.K.; Krausz, K.W.; Bonzo, J.A.; Idle, J.; Gonzalez, F.J. Metabolomics Reveals Aging-associated Attenuation of Noninvasive Radiation Biomarkers in Mice: Potential Role of Polyamine Catabolism and Incoherent DNA Damage-repair. J. Proteome Res. 2013, 12, 2269–2281. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.H.; Patterson, A.D.; Krausz, K.W.; Lanz, C.; Kang, N.W.; Luecke, H.; Gonzalez, F.J.; Idle, J. Radiation metabolomics. 4. UPLC-ESI-QTOFMS-Based metabolomics for urinary biomarker discovery in gamma-irradiated rats. Radiat. Res. 2011, 175, 473–484. [Google Scholar] [CrossRef]
- Ou, Y.; Wang, S.-J.; Li, D.; Chu, B.; Gu, W. Activation ofSAT1engages polyamine metabolism with p53-mediated ferroptotic responses. Proc. Natl. Acad. Sci. USA 2016, 113, E6806–E6812. [Google Scholar] [CrossRef]
- Haberland, G.L.; Schreier, K.; Bruns, F.; Altman, K.I.; Hempelmann, L.H.; Haberland, K.S.G.L. Creatine–Creatinine Metabolism in Radiation Myopathy. Nature 1955, 175, 1039–1040. [Google Scholar] [CrossRef] [PubMed]
- Zambetti, G.P.; Bargonetti, J.; Walker, K.; Prives, C.; Levine, A.J. Wild-type p53 mediates positive regulation of gene expression through a specific DNA sequence element. Genes Dev. 1992, 6, 1143–1152. [Google Scholar] [CrossRef]
- Ebert, S.M.; Dierdorff, J.M.; Meyerholz, D.K.; Bullard, S.A.; Al-Zougbi, A.; Delau, A.D.; Tomcheck, K.C.; Skopec, Z.P.; Marcotte, G.R.; Bodine, S.C.; et al. An investigation of p53 in skeletal muscle aging. J. Appl. Physiol. 2019, 127, 1075–1084. [Google Scholar] [CrossRef]
- Stocks, B.; Dent, J.R.; Joanisse, S.; McCurdy, C.; Philp, A. Skeletal Muscle Fibre-Specific Knockout of p53 Does Not Reduce Mitochondrial Content or Enzyme Activity. Front. Physiol. 2017, 8, 941. [Google Scholar] [CrossRef]
- Brett-Morris, A.; Wright, B.M.; Seo, Y.; Pasupuleti, V.; Zhang, J.; Lu, J.; Spina, R.; Bar, E.E.; Gujrati, M.; Schur, R.; et al. The polyamine catabolic enzyme SAT1 modulates tumorigenesis and radiation response in GBM. Cancer Res. 2014, 74, 6925–6934. [Google Scholar] [CrossRef]

| Metabolite | Adduct | RT (min) | Experimental (m/z) | Calculated (m/z) | Mass Error (ppm) | Formula | HMDB ID | Metabolic Pathway |
|---|---|---|---|---|---|---|---|---|
| Carnitine | [M + H]+ | 0.29 | 162.1137 | 162.1130 | 4.3 | C7H15NO3 | 0000062 | Fatty acid β oxidation |
| Kynurenic acid | [M + H]+ | 1.36 | 190.0512 | 190.0504 | 4.2 | C10H7NO3 | 0000715 | Tryptophan metabolism |
| Hippuric acid | [M + H]+ | 1.75 | 180.0669 | 180.0661 | 4.4 | C9H9NO3 | 0000714 | Phenylalanine metabolism/Glycine conjugation |
| Phenylacetylglycine | [M + H]+ | 2.33 | 194.0817 | 194.0817 | 0.0 | C10H11NO3 | 0000821 | Phenylalanine metabolism/Glycine conjugation |
| Creatine | [M + H]+ | 0.29 | 132.0780 | 132.0773 | 5.3 | C4H9N3O2 | 0000064 | Amino acid metabolism |
| 4-Pyridoxic acid | [M + H]+ | 0.34 | 184.0603 | 184.0610 | 3.8 | C8H9NO4 | 0000017 | Vitamin B6 metabolism |
| MHPG-SO4 | [M − H]− | 0.39 | 263.0224 | 263.0226 | 0.6 | C9H12O7S | 0000559 | Noradrenaline metabolism |
| 1-Methylhistamine | [M + H]+ | 0.25 | 126.1033 | 126.1031 | 1.4 | C6H11N3 | 0000898 | Histidine metabolism |
| 1-Methylnicotinamide | [M + H]+ | 0.28 | 137.0722 | 137.0715 | 5.2 | C7H8N2O | 0000699 | Nicotinate and nicotinamide metabolism |
| N1-Acetylspermidine | [M + H]+ | 0.25 | 188.1769 | 188.1763 | 3.3 | C9H21N3O | 0001276 | Polyamine metabolism |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pannkuk, E.L.; Laiakis, E.C.; Ake, P.; Strawn, S.J.; Wang, Y.-W.; Fornace, A.J. Effects of Genetic Variation on Urinary Small Molecule Signatures of Mice after Exposure to Ionizing Radiation: A Study of p53 Deficiency. Metabolites 2020, 10, 234. https://doi.org/10.3390/metabo10060234
Pannkuk EL, Laiakis EC, Ake P, Strawn SJ, Wang Y-W, Fornace AJ. Effects of Genetic Variation on Urinary Small Molecule Signatures of Mice after Exposure to Ionizing Radiation: A Study of p53 Deficiency. Metabolites. 2020; 10(6):234. https://doi.org/10.3390/metabo10060234
Chicago/Turabian StylePannkuk, Evan L., Evagelia C. Laiakis, Pelagie Ake, Steven J. Strawn, Yi-Wen Wang, and Albert J. Fornace. 2020. "Effects of Genetic Variation on Urinary Small Molecule Signatures of Mice after Exposure to Ionizing Radiation: A Study of p53 Deficiency" Metabolites 10, no. 6: 234. https://doi.org/10.3390/metabo10060234
APA StylePannkuk, E. L., Laiakis, E. C., Ake, P., Strawn, S. J., Wang, Y.-W., & Fornace, A. J. (2020). Effects of Genetic Variation on Urinary Small Molecule Signatures of Mice after Exposure to Ionizing Radiation: A Study of p53 Deficiency. Metabolites, 10(6), 234. https://doi.org/10.3390/metabo10060234