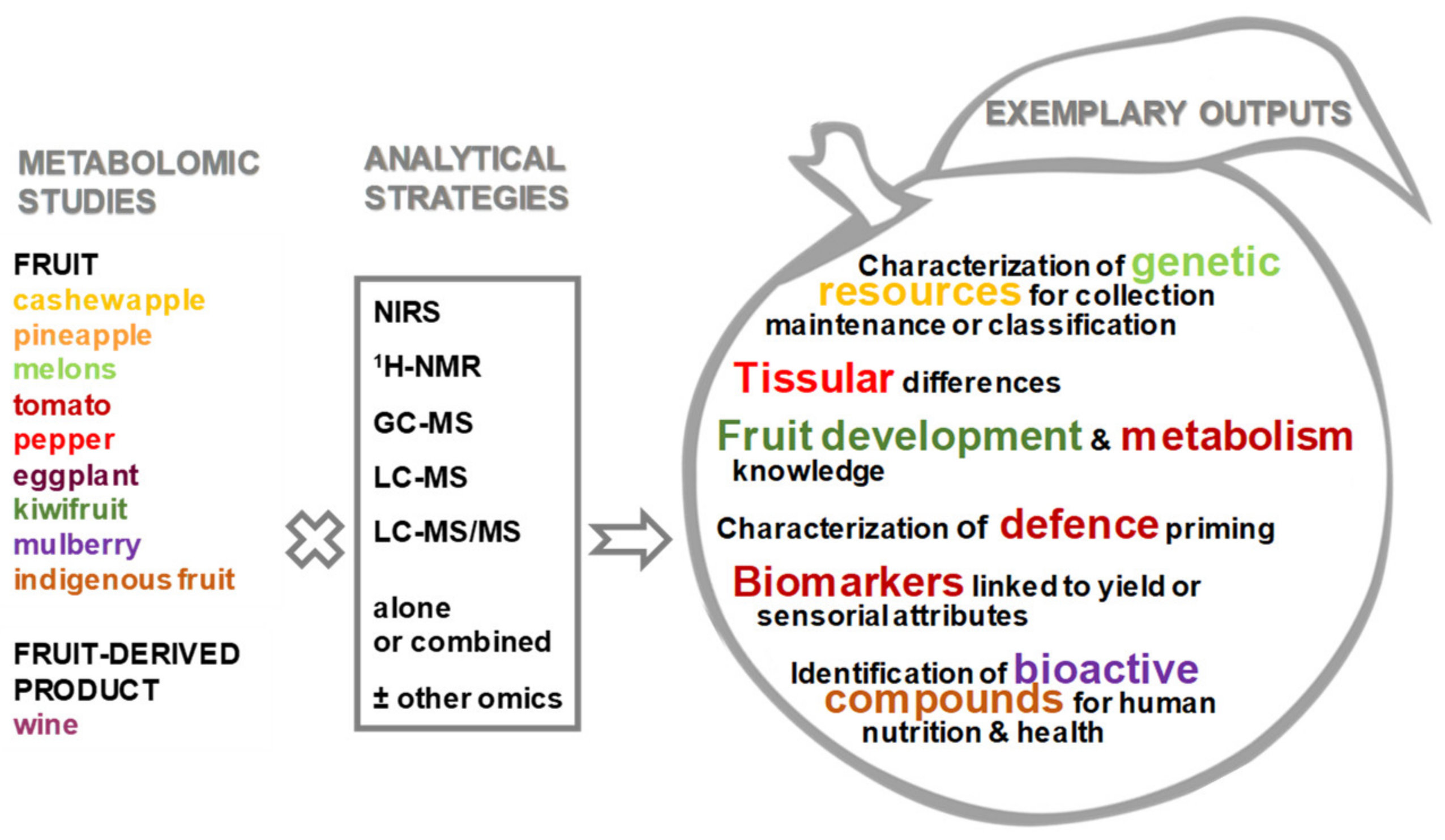
Special Issue on “Fruit Metabolism and Metabolomics”
Abstract
Funding
Conflicts of Interest
References
- van der Knaap, E.; Østergaard, L. Shaping a fruit: Developmental pathways that impact growth patterns. Semin. Cell Dev. Biol. 2018, 79, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Pott, D.M.; Osorio, S.; Vallarino, J.G. From central to specialized metabolism: An overview of some secondary compounds derived from the primary metabolism for their role in conferring nutritional and organoleptic characteristics to fruit. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Casado, A. The health potential of fruits and vegetables phytochemicals: Notable examples. Crit. Rev. Food Sci. Nutr. 2016, 56, 1097–1107. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Schuurink, R.C.; Caissard, J.-C.; Hugueney, P.; Baudino, S. My way: Noncanonical biosynthesis pathways for plant volatiles. Trends Plant Sci. 2016, 21, 884–894. [Google Scholar] [CrossRef]
- Beauvoit, B.; Belouah, I.; Bertin, N.; Cakpo, C.B.; Colombié, S.; Dai, Z.; Gautier, H.; Génard, M.; Moing, A.; Roch, L.; et al. Putting primary metabolism into perspective to obtain better fruits. Ann. Bot. 2018, 122, 1–21. [Google Scholar] [CrossRef]
- Ilahy, R.; Tlili, I.; Siddiqui, M.W.; Hdider, C.; Lenucci, M.S. Inside and beyond color: Comparative overview of functional quality of tomato and watermelon fruits. Front. Plant Sci. 2019, 10, 769. [Google Scholar] [CrossRef]
- Hanhineva, K.; Aharoni, A. Metabolomics in Fruit Development. In Molecular Techniques in Crop Improvement, 2nd ed.; Jain, S.M., Brar, D.S., Eds.; Springer: Heidelberg, Germany, 2010; pp. 675–693. [Google Scholar]
- de Vos, R.C.H.; Hall, R.; Moing, A. Metabolomics of a model fruit: Tomato In Biology of Plant Metabolomics; Hall, R., Ed.; Wiley-Blackwell Ltd.: Oxford, UK, 2011; Volume 43, pp. 109–155. [Google Scholar]
- Tohge, T.; Fernie, A.R. Metabolomics-inspired insight into developmental, environmental and genetic aspects of tomato fruit chemical composition and quality. Plant Cell Physiol. 2015, 56, 1681–1696. [Google Scholar] [CrossRef]
- Yu, G.; Li, C.; Zhang, L.; Zhu, G.; Munir, S.; Shi, C.; Zhang, H.; Ai, G.; Gao, S.; Zhang, Y.; et al. An allelic variant of GAME9 determines its binding capacity with the GAME17 promoter in the regulation of steroidal glycoalkaloid biosynthesis in tomato. J. Exp. Bot. 2020, 71, 2527–2536. [Google Scholar] [CrossRef]
- Tietel, Z.; Srivastava, S.; Fait, A.; Tel-Zur, N.; Carmi, N.; Raveh, E. Impact of scion/rootstock reciprocal effects on metabolomics of fruit juice and phloem sap in grafted Citrus reticulata. PLoS ONE 2020, 15, e0227192. [Google Scholar] [CrossRef]
- Dono, G.; Rambla, J.L.; Frusciante, S.; Granell, A.; Diretto, G.; Mazzucato, A. Color mutations alter the biochemical composition in the San Marzano tomato fruit. Metabolites 2020, 10, 110. [Google Scholar] [CrossRef]
- Kuhalskaya, A.; Wijesingha Ahchige, M.; Perez de Souza, L.; Vallarino, J.; Brotman, Y.; Alseekh, S. Network analysis provides insight into tomato lipid metabolism. Metabolites 2020, 10, 152. [Google Scholar] [CrossRef] [PubMed]
- Luna, E.; Flandin, A.; Cassan, C.; Prigent, S.; Chevanne, C.; Kadiri, C.F.; Gibon, Y.; Pétriacq, P. Metabolomics to exploit the primed immune system of tomato fruit. Metabolites 2020, 10, 96. [Google Scholar] [CrossRef] [PubMed]
- Nardozza, S.; Cooney, J.; Boldingh, H.L.; Hewitt, K.G.; Trower, T.; Jones, D.; Thrimawithana, A.H.; Allan, A.C.; Richardson, A.C. Phytohormone and transcriptomic analysis reveals endogenous cytokinins affect kiwifruit growth under restricted carbon supply. Metabolites 2020, 10, 23. [Google Scholar] [CrossRef] [PubMed]
- D’Urso, G.; Mes, J.J.; Montoro, P.; Hall, R.D.; de Vos, R.C.H. Identification of bioactive phytochemicals in mulberries. Metabolites 2020, 10, 7. [Google Scholar] [CrossRef]
- Ikram, M.M.M.; Ridwani, S.; Putri, S.P.; Fukusaki, E. GC-MS based metabolite profiling to monitor ripening-specific metabolites in pineapple (Ananas comosus). Metabolites 2020, 10, 134. [Google Scholar] [CrossRef]
- Alves Filho, E.; Silva, L.M.; Lima, Y.; Ribeiro, P.; Silva, E.; Zocolo, G.; Canuto, K.; Morais, S.; Castro, A.C.; de Brito, E. Metabolomic variability of different genotypes of cashew by LC-MS and correlation with near-infrared spectroscopy as a tool for fast phenotyping. Metabolites 2019, 9, 121. [Google Scholar] [CrossRef]
- Moing, A.; Allwood, J.W.; Aharoni, A.; Baker, J.; Beale, M.H.; Ben-Dor, S.; Biais, B.; Brigante, F.; Burger, Y.; Deborde, C.; et al. Comparative metabolomics and molecular phylogenetics of melon (Cucumis melo, Cucurbitaceae) biodiversity. Metabolites 2020, 10, 121. [Google Scholar] [CrossRef]
- Lim, V.; Gorji, S.G.; Daygon, V.D.; Fitzgerald, M. Untargeted and targeted metabolomic profiling of Australian indigenous fruits. Metabolites 2020, 10, 114. [Google Scholar] [CrossRef]
- Calumpang, C.L.F.; Saigo, T.; Watanabe, M.; Tohge, T. Cross-species comparison of fruit-metabolomics to elucidate metabolic regulation of fruit polyphenolics among Solanaceous crops. Metabolites 2020, 10, 209. [Google Scholar] [CrossRef]
- Cervantes-Hernández, F.; Alcalá-González, P.; Martínez, O.; Ordaz-Ortiz, J.J. Placenta, pericarp, and seeds of tabasco chili pepper fruits show a contrasting diversity of bioactive metabolites. Metabolites 2019, 9, 206. [Google Scholar] [CrossRef]
- Pott, D.M.; Vallarino, J.G.; Osorio, S. Metabolite changes during postharvest storage: Effects on fruit quality traits. Metabolites 2020, 10, 187. [Google Scholar] [CrossRef] [PubMed]
- Cozzolino, D. Metabolomics in grape and wine: Definition, current status and future prospects. Food Anal. Methods 2016, 9, 2986–2997. [Google Scholar] [CrossRef]
- Pinu, F.R. Grape and wine metabolomics to develop new insights using untargeted and targeted approaches. Fermentation 2018, 4, 92. [Google Scholar] [CrossRef]
- Cuadros-Inostroza, A.; Verdugo-Alegría, C.; Willmitzer, L.; Moreno, Y.; Vallarino, J.G. Non-targeted metabolite profiles and sensory properties elucidate commonalities and differences of wines made with the same variety but different cultivar clones. Metabolites 2020, 10, 220. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moing, A.; Pétriacq, P.; Osorio, S. Special Issue on “Fruit Metabolism and Metabolomics”. Metabolites 2020, 10, 230. https://doi.org/10.3390/metabo10060230
Moing A, Pétriacq P, Osorio S. Special Issue on “Fruit Metabolism and Metabolomics”. Metabolites. 2020; 10(6):230. https://doi.org/10.3390/metabo10060230
Chicago/Turabian StyleMoing, Annick, Pierre Pétriacq, and Sonia Osorio. 2020. "Special Issue on “Fruit Metabolism and Metabolomics”" Metabolites 10, no. 6: 230. https://doi.org/10.3390/metabo10060230
APA StyleMoing, A., Pétriacq, P., & Osorio, S. (2020). Special Issue on “Fruit Metabolism and Metabolomics”. Metabolites, 10(6), 230. https://doi.org/10.3390/metabo10060230






