Identification of the Main Metabolites of a Marine-Derived Strain of Penicillium brevicompactum Using LC and GC MS Techniques
Abstract
1. Introduction
2. Results
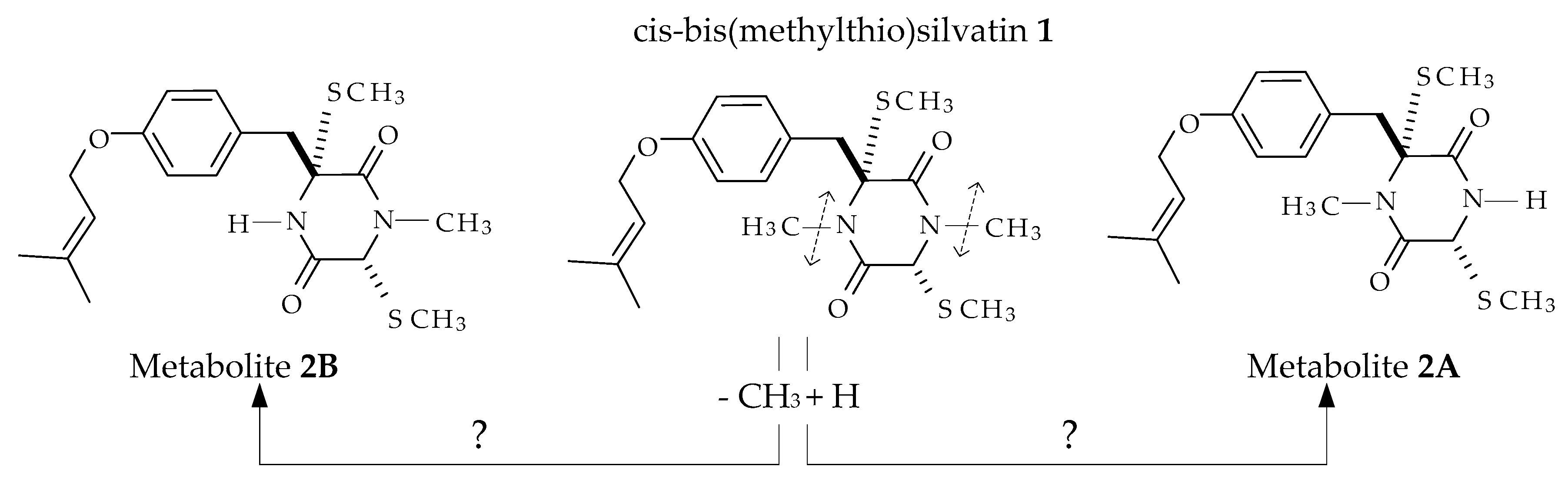
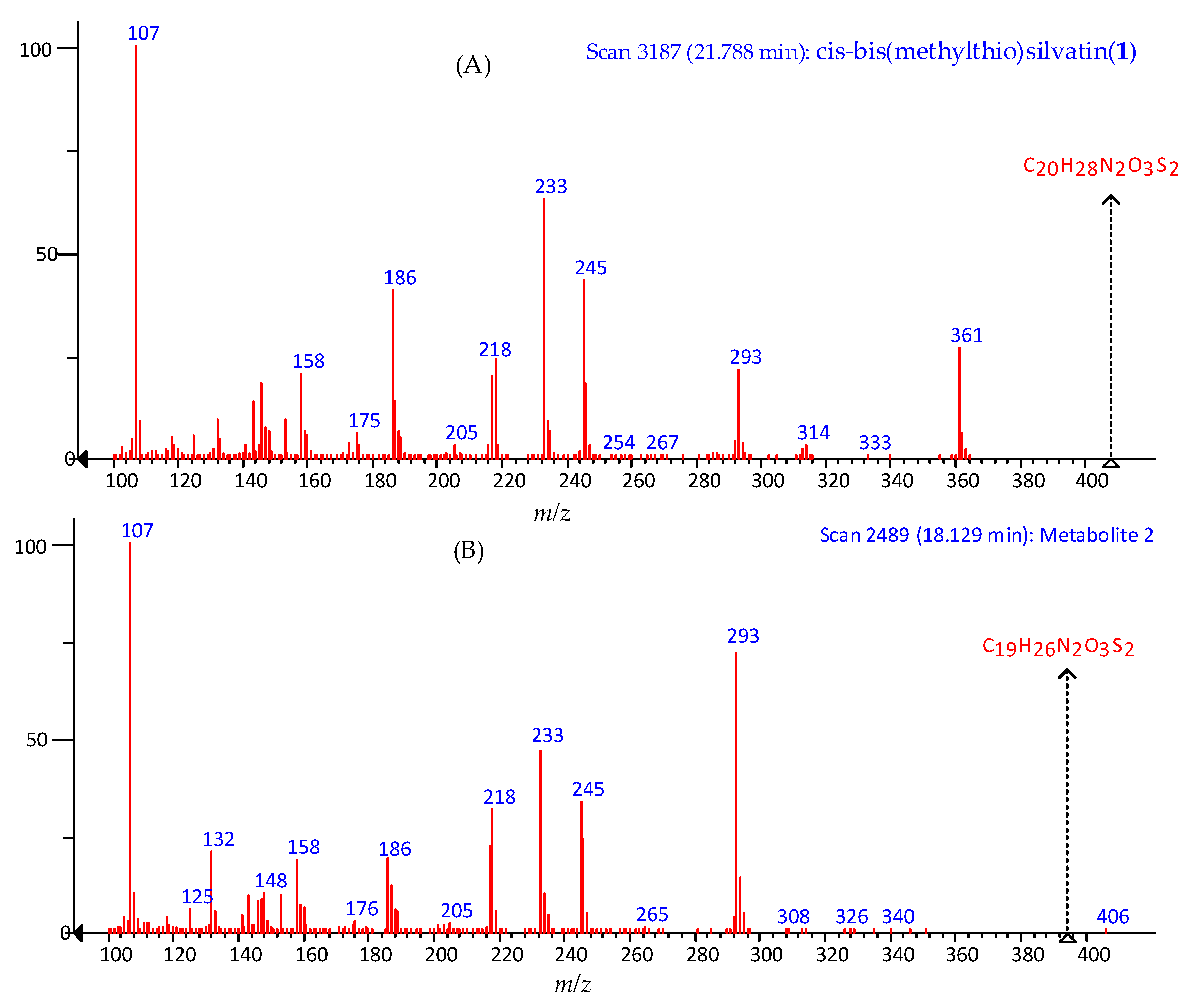
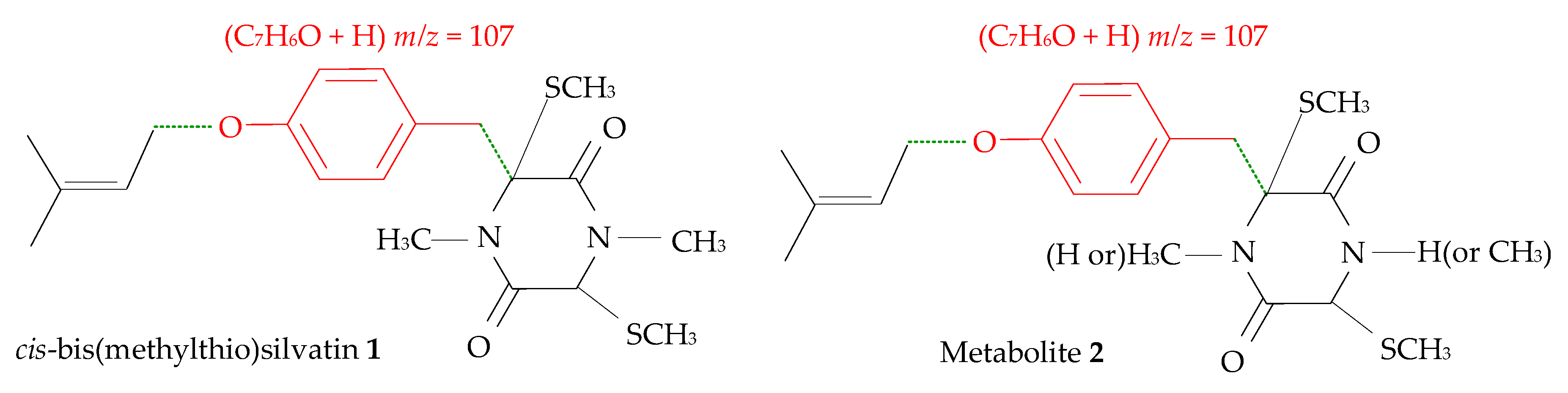
2.1. Profiling and Structural Elucidation of Secondary Metabolites from Penicillium brevicompactum AN4
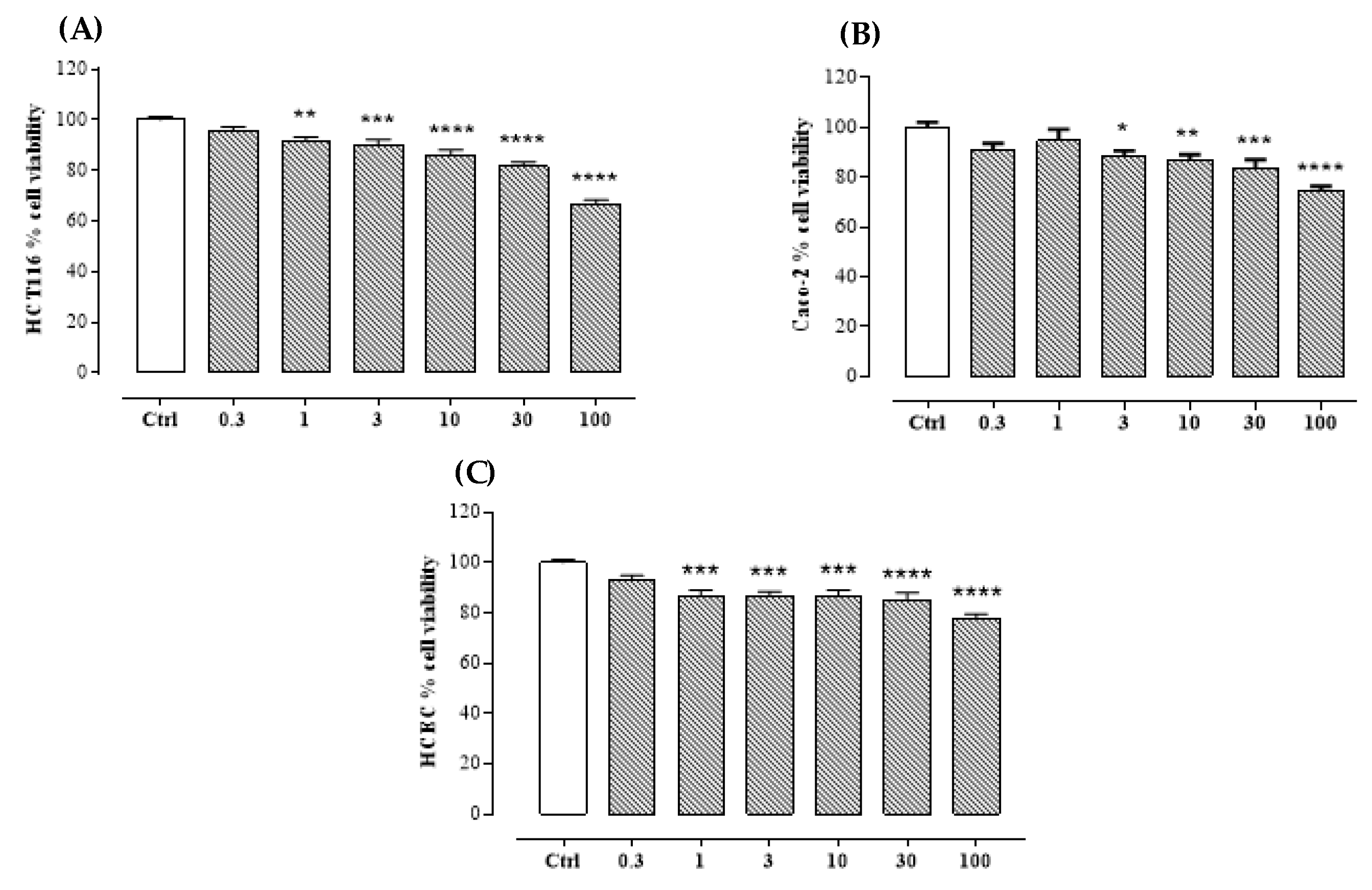
2.2. Bioassay
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Isolation and Identification of Strain AN4
4.3. Production and Fractionation of Culture Filtrates
4.4. LC-MS Analysis
4.5. GC-MS Analysis
4.6. Bioassay
4.6.1. Cell Lines
4.6.2. Cell Viability Assay
4.6.3. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Silber, J.; Kramer, A.; Labes, A.; Tasdemir, D. From discovery to production: Biotechnology of marine fungi for the production of new antibiotics. Mar. Drugs. 2016, 14, 137. [Google Scholar] [CrossRef] [PubMed]
- Nicoletti, R.; Andolfi, A. The marine-derived filamentous fungi in biotechnology. In Grand Challenges in Marine Biotechnology; Springer: Cham, Switzerland, 2018; pp. 157–189. [Google Scholar]
- Nicoletti, R.; Trincone, A. Bioactive compounds produced by strains of Penicillium and Talaromyces of marine origin. Mar. Drugs. 2016, 14, 37. [Google Scholar] [CrossRef] [PubMed]
- Koul, M.; Singh, S. Penicillium spp.: Prolific producer for harnessing cytotoxic secondary metabolites. Anti-Cancer Drugs. 2017, 28, 11–30. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Su, M.; Song, S.J.; Jung, J.H. Marine-derived Penicillium species as producers of cytotoxic metabolites. Mar. Drugs. 2017, 15, 329. [Google Scholar] [CrossRef]
- Nicoletti, R.; Vinale, F. Bioactive compounds from marine-derived Aspergillus, Penicillium, Talaromyces and Trichoderma species. Mar. Drugs. 2018, 16, 408. [Google Scholar] [CrossRef]
- Salvatore, M.M.; Nicoletti, R.; Salvatore, F.; Naviglio, D.; Andolfi, A. GC–MS approaches for the screening of metabolites produced by marine-derived. Aspergillus. Mar. Chem. 2018, 206, 19–33. [Google Scholar] [CrossRef]
- Marra, R.; Nicoletti, R.; Pagano, E.; DellaGreca, M.; Salvatore, M.M.; Borrelli, F.; Lombardi, N.; Vinale, F.; Woo, S.L.; Andolfi, A. Inhibitory effect of trichodermanone C, a sorbicillinoid produced by Trichoderma citrinoviride associated to the green alga Cladophora sp., on nitrite production in LPS-stimulated macrophages. Nat. Prod. Res. 2018, 1–9. [Google Scholar] [CrossRef]
- Hanson, J.R.; O’Leary, M.A. New piperazinedione metabolites of Gliocladium deliquescens. J. Chem. Soc. Perkin Trans. 1 1981, 218–220. [Google Scholar] [CrossRef]
- Lu, X.; Zheng, Z.; Zhang, H.; Huo, C.; Dong, Y.; Ma, Y.; Ren, X.; He, J.; Gu, Y.; Shi, Q. Two new members of mycophenolic acid family from Penicillium brevicompactum Dierckx. J. Antibiot. 2009, 62, 527. [Google Scholar] [CrossRef]
- Sparkman, O.D.; Penton, Z.E.; Kitson, F.G. Gas Chromatography and Mass Spectrometry: A Practical Guide, 2nd ed.; Elsevier Inc.: Burlington, MA, USA, 2011. [Google Scholar]
- Salvatore, M.M.; Nicoletti, R.; DellaGreca, M.; Andolfi, A. Occurrence and proprieties of thiosilvatins. Mar. Drugs. 2019, 17, 664. [Google Scholar] [CrossRef]
- NIST MS Interpreter. Mass Spectrum Interpreter Ver. 3.4. Available online: https://chemdata.nist.gov/dokuwiki/doku.php?id=chemdata:interpreter (accessed on 14 December 2019).
- Li, F.; Guo, W.; Wu, L.; Zhu, T.; Gu, Q.; Li, D.; Che, Q. Saroclazines A–C, thio-diketopiperazines from mangrove-derived fungi Sarocladium kiliense HDN11-84. Arch. Pharm. Res. 2018, 41, 30–34. [Google Scholar] [CrossRef] [PubMed]
- Andersen, B. Consistent production of phenolic compounds by Penicillium brevicompactum for chemotaxonomic characterization. Antonie Leeuwenhoek 1991, 60, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Frisvad, J.C.; Smedsgaard, J.; Larsen, T.O.; Samson, R.A. Mycotoxins, drugs and other extrolites produced by species in Penicillium subgenus. Penicillium. Stud. Mycol. 2004, 49, 201–241. [Google Scholar]
- Frisvad, J.C.; Houbraken, J.; Popma, S.; Samson, R.A. Two new Penicillium species Penicillium buchwaldii and Penicillium spathulatum, producing the anticancer compound asperphenamate. Fems Microbiol. Lett. 2013, 339, 77–92. [Google Scholar] [CrossRef]
- Ayer, W.A.; Altena, I.V.; Browne, L.M. Three piperazinediones and a drimane diterpenoid from Penicillium brevi-compactum. Phytochemistry 1990, 29, 1661–1665. [Google Scholar] [CrossRef]
- Capon, R.J.; Stewart, M.; Ratnayake, R.; Lacey, E.; Gill, J.H. Citromycetins and bilains A–C: New aromatic polyketides and diketopiperazines from Australian marine-derived and terrestrial Penicillium spp. J. Nat. Prod. 2007, 70, 1746–1752. [Google Scholar] [CrossRef]
- Ahmed, D.; Eide, P.W.; Eilertsen, I.A.; Danielsen, S.A.; Eknaes, M.; Hektoen, M.; Hektoen, M.; Lothe, R.A. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis 2013, 2, 71. [Google Scholar] [CrossRef]
- Berger, S.; Braun, S. 200 and More NMR Experiments; Weinheim: Wiley-Vch, NJ, USA, 2004; pp. 305–307. [Google Scholar]
- Frisvad, J.C.; Samson, R.A. Polyphasic taxonomy of Penicillium subgenus Penicillium. A guide to identification of food and air-borne terverticillate Penicillia and their mycotoxins. Stud. Mycol. 2004, 49, 1–174. [Google Scholar]
- Manganiello, G.; Marra, R.; Staropoli, A.; Lombardi, N.; Vinale, F.; Nicoletti, R. The shifting mycotoxin profiles of endophytic Fusarium strains: A case study. Agriculture 2019, 9, 143. [Google Scholar] [CrossRef]
- Vinale, F.; Arjona Girona, I.; Nigro, M.; Mazzei, P.; Piccolo, A.; Ruocco, M.; Woo, S.; Ruano Rosa, D.; López Herrera, C.; Lorito, M. Cerinolactone, a hydroxy-lactone derivative from Trichoderma cerinum. J. Nat. Prod. 2011, 75, 103–106. [Google Scholar] [CrossRef]
- Vinale, F.; Strakowska, J.; Mazzei, P.; Piccolo, A.; Marra, R.; Lombardi, N.; Manganiello, G.; Pascale, A.; Woo, S.L.; Lorito, M. Cremenolide, a new antifungal, 10-member lactone from Trichoderma cremeum with plant growth promotion activity. Nat. Prod. Res. 2016, 31, 2207. [Google Scholar] [CrossRef] [PubMed]
- Vinale, F.; Nicoletti, R.; Lacatena, F.; Marra, R.; Sacco, A.; Lombardi, N.; d’Errico, G.; Digilio, M.C.; Woo, S.L. Secondary metabolites from the endophytic fungus Talaromyces pinophilus. Nat. Prod. Res. 2017, 31, 1778–1785. [Google Scholar] [CrossRef] [PubMed]
- Guida, M.; Salvatore, M.M.; Salvatore, F. A strategy for GC/MS quantification of polar compounds via their silylated surrogates: Silylation and quantification of biological amino acids. J. Anal. Bioanal. Tech. 2015, 6, 263–279. [Google Scholar]
- NIST Standard Reference Data. Available online: http://www.nist.gov/srd/nist1a.cfm (accessed on 14 December 2018).


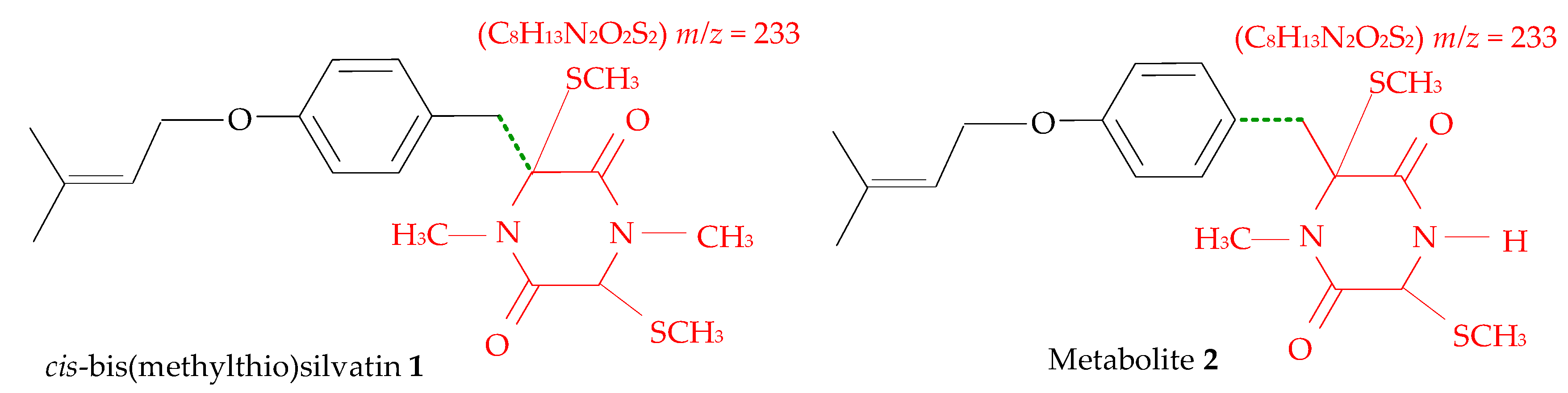
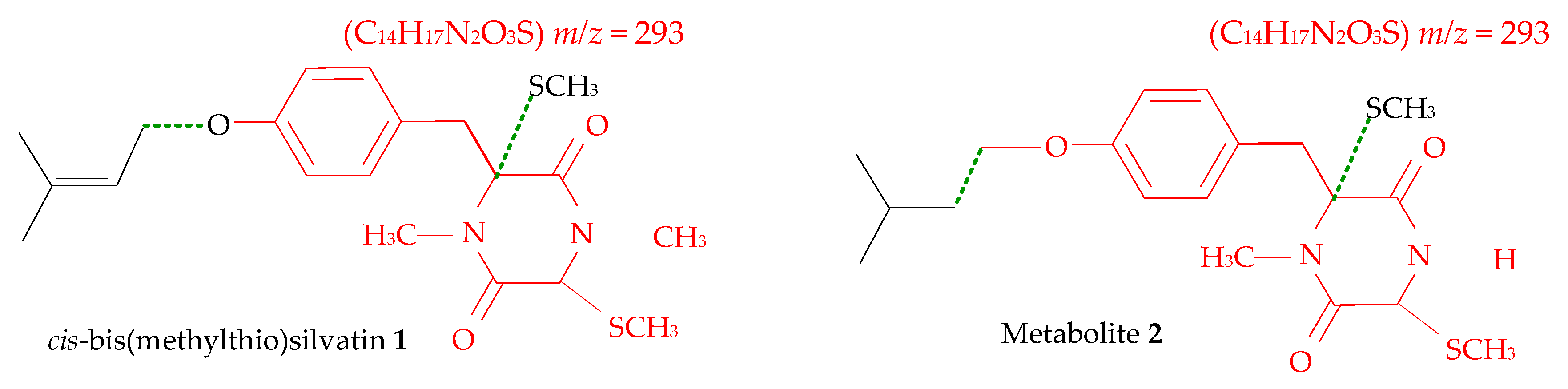
| Code | Compound | GC-EI-MS Data and RI | Formula | Accurate Mass (u) | RDB |
|---|---|---|---|---|---|
| 1 | cis-bis(methylthio)silvatin | Compound (RI = 3229) | C20H28N2O3S2 | 408.1538 | 8 |
| 2 | Metabolite 2 | Compound (RI = 2892) | C19H26N2O3S2 | 394.1388 | 8 |
| 3 | Metabolite 3 | Compound (RI = 2083) | C19H24N2O3S | 360.1510 | 9 |
| 4 | Metabolite 4 | Compound (RI = 3203) | C19H24N2O4S | 376.1462 | 9 |
| 5 | Metabolite 5 | TMS derivative (RI = 3047) | C20H30N2O5S2 | 442.1593 | 7 |
| 6 | Metabolite 6 | TMS derivative (RI = 2014) | C15H20N2O3S2 | 340.0919 | 7 |
| 7 | Metabolite 7 | TMS derivative (RI = 1876) | C13H16N2O3S2 | 312.0600 | 7 |
| 8 | mycophenolic acid | Compound (RI = 2871) | C17H20O6 | 320.1268 | 8 |
| 9 | Metabolite 9 | TMS derivative (RI = 3227) | C21H23N3O3 | 365.1725 | 12 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vinale, F.; Salvatore, M.M.; Nicoletti, R.; Staropoli, A.; Manganiello, G.; Venneri, T.; Borrelli, F.; DellaGreca, M.; Salvatore, F.; Andolfi, A. Identification of the Main Metabolites of a Marine-Derived Strain of Penicillium brevicompactum Using LC and GC MS Techniques. Metabolites 2020, 10, 55. https://doi.org/10.3390/metabo10020055
Vinale F, Salvatore MM, Nicoletti R, Staropoli A, Manganiello G, Venneri T, Borrelli F, DellaGreca M, Salvatore F, Andolfi A. Identification of the Main Metabolites of a Marine-Derived Strain of Penicillium brevicompactum Using LC and GC MS Techniques. Metabolites. 2020; 10(2):55. https://doi.org/10.3390/metabo10020055
Chicago/Turabian StyleVinale, Francesco, Maria Michela Salvatore, Rosario Nicoletti, Alessia Staropoli, Gelsomina Manganiello, Tommaso Venneri, Francesca Borrelli, Marina DellaGreca, Francesco Salvatore, and Anna Andolfi. 2020. "Identification of the Main Metabolites of a Marine-Derived Strain of Penicillium brevicompactum Using LC and GC MS Techniques" Metabolites 10, no. 2: 55. https://doi.org/10.3390/metabo10020055
APA StyleVinale, F., Salvatore, M. M., Nicoletti, R., Staropoli, A., Manganiello, G., Venneri, T., Borrelli, F., DellaGreca, M., Salvatore, F., & Andolfi, A. (2020). Identification of the Main Metabolites of a Marine-Derived Strain of Penicillium brevicompactum Using LC and GC MS Techniques. Metabolites, 10(2), 55. https://doi.org/10.3390/metabo10020055











