EFMviz: A COBRA Toolbox Extension to Visualize Elementary Flux Modes in Genome-Scale Metabolic Models
Abstract
1. Introduction
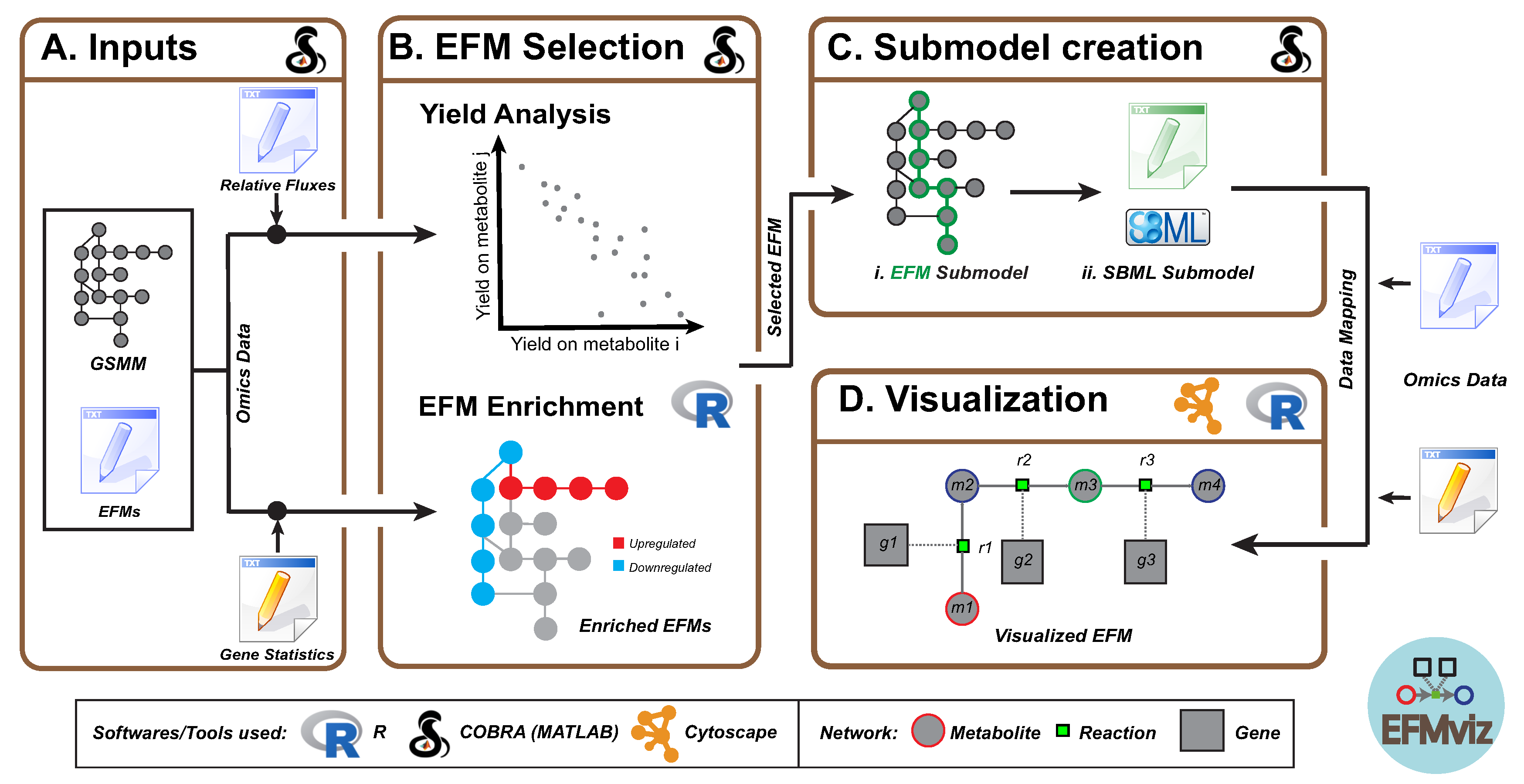
2. Materials and Methods
2.1. Inputs
2.2. EFM Selection
2.2.1. Yield Analysis
2.2.2. EFM Enrichment
2.3. Submodel Creation
2.4. Visualization
3. Results
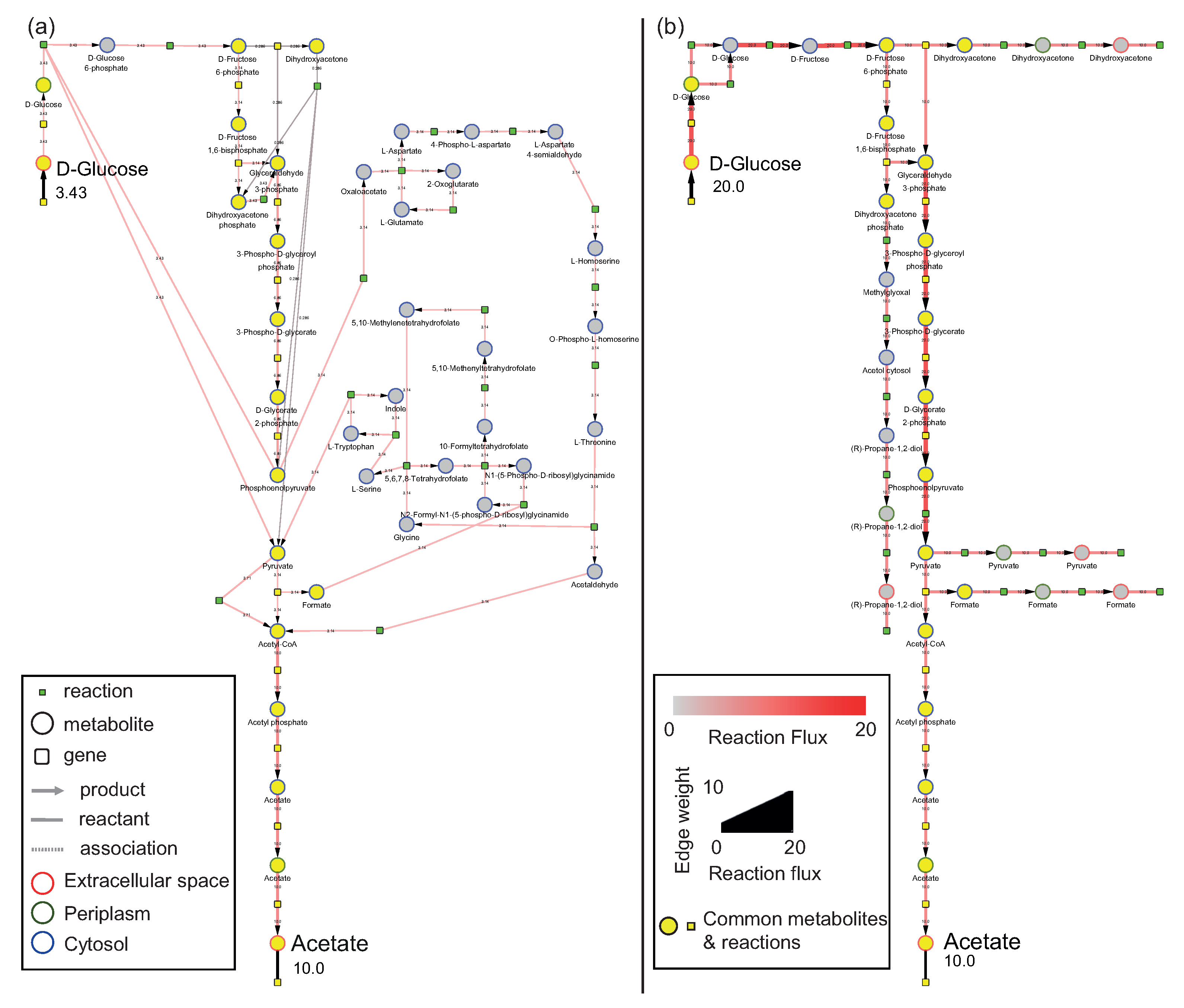
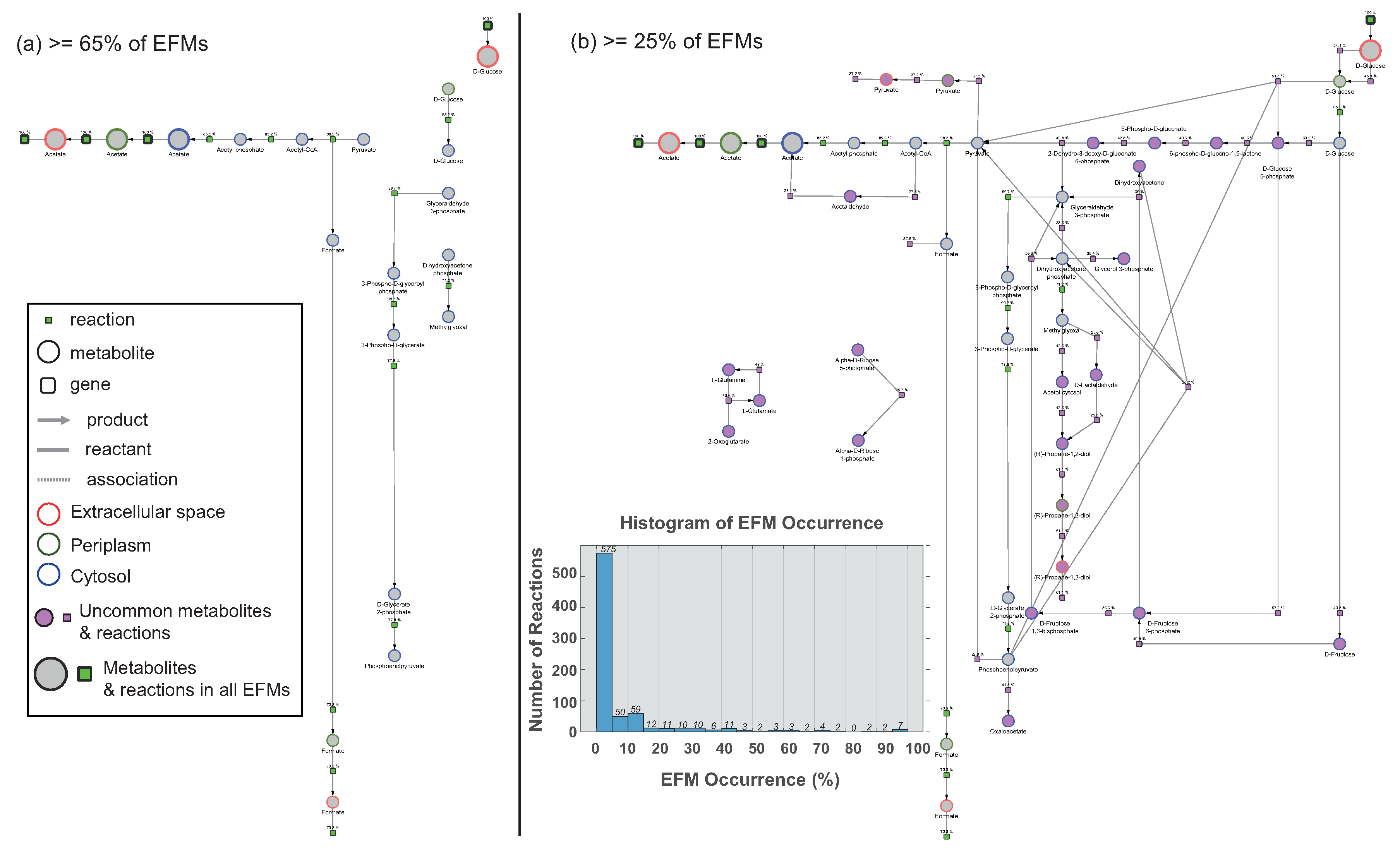
3.1. Use Case 1: Visualizing Elementary Flux Modes from the E. coli Model
3.1.1. Model
3.1.2. Analysis
Comparison of EFMs Using Preserved Visual Arrangement
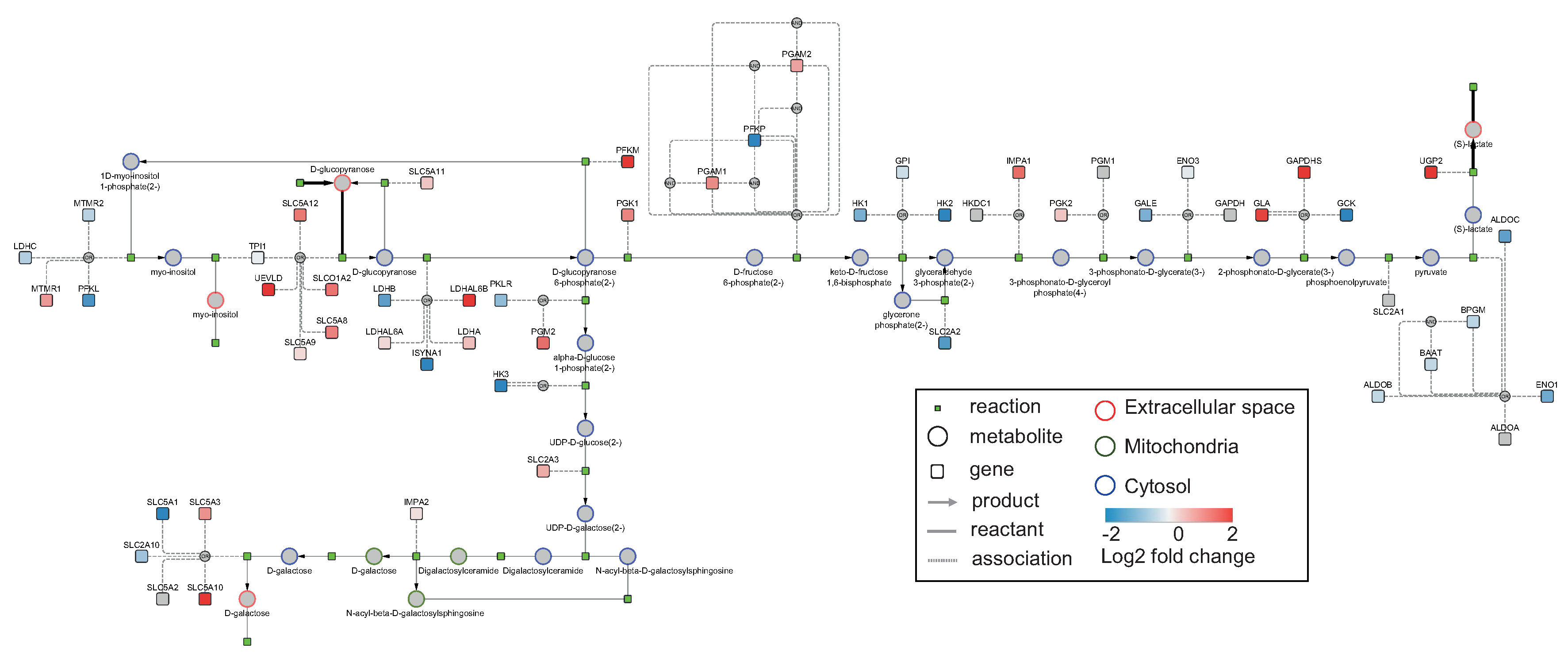
3.2. Use Case 2: Visualizing Elementary Flux Modes from the Human Model
3.2.1. Data
3.2.2. Model
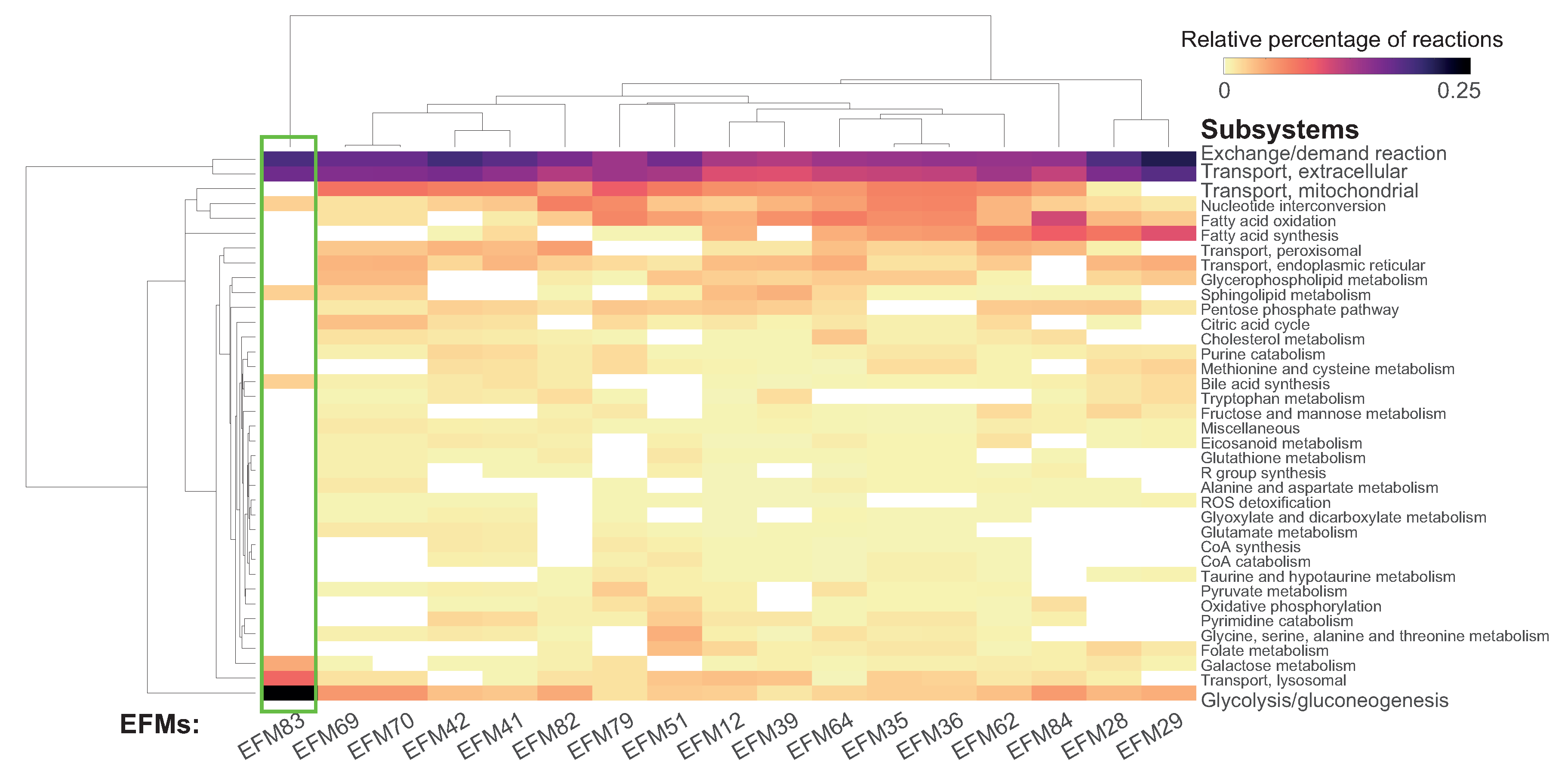
3.2.3. Analysis
Comparing EFMs Using Subsystem Occurrence
3.3. Assessing Overlap between EFMs: Backbone Identification
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Robinson, J.L.; Nielsen, J. Integrative analysis of human omics data using biomolecular networks. Mol. BioSyst. 2016, 12, 2953–2964. [Google Scholar] [CrossRef] [PubMed]
- Schuster, S.; Dandekar, T.; Fell, D.A.; Schuster, S.; Dandekar, T.; Fell, D.; Schuster, S.; Dandekar, T.; Fell, D.; Schuster, S.; et al. Detection of elementary flux modes in biochemical networks: A promising tool for pathway analysis and metabolic engineering. Trends Biotechnol. 1999, 17, 53–60. [Google Scholar] [CrossRef]
- Schuster, S.; Fell, D.A.; Dandekar, T. A general definition of metabolic pathways useful for systematic organization and analysis of complex metabolic networks. Nat. Biotechnol. 2000, 18, 326. [Google Scholar] [CrossRef]
- Carlson, R.; Srienc, F. FundamentalEscherichia coli biochemical pathways for biomass and energy production: Identification of reactions. Biotechnol. Bioeng. 2004, 85, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Trinh, C.T.; Wlaschin, A.; Srienc, F. Elementary mode analysis: A useful metabolic pathway analysis tool for characterizing cellular metabolism. Appl. Microbiol. Biotechnol. 2009, 81, 813–826. [Google Scholar] [CrossRef] [PubMed]
- Carlson, R. Metabolic systems cost-benefit analysis for interpreting network structure and regulation. Bioinformatics 2007, 23, 2202. [Google Scholar] [CrossRef]
- Trinh, C.T.; Srienc, F. Metabolic engineering of Escherichia coli for efficient conversion of glycerol to ethanol. Appl. Environ. Microbiol. 2009, 75, 6696–6705. [Google Scholar] [CrossRef]
- Trinh, C.T.; Unrean, P.; Srienc, F. Minimal Escherichia coli cell for the most efficient production of ethanol from hexoses and pentoses. Appl. Environ. Microbiol. 2008, 74, 3634–3643. [Google Scholar] [CrossRef]
- Klamt, S.; Gilles, E.D. Minimal cut sets in biochemical reaction networks. Bioinformatics 2004, 20, 226–234. [Google Scholar] [CrossRef]
- Stelling, J.; Klamt, S.; Bettenbrock, K.; Schuster, S.; Gilles, E.D. Metabolic network structure determines key aspects of functionality and regulation. Nature 2002, 420, 190–193. [Google Scholar] [CrossRef]
- Gebauer, J.; Schuster, S.; de Figueiredo, L.F.; Kaleta, C. Detecting and investigating substrate cycles in a genome-scale human metabolic network. FEBS J. 2012, 279, 3192–3202. [Google Scholar] [CrossRef]
- Klamt, S.; Stelling, J. Combinatorial Complexity of Pathway Analysis in Metabolic Networks. Mol. Biol. Rep. 2002, 29, 233–236. [Google Scholar] [CrossRef] [PubMed]
- Gagneur, J.; Klamt, S. Computation of elementary modes: A unifying framework and the new binary approach. BMC Bioinform. 2004, 5, 175. [Google Scholar] [CrossRef] [PubMed]
- Schilling, C.H.; Letscher, D.; Palsson, B.Ø. Theory for the Systemic Definition of Metabolic Pathways and their use in Interpreting Metabolic Function from a Pathway-Oriented Perspective. J. Theor. Biol. 2000, 203, 229–248. [Google Scholar] [CrossRef] [PubMed]
- Yeung, M.; Thiele, I.; Palsson, B.Ø. Estimation of the number of extreme pathways for metabolic networks. BMC Bioinform. 2007, 8, 363. [Google Scholar] [CrossRef] [PubMed]
- Croes, D.; Couche, F.; Wodak, S.J.; van Helden, J. Inferring meaningful pathways in weighted metabolic networks. J. Mol. Biol. 2006, 356, 222–236. [Google Scholar] [CrossRef] [PubMed]
- Blum, T.; Kohlbacher, O. Using Atom Mapping Rules for an Improved Detection of Relevant Routes in Weighted Metabolic Networks. J. Comput. Biol. 2008, 15, 565–576. [Google Scholar] [CrossRef]
- de Figueiredo, L.F.; Podhorski, A.; Rubio, A.; Kaleta, C.; Beasley, J.E.; Schuster, S.; Planes, F.J. Computing the shortest elementary flux modes in genome-scale metabolic networks. Bioinformatics 2009, 25, 3158–3165. [Google Scholar] [CrossRef]
- Machado, D.; Soons, Z.; Patil, K.R.; Ferreira, E.C.; Rocha, I. Random sampling of elementary flux modes in large-scale metabolic networks. Bioinformatics 2012, 28, i515–i521. [Google Scholar] [CrossRef]
- Kelk, S.M.; Olivier, B.G.; Stougie, L.; Bruggeman, F.J. Optimal flux spaces of genome-scale stoichiometric models are determined by a few subnetworks. Sci. Rep. 2012, 2, 580. [Google Scholar] [CrossRef]
- Rezola, A.; Pey, J.; de Figueiredo, L.F.; Podhorski, A.; Schuster, S.; Rubio, A.; Planes, F.J. Selection of human tissue-specific elementary flux modes using gene expression data. Bioinformatics 2013, 29, 2009–2016. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hunt, K.A.; Folsom, J.P.; Taffs, R.L.; Carlson, R.P. Complete enumeration of elementary flux modes through scalable demand-based subnetwork definition. Bioinformatics 2014, 30, 1569–1578. [Google Scholar] [CrossRef] [PubMed]
- Gerstl, M.P.; Ruckerbauer, D.E.; Mattanovich, D.; Jungreuthmayer, C.; Zanghellini, J. Metabolomics integrated elementary flux mode analysis in large metabolic networks. Sci. Rep. 2015, 5, 8930. [Google Scholar] [CrossRef] [PubMed]
- Rezola, A.; Pey, J.; Rubio, Á.; Planes, F.J. In-Silico Prediction of Key Metabolic Differences between Two Non-Small Cell Lung Cancer Subtypes. PLoS ONE 2014, 9, e103998. [Google Scholar] [CrossRef] [PubMed]
- Orth, J.D.; Thiele, I.; Palsson, B.Ø. What is flux balance analysis? Nat. Biotechnol. 2010, 28, 245–248. [Google Scholar] [CrossRef]
- Rowe, E.; Palsson, B.O.; King, Z.A. Escher-FBA: A web application for interactive flux balance analysis. BMC Syst. Biol. 2018, 12, 84. [Google Scholar] [CrossRef]
- König, M.; Holzhütter, H.G. Fluxviz—Cytoscape plug-in for visualization of flux distributions in networks. In Genome Informatics 2010; Imperial College Press: London, UK, 2010; pp. 96–103. [Google Scholar] [CrossRef]
- Hoppe, A.; Hoffmann, S.; Gerasch, A.; Gille, C.; Holzhütter, H.G. FASIMU: Flexible software for flux-balance computation series in large metabolic networks. BMC Bioinform. 2011, 12, 28. [Google Scholar] [CrossRef]
- Junker, B.; Klukas, C.; Schreiber, F. VANTED: A system for advanced data analysis and visualization in the context of biological networks. BMC Bioinform. 2006, 7, 109. [Google Scholar] [CrossRef]
- Kostromins, A.; Stalidzans, E. Paint4Net: COBRA Toolbox extension for visualization of stoichiometric models of metabolism. Biosystems 2012, 109, 233–239. [Google Scholar] [CrossRef]
- Heirendt, L.; Arreckx, S.; Pfau, T.; Mendoza, S.N.; Richelle, A.; Heinken, A.; Haraldsdóttir, H.S.; Wachowiak, J.; Keating, S.M.; Vlasov, V.; et al. Creation and analysis of biochemical constraint-based models: The COBRA Toolbox v3.0. arXiv 2017, arXiv:1710.04038. [Google Scholar] [CrossRef]
- Klamt, S.; Saez-Rodriguez, J.; Gilles, E.D. Structural and functional analysis of cellular networks with CellNetAnalyzer. BMC Syst. Biol. 2007, 1, 2. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.A.; Papin, J.A. MetDraw: Automated visualization of genome-scale metabolic network reconstructions and high-throughput data. Bioinformatics 2014, 30, 1327–1328. [Google Scholar] [CrossRef] [PubMed]
- Noronha, A.; Daníelsdóttir, A.D.; Gawron, P.; Jóhannsson, F.; Jónsdóttir, S.; Jarlsson, S.; Gunnarsson, J.P.; Brynjólfsson, S.; Schneider, R.; Thiele, I.; et al. ReconMap: An interactive visualization of human metabolism. Bioinformatics 2016, 33, btw667. [Google Scholar] [CrossRef] [PubMed]
- Rose, T.; Mazat, J.P.; Rose, T.D.; Mazat, J.P. FluxVisualizer, a Software to Visualize Fluxes through Metabolic Networks. Processes 2018, 6, 39. [Google Scholar] [CrossRef]
- Chazalviel, M.; Frainay, C.; Poupin, N.; Vinson, F.; Merlet, B.; Gloaguen, Y.; Cottret, L.; Jourdan, F. MetExploreViz: Web component for interactive metabolic network visualization. Bioinformatics 2018, 34, 312–313. [Google Scholar] [CrossRef]
- Noronha, A.; Vilaça, P.; Rocha, M. An integrated network visualization framework towards metabolic engineering applications. BMC Bioinform. 2014, 15, 420. [Google Scholar] [CrossRef]
- Feist, A.M.; Henry, C.S.; Reed, J.L.; Krummenacker, M.; Joyce, A.R.; Karp, P.D.; Broadbelt, L.J.; Hatzimanikatis, V.; Palsson, B.Ø. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol. Syst. Biol. 2007, 3, 121. [Google Scholar] [CrossRef]
- Swainston, N.; Smallbone, K.; Hefzi, H.; Dobson, P.D.; Brewer, J.; Hanscho, M.; Zielinski, D.C.; Ang, K.S.; Gardiner, N.J.; Gutierrez, J.M.; et al. Recon 2.2: From reconstruction to model of human metabolism. Metabolomics 2016, 12, 109. [Google Scholar] [CrossRef]
- Thiele, I.; Palsson, B.Ø. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 2010, 5, 93–121. [Google Scholar] [CrossRef]
- Pey, J.; Villar, J.A.; Tobalina, L.; Rezola, A.; García, J.M.; Beasley, J.E.; Planes, F.J. TreeEFM: Calculating elementary flux modes using linear optimization in a tree-based algorithm. Bioinformatics 2015, 31, 897–904. [Google Scholar] [CrossRef]
- Song, H.S.; Ramkrishna, D. Reduction of a set of elementary modes using yield analysis. Biotechnol. Bioeng. 2009, 102, 554–568. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Väremo, L.; Nielsen, J.; Nookaew, I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 2013, 41, 4378–4391. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- König, M.; Drager, A.; Holzhutter, H.G. CySBML: A Cytoscape plugin for SBML. Bioinformatics 2012, 28, 2402–2403. [Google Scholar] [CrossRef][Green Version]
- Novère, N.L.; Hucka, M.; Mi, H.; Moodie, S.; Schreiber, F.; Sorokin, A.; Demir, E.; Wegner, K.; Aladjem, M.I.; Wimalaratne, S.M.; et al. The Systems Biology Graphical Notation. Nat. Biotechnol. 2009, 27, 735–741. [Google Scholar] [CrossRef]
- yWorks GmbH. yFiles Layout Algorithms for Cytoscape app; yWorks GmbH: Tübingen, Germany, 2018. [Google Scholar]
- Ono, K.; Muetze, T.; Kolishovski, G.; Shannon, P.; Demchak, B. CyREST: Turbocharging Cytoscape Access for External Tools via a RESTful API. F1000Research 2015, 4, 478. [Google Scholar] [CrossRef]
- Pratt, D.; Chen, J.; Welker, D.; Rivas, R.; Pillich, R.; Rynkov, V.; Ono, K.; Miello, C.; Hicks, L.; Szalma, S.; et al. NDEx, the network data exchange. Cell Syst. 2015, 1, 302–305. [Google Scholar] [CrossRef]
- Eiteman, M.A.; Altman, E. Overcoming acetate in Escherichia coli recombinant protein fermentations. Trends Biotechnol. 2006, 24, 530–536. [Google Scholar] [CrossRef]
- Clomburg, J.M.; Gonzalez, R. Biofuel production in Escherichia coli: The role of metabolic engineering and synthetic biology. Appl. Microbiol. Biotechnol. 2010, 86, 419–434. [Google Scholar] [CrossRef]
- Nakano, K.; Rischke, M.; Sato, S.; Märkl, H. Influence of acetic acid on the growth of Escherichia coli K12 during high-cell-density cultivation in a dialysis reactor. Appl. Microbiol. Biotechnol. 1997, 48, 597–601. [Google Scholar] [CrossRef]
- Contiero, J.; Beatty, C.; Kumari, S.; DeSanti, C.L.; Strohl, W.R.; Wolfe, A. Effects of mutations in acetate metabolism on high-cell-density growth of Escherichia coli. J. Ind. Microbiol. Biotechnol. 2000, 24, 421–430. [Google Scholar] [CrossRef]
- Warburg, O.; Wind, F.; Negelein, E. The metabolism of tumors in the body. J. Gen. Physiol. 1927, 8, 519. [Google Scholar] [CrossRef] [PubMed]
- CGAN. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarathy, C.; Kutmon, M.; Lenz, M.; Adriaens, M.E.; Evelo, C.T.; Arts, I.C.W. EFMviz: A COBRA Toolbox Extension to Visualize Elementary Flux Modes in Genome-Scale Metabolic Models. Metabolites 2020, 10, 66. https://doi.org/10.3390/metabo10020066
Sarathy C, Kutmon M, Lenz M, Adriaens ME, Evelo CT, Arts ICW. EFMviz: A COBRA Toolbox Extension to Visualize Elementary Flux Modes in Genome-Scale Metabolic Models. Metabolites. 2020; 10(2):66. https://doi.org/10.3390/metabo10020066
Chicago/Turabian StyleSarathy, Chaitra, Martina Kutmon, Michael Lenz, Michiel E. Adriaens, Chris T. Evelo, and Ilja C.W. Arts. 2020. "EFMviz: A COBRA Toolbox Extension to Visualize Elementary Flux Modes in Genome-Scale Metabolic Models" Metabolites 10, no. 2: 66. https://doi.org/10.3390/metabo10020066
APA StyleSarathy, C., Kutmon, M., Lenz, M., Adriaens, M. E., Evelo, C. T., & Arts, I. C. W. (2020). EFMviz: A COBRA Toolbox Extension to Visualize Elementary Flux Modes in Genome-Scale Metabolic Models. Metabolites, 10(2), 66. https://doi.org/10.3390/metabo10020066





