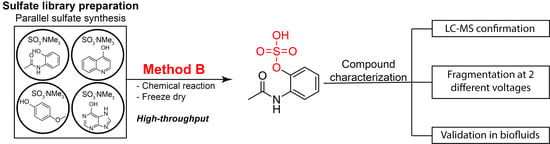
Rapid Preparation of a Large Sulfated Metabolite Library for Structure Validation in Human Samples
Abstract
1. Introduction
2. Results and Discussion
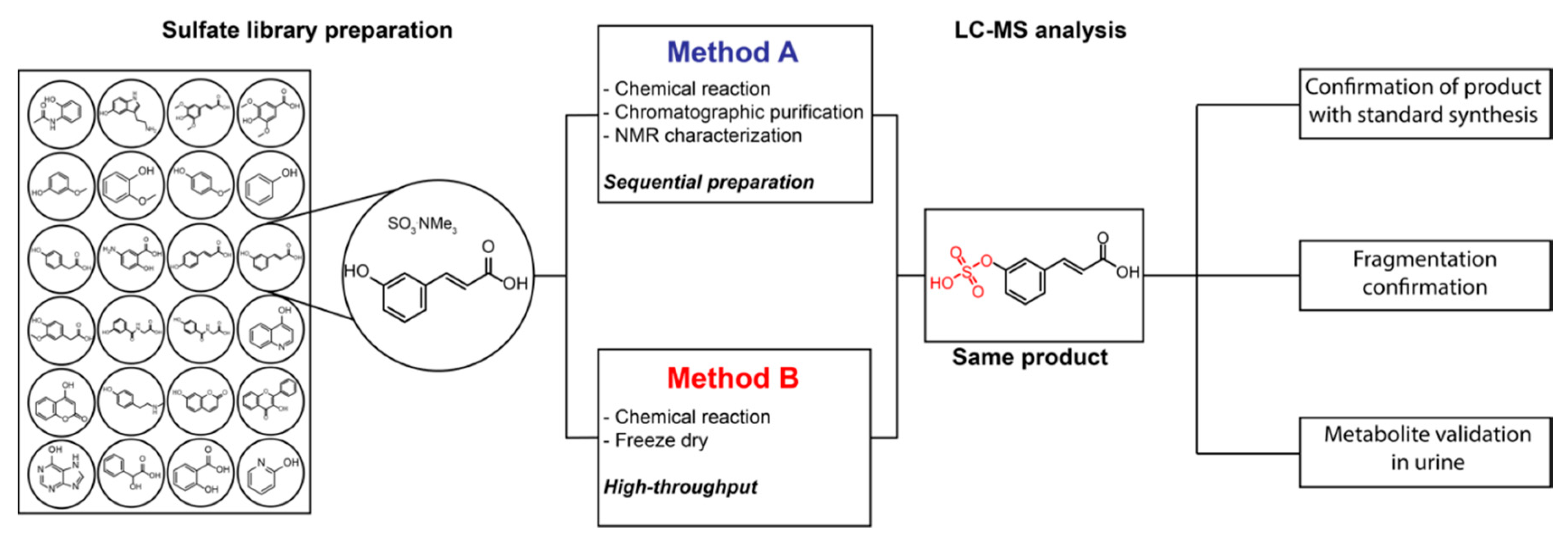
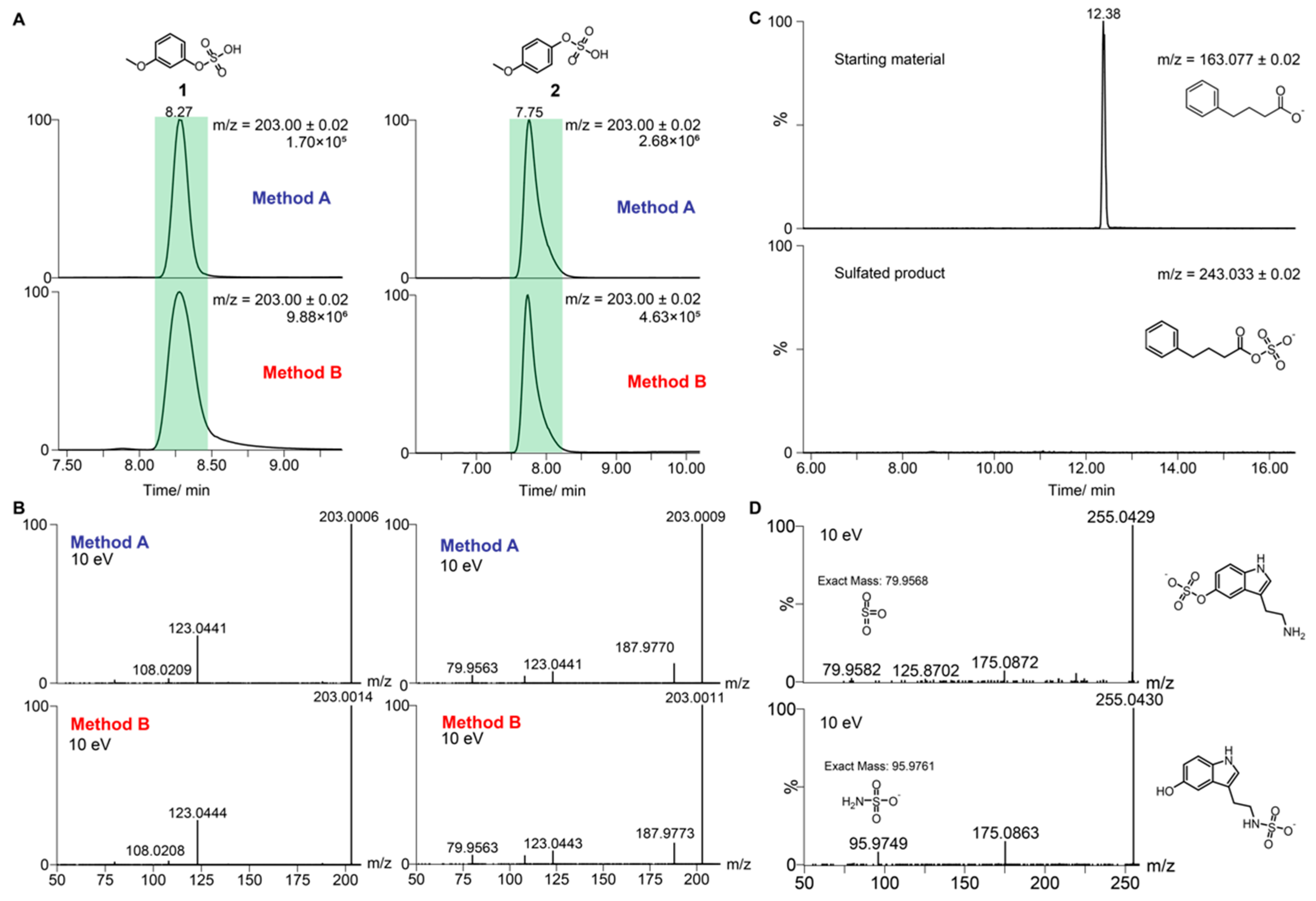
2.1. Validation of Standard Preparation Procedure
2.2. Construction of the Sulfate Library
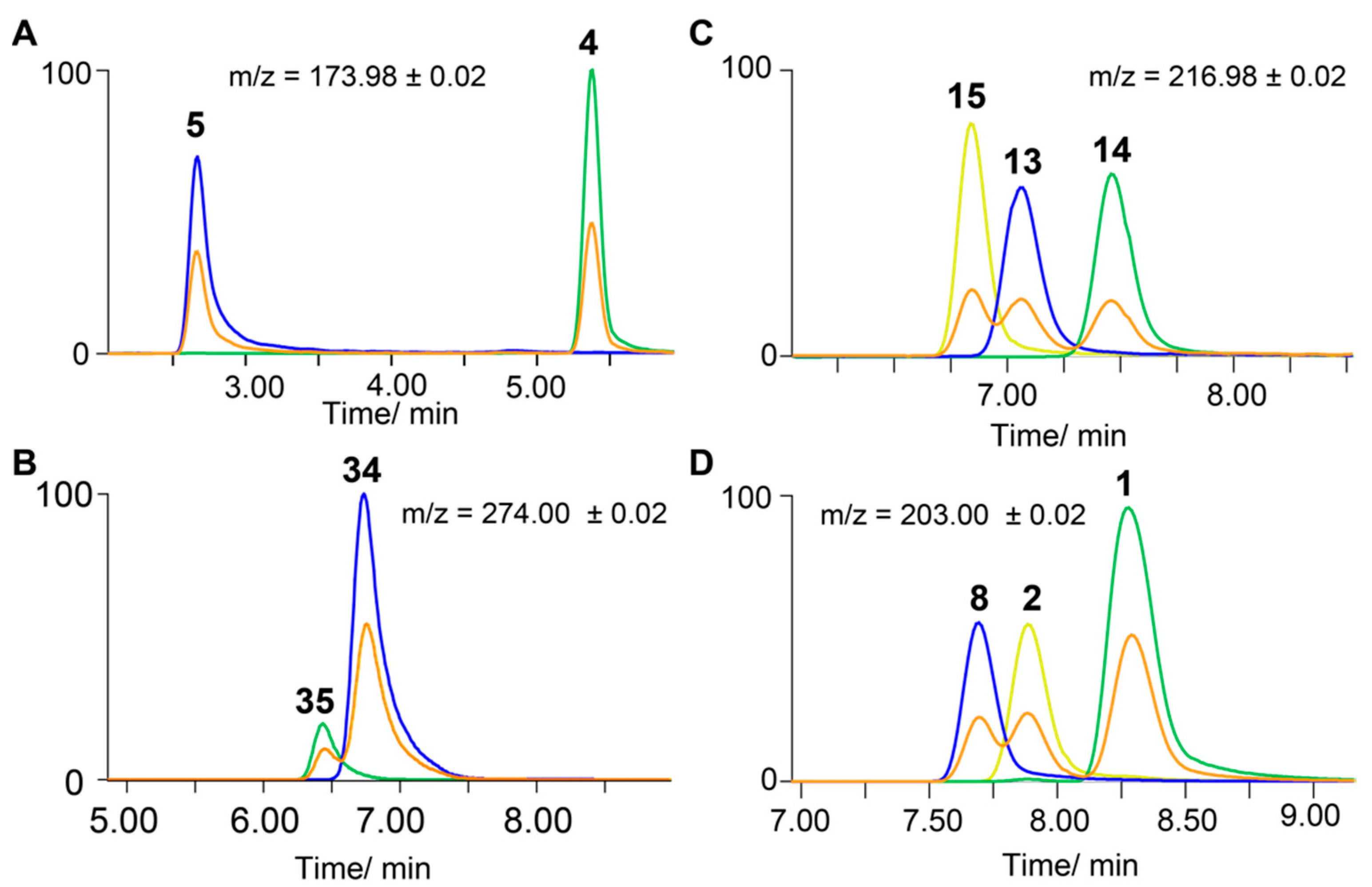
2.3. Separation of Regioisomers
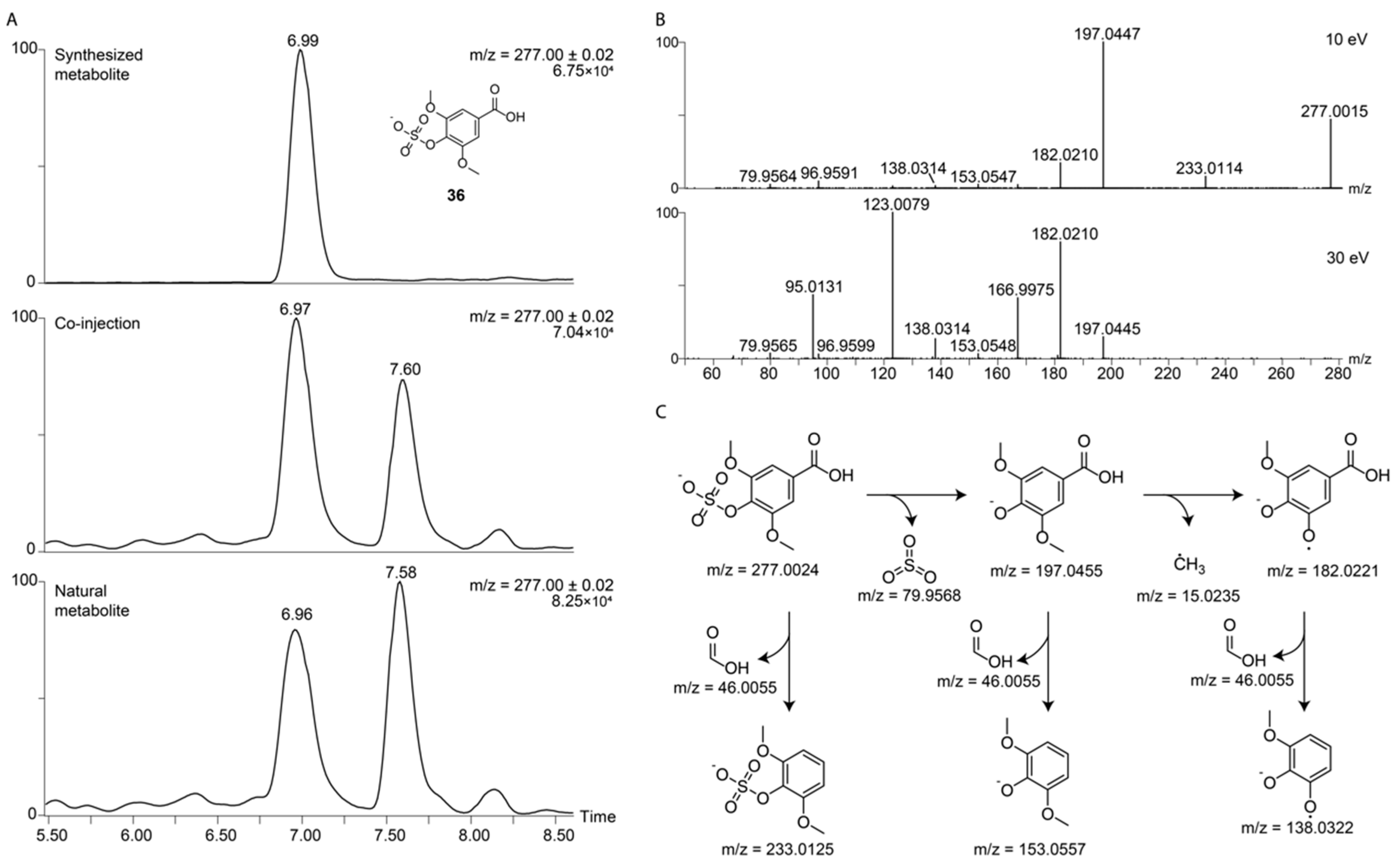
2.4. Metabolite Library and Significance
3. Materials and Methods
3.1. Materials and Equipment
3.2. Human Samples
3.3. Chemical Synthesis: Method A
3.4. Preparation of Sulfates: Method B
3.5. UPLC-MS/MS Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kuehnbaum, N.L.; Britz-McKibbin, P. New advances in separation science for metabolomics: Resolving chemical diversity in a post-genomic era. Chem. Rev. 2013, 113, 2437–2468. [Google Scholar] [CrossRef]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. Mass spectrometry-based metabolomics. Mass Spectrom. Rev. 2007, 26, 51–78. [Google Scholar] [CrossRef] [PubMed]
- Smirnov, K.S.; Maier, T.V.; Walker, A.; Heinzmann, S.S.; Forcisi, S.; Martinez, I.; Walter, J.; Schmitt-Kopplin, P. Challenges of metabolomics in human gut microbiota research. Int. J. Med. Microbiol. 2016, 306, 266–279. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, W.J.; Koal, T.; Wang, Y.; Kohl, M.; Enot, D.P.; Deigner, H.-P. Targeted metabolomics for biomarker discovery. Angew. Chem. Int. Ed. 2010, 49, 5426–5445. [Google Scholar] [CrossRef]
- Khamis, M.M.; Adamko, D.J.; El-Aneed, A. Mass spectrometric based approaches in urine metabolomics and biomarker discovery. Mass Spectrom. Rev. 2017, 36, 115–134. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Vargas, F.; Weldon, K.C.; Sikora, N.; Wang, M.; Zhang, Z.; Gentry, E.C.; Panitchpakdi, M.W.; Caraballo-Rodriguez, A.M.; Dorrestein, P.C.; Jarmusch, A.K. Protocol for community-created public ms/ms reference spectra within the global natural products social molecular networking infrastructure. Rapid Commun. Mass Spectrom. 2020, 34, e8725. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. Hmdb 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef]
- Guijas, C.; Montenegro-Burke, J.R.; Domingo-Almenara, X.; Palermo, A.; Warth, B.; Hermann, G.; Koellensperger, G.; Huan, T.; Uritboonthai, W.; Aisporna, A.E.; et al. Metlin: A technology platform for identifying knowns and unknowns. Anal. Chem. 2018, 90, 3156–3164. [Google Scholar] [CrossRef]
- Simon-Manso, Y.; Marupaka, R.; Yan, X.; Liang, Y.; Telu, K.H.; Mirokhin, Y.; Stein, S.E. Mass spectrometry fingerprints of small-molecule metabolites in biofluids: Building a spectral library of recurrent spectra for urine analysis. Anal. Chem. 2019, 91, 12021–12029. [Google Scholar] [CrossRef]
- Kind, T.; Tsugawa, H.; Cajka, T.; Ma, Y.; Lai, Z.; Mehta, S.S.; Wohlgemuth, G.; Barupal, D.K.; Showalter, M.R.; Arita, M.; et al. Identification of small molecules using accurate mass ms/ms search. Mass Spectrom. Rev. 2018, 37, 513–532. [Google Scholar] [CrossRef]
- Duhrkop, K.; Fleischauer, M.; Ludwig, M.; Aksenov, A.A.; Melnik, A.V.; Meusel, M.; Dorrestein, P.C.; Rousu, J.; Bocker, S. Sirius 4: A rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 2019, 16, 299–302. [Google Scholar] [CrossRef] [PubMed]
- da Silva, R.R.; Dorrestein, P.C.; Quinn, R.A. Illuminating the dark matter in metabolomics. Proc. Natl. Acad. Sci. USA 2015, 112, 12549–12550. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Hayashi, H.; Chong Teo, S. Chemical toolbox to decode the microbiota lexicon. Chem. Asian J. 2020, 15, 2117–2128. [Google Scholar] [CrossRef] [PubMed]
- Schymanski, E.L.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.P.; Hollender, J. Identifying small molecules via high resolution mass spectrometry: Communicating confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef]
- Correia, M.S.P.; Rao, M.; Ballet, C.; Globisch, D. Coupled enzymatic treatment and mass spectrometric analysis for identification of glucuronidated metabolites in human samples. ChemBioChem 2019, 20, 1678–1683. [Google Scholar] [CrossRef]
- Ciejka, M.; Nguyen, K.; Bluth, M.H.; Dubey, E. Drug toxicities of common analgesic medications in the emergency department. Clin. Lab. Med. 2016, 36, 761–776. [Google Scholar] [CrossRef]
- Wang, S.; Bilal, M.; Hu, H.; Wang, W.; Zhang, X. 4-hydroxybenzoic acid—a versatile platform intermediate for value-added compounds. Appl. Microbiol. Biotechnol. 2018, 102, 3561–3571. [Google Scholar] [CrossRef]
- Ballet, C.; Correia, M.S.P.; Conway, L.P.; Locher, T.L.; Lehmann, L.C.; Garg, N.; Vujasinovic, M.; Deindl, S.; Lohr, J.M.; Globisch, D. New enzymatic and mass spectrometric methodology for the selective investigation of gut microbiota-derived metabolites. Chem. Sci. 2018, 9, 6233–6239. [Google Scholar] [CrossRef]
- Garg, N.; Conway, L.P.; Ballet, C.; Correia, M.S.P.; Olsson, F.K.S.; Vujasinovic, M.; Lohr, J.M.; Globisch, D. Chemoselective probe containing a unique bioorthogonal cleavage site for investigation of gut microbiota metabolism. Angew. Chem. Int. Ed. 2018, 57, 13805–13809. [Google Scholar] [CrossRef]
- Conway, L.P.; Garg, N.; Lin, W.; Vujasinovic, M.; Lohr, J.M.; Globisch, D. Chemoselective probe for detailed analysis of ketones and aldehydes produced by gut microbiota in human samples. Chem. Commun. 2019, 55, 9080–9083. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Conway, L.P.; Block, A.; Sommi, G.; Vujasinovic, M.; Lohr, J.M.; Globisch, D. Sensitive mass spectrometric analysis of carbonyl metabolites in human urine and fecal samples using chemoselective modification. Analyst 2020, 145, 3822–3831. [Google Scholar] [CrossRef] [PubMed]
- Wikoff, W.R.; Anfora, A.T.; Liu, J.; Schultz, P.G.; Lesley, S.A.; Peters, E.C.; Siuzdak, G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA 2009, 106, 3698–3703. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Huang, J.; Chen, Z.; Jiang, Z.; Li, X.; Chen, Z. Metabolomics in gut microbiota: Applications and challenges. Sci. Bull. 2016, 61, 1151–1153. [Google Scholar] [CrossRef]
- Johnson, C.H.; Patterson, A.D.; Idle, J.R.; Gonzalez, F.J. Xenobiotic metabolomics: Major impact on the metabolome. Annu. Rev. Pharmacol. Toxicol. 2012, 52, 37–56. [Google Scholar] [CrossRef]
- Clarke, G.; Sandhu, K.V.; Griffin, B.T.; Dinan, T.G.; Cryan, J.F.; Hyland, N.P. Gut reactions: Breaking down xenobiotic-microbiome interactions. Pharmacol. Rev. 2019, 71, 198–224. [Google Scholar] [CrossRef]
- Correia, M.S.P.; Jain, A.; Alotaibi, W.; Young Tie Yang, P.; Rodriguez-Mateos, A.; Globisch, D. Comparative dietary sulfated metabolome analysis reveals unknown metabolic interactions of the gut microbiome and the human host. Free Radic. Biol. Med. 2020, 160, 745–754. [Google Scholar] [CrossRef]
- Al-Horani, R.A.; Desai, U.R. Chemical sulfation of small molecules - advances and challenges. Tetrahedron 2010, 66, 2907–2918. [Google Scholar] [CrossRef]
- Simpson, L.S.; Widlanski, T.S. A comprehensive approach to the synthesis of sulfate esters. J. Am. Chem. Soc. 2006, 128, 1605–1610. [Google Scholar] [CrossRef]
- Globisch, D.; Moreno, A.Y.; Hixon, M.S.; Nunes, A.A.K.; Denery, J.R.; Specht, S.; Hoerauf, A.; Janda, K.D. Onchocerca volvulus-neurotransmitter tyramine is a biomarker for river blindness. Proc. Natl. Acad. Sci. USA 2013, 110, 4218–4223. [Google Scholar] [CrossRef]
- Garg, N.; Hansson, A.; Knych, H.K.; Stanley, S.D.; Thevis, M.; Bondesson, U.; Hedeland, M.; Globisch, D. Structural elucidation of major selective androgen receptor modulator (sarm) metabolites for doping control. Org. Biomol. Chem. 2018, 16, 698–702. [Google Scholar] [CrossRef]
- Yi, L.; Dratter, J.; Wang, C.; Tunge, J.A.; Desaire, H. Identification of sulfation sites of metabolites and prediction of the compounds’ biological effects. Anal. Bioanal. Chem. 2006, 386, 666–674. [Google Scholar] [CrossRef] [PubMed]
- Correia, M.S.P.; Ballet, C.; Meistermann, H.; Conway, L.P.; Globisch, D. Comprehensive kinetic and substrate specificity analysis of an arylsulfatase from helix pomatia using mass spectrometry. Bioorg. Med. Chem. 2019, 27, 955–962. [Google Scholar] [CrossRef] [PubMed]
- Witting, M.; Bocker, S. Current status of retention time prediction in metabolite identification. J. Sep. Sci. 2020, 43, 1746–1754. [Google Scholar] [CrossRef] [PubMed]
- Van Rymenant, E.; Grootaert, C.; Beerens, K.; Needs, P.W.; Kroon, P.A.; Kerimi, A.; Williamson, G.; García-Villalba, R.; González-Sarrías, A.; Tomas-Barberan, F.; et al. Vasorelaxant activity of twenty-one physiologically relevant (poly)phenolic metabolites on isolated mouse arteries. Food Funct. 2017, 8, 4331–4335. [Google Scholar] [CrossRef]
- Pallister, T.; Jackson, M.A.; Martin, T.C.; Zierer, J.; Jennings, A.; Mohney, R.P.; MacGregor, A.; Steves, C.J.; Cassidy, A.; Spector, T.D.; et al. Hippurate as a metabolomic marker of gut microbiome diversity: Modulation by diet and relationship to metabolic syndrome. Sci. Rep. 2017, 7, 13670. [Google Scholar] [CrossRef]
- van den Brand, J.A.J.G.; Mutsaers, H.A.M.; van Zuilen, A.D.; Blankestijn, P.J.; van den Broek, P.H.; Russel, F.G.M.; Masereeuw, R.; Wetzels, J.F.M. Uremic solutes in chronic kidney disease and their role in progression. PLoS ONE 2016, 11, e0168117. [Google Scholar] [CrossRef]
- Kaiser, J.P.; Feng, Y.; Bollag, J.M. Microbial metabolism of pyridine, quinoline, acridine, and their derivatives under aerobic and anaerobic conditions. Microbiol. Rev. 1996, 60, 483–498. [Google Scholar] [CrossRef]
- D’arrigo, P.; Pedrocchi-Fantoni, G.; Servi, S. Old and new synthetic capacities of baker’s yeast. Adv. Appl. Microbiol. 1997, 44, 81–123. [Google Scholar] [PubMed]
- Zorn, H.; Fischer-Zorn, M.; Berger, R.G. A labeling study to elucidate the biosynthesis of 4-(4-hydroxyphenyl)-butan-2-one (raspberry ketone) by nidula niveo-tomentosa. Appl. Environ. Microbiol. 2003, 69, 367. [Google Scholar] [CrossRef]
- Rechner, A.R.; Spencer, J.P.E.; Kuhnle, G.; Hahn, U.; Rice-Evans, C.A. Novel biomarkers of the metabolism of caffeic acid derivatives in vivo. Free Radic. Biol. Med. 2001, 30, 1213–1222. [Google Scholar] [CrossRef]
- Meyer, F.; Netzer, J.; Meinert, C.; Voigt, B.; Riedel, K.; Steinbüchel, A. A proteomic analysis of ferulic acid metabolism in amycolatopsis sp. Atcc 39116. Appl. Microbiol. Biotechnol. 2018, 102, 6119–6142. [Google Scholar] [CrossRef]
- Kimura, T.; Iwasaki, N.; Yokoe, J.I.; Haruta, S.; Yokoo, Y.; Ogawara, K.I.; Higaki, K. Analysis and prediction of absorption profile including hepatic first-pass metabolism of n-methyltyramine, a potent stimulant of gastrin release present in beer, after oral ingestion in rats by gastrointestinal-transit-absorption model. Drug Metab. Dispos. 2000, 28, 577–581. [Google Scholar]
- Xiong, X.; Liu, D.; Wang, Y.; Zeng, T.; Peng, Y. Urinary 3-(3-hydroxyphenyl)-3-hydroxypropionic acid, 3-hydroxyphenylacetic acid, and 3-hydroxyhippuric acid are elevated in children with autism spectrum disorders. BioMed. Res. Int. 2016, 2016, 9485412. [Google Scholar] [CrossRef] [PubMed]
- Whiting, M.J.; Doogue, M.P. Advances in biochemical screening for phaeochromocytoma using biogenic amines. Clin. Biochem. Rev. 2009, 30, 3–17. [Google Scholar] [PubMed]
- Born, S.L.; Caudill, D.; Fliter, K.L.; Purdon, M.P. Identification of the cytochromes p450 that catalyze coumarin 3,4-epoxidation and 3-hydroxylation. Drug Metab. Dispos. 2002, 30, 483. [Google Scholar] [CrossRef] [PubMed]
- Markus, B.; Kwon, C.H. In vitro metabolism of aromatic nitriles. J. Pharm. Sci. 1994, 83, 1729–1734. [Google Scholar] [CrossRef]
| Metabolite | Chemical Formula | m/z | Retention Time/min | Fragmentation m/z (% of Maximum Intensity) |
|---|---|---|---|---|
| Phenol sulfate (3) | C6H5O4S− | 172.9914 | 7.38 | 10 eV—172.9893 (100), 93.0324 (38), 79.9551 (4) 30 eV—93.0337 (100), 79.9563 (6) |
| 2-Hydroxypyridine sulfate (4) | C5H4NO4S− | 173.9867 | 5.15 | 10 eV—173.9857 (100), 94.0289 (89) 30 eV—94.0288 (100) |
| 3-Hydroxypyridine sulfate (5) | C5H4NO4S− | 173.9867 | 2.53 | 10 eV—173.9860 (100), 94.0289 (53) 30 eV—94.0288 (100) |
| Benzyl alcohol sulfate (6) | C7H7O4S− | 187.0071 | 8.21 | 10 eV—187.0061 (100), 95.9514 (6) 30 eV - 95.9511 (100), 187.0056 (11), 79.9562 (10), 80.9639 (8), 77.0384 (7) |
| 4-Cyanophenol sulfate (7) | C7H4NO4S- | 197.9867 | 7.54 | 10 eV—118.0290 (100), 197.9854 (55) 30 eV—118.0287 (100), 90.0340 (4) |
| 2-Methoxyphenol sulfate (8) | C7H7O5S− | 203.0020 | 7.67 | 10 eV—203.0009 (100), 123.0445 (30), 108.0207 (6), 79.9561 (5) 30 eV—108.0205 (100), 123.0442 (15), 79.9562 (9) |
| 3-Methoxyphenol sulfate (1) | C7H7O5S− | 203.0020 | 8.51 | 10 eV—203.0015 (100), 123.0444 (28) 30 eV—108.0213 (100), 123.0444 (59), 79.9564 (5) |
| 4-Methoxyphenol sulfate (2) | C7H7O5S− | 203.0020 | 8.15 | 10 eV—203.0011 (100), 187.9773 (13), 123.0443 (8), 108.0208 (5), 79.9563 (5) 30 eV—108.0212 (100), 79.9564 (19), 123.0442 (4) |
| 3-Hydroxy-2-methyl-4-pyrone sulfate (9) | C6H5O6S− | 204.9812 | 5.30 | 10 eV—204.9790 (100), 125.0223 (50), 80.9630 (4) 30 eV—125.0236 (100), 97.0285 (62), 79.9561 (38), 69.0330 (13), 80.9640 (12) |
| 4,5-Dimethyl-3-hydroxy-2,5-dihydrofuran-2-one sulfate (10) | C6H7O6S− | 206.9969 | 7.28 | 10 eV—206.9946 (100), 127.0381 (24), 135.0099 (8) 30 eV—127.0391 (100), 135.0110 (36), 79.9562 (36), 80.9641 (28), 99.0443 (21) |
| 2-Hydroxyacetophenone sulfate (11) | C8H7O5S− | 215.0020 | 8.28 | 10 eV—215.0006 (100), 135.0441 (99) 30 eV—135.0438 (100), 93.0331 (44) |
| Hypoxanthine sulfate (12) | C5H3N4O4S− | 215.9953 | 2.58 | 10 eV—136.0325 (100), 215.9891 (99), 135.0298 (13) 30 eV—136.0331 (100), 93.0268 (41), 92.0240 (16), 135.0310 (13), 109.0228 (4) |
| 2-Hydroxybenzoic acid sulfate (13) | C7H5O6S− | 216.9812 | 7.00 | 10 eV—137.0223 (100), 93.0325 (15), 216.9792 (15); |
| 3-Hydroxybenzoic acid sulfate (14) | C7H5O6S− | 216.9812 | 7.19 | 10 eV—216.9789 (100), 137.0223 (49), 93.0326 (5) 30 eV—93.0337 (100), 137.0232 (44) |
| 4-Hydroxybenzoic acid sulfate (15) | C7H5O6S− | 216.9812 | 6.80 | 10 eV—216.9801 (100), 137.0225 (87), 93.0336 (12), 172.9905 (8), 96.9591 (5) 30 eV—93.0337 (100), 137.0235 (27) |
| 4-Hydroxyquinoline sulfate (16) | C9H9NO4S− | 224.0023 | 8.09 | 10 eV—224.000 (100), 144.0433 (99) 30 eV—144.0447 (100) |
| 4-Acetamidophenol sulfate (17) | C8H8NO5S− | 230.0129 | 5.65 | 10 eV—230.0120 (100), 150.0556 (42) 30 eV—150.0552 (100), 107.0368 (53), 108.0448 (5) |
| N-Methyltyramine sulfate (18) | C9H12NO4S− | 230.0493 | 7.56 | 10 eV—230.0467 (100), 109.9893 (8) 30 eV—79.9561 (100), 109.9906 (21), 150.0916 (8), 80.9639 (5) |
| 4-Hydroxyphenylacetic acid sulfate (19) | C8H7O6S− | 230.9969 | 7.09 | 10 eV—230.9966 (100), 151.0398 (31) 30 eV—107.0497 (100),151.0393 (48), 105.0337 (7), 79.9564 (6) |
| Mandelic acid sulfate (20) | C8H7O6S− | 230.9969 | 7.33 | 10 eV—230.9959 (100), 96.9591 (75), 151.0391 (34) 30 eV—96.9590 (100), 151.0390 (13), 107.0492 (8) |
| 3-Hydroxyphenylacetic acid sulfate (21) | C8H7O6S− | 230.9969 | 7.18 | 10 eV—230.9948 (100), 187.0047 (70), 107.0481 (22), 79.9553 (5) 30 eV—107.0492 (100), 79.9564 (16) |
| Vanillin sulfate (22) | C8H7O6S− | 230.9969 | 7.42 | 10 eV—151.0392 (100), 230.9958 (44), 136.0157 (23) 30 eV—136.0157 (100), 92.0259 (15). 151.0391 (13), 108.0206 (12) |
| 5-Aminosalicylic acid sulfate (23) | C7H6NO6S− | 231.9921 | 5.98 | 10 eV—231.9896 (100), 152.0331 (85), 150.0174 (57), 213.9791 (49), 106.0280 (17) 30 eV—79.9565 (100), 106.0289 (48), 78.0341 (40), 80.9642 (38), 108.0447 (35) |
| 4-Hydroxycoumarin sulfate (24) | C6H5O6S− | 240.9812 | 9.36 | 10 eV—161.0228 (100), 204.9792 (15), 117.0323 (6) 30 eV—117.0338 (100), 161.0236 (55) |
| Umbelliferone sulfate (25) | C6H5O6S− | 240.9812 | 7.89 | 10 eV—161.0226 (100), 240.9791 (40) 30 eV—161.0237 (100), 133.0286 (21), 105.0337 (7), 77.0385 (5), 89.0385 (4) |
| trans-3-Hydroxycinnamic acid sulfate (26) | C6H7O6S− | 242.9969 | 8.48 | 10 eV—242.9962 (100), 163.0393 (54) 30 eV—119.0497 (100), 163.0395 (67) |
| trans-4-Hydroxycinnamic acid sulfate (27) | C6H7O6S− | 242.9969 | 7.99 | 10 eV—163.0379 (100), 242.9947 (64), 119.0481 (16) 30 eV—119.0495 (100), 163.0393 (10) |
| Raspberry ketone sulfate (28) | C10H11O5S− | 243.0333 | 8.34 | 10 eV—243.0327 (100), 163.0750 (13) 30 eV—163.0755 (100), 57.0329 (81), 79.9561 (45), 80.9639 (4) |
| D-3-Phenyllactic acid sulfate (29) | C9H9O6S− | 245.0125 | 10.20 | 10 eV—245.011 (100), 96.9578 (59), 165.0534 (21), 244.9520 (6) 30 eV—96.9591 (100), 147.0442 (43), 103.0542 (20), 165.0549 (16), 128.9950 (7) |
| 3-(3-Hydroxyphenyl)propionic acid sulfate (30) | C9H9O6S− | 245.0125 | 8.08 | 10 eV—245.0101 (100), 96.9578 (58), 165.0534 (20) 30 eV—96.9591 (100), 147.0443 (42), 103.0542 (19), 165.0552 (18), 119.0493 (15) |
| 5-Methoxysalicylic acid sulfate (31) | C8H7O7S− | 246.9918 | 7.58 | 10 eV—167.0325 (100), 152.0090 (16), 246.9891 (15), 108.0195 (11) 30 eV—108.0207 (100), 152.0104 (27), 167.0341 (7) |
| Serotonin sulfate (32) | C10H11N2O4S− | 255.0445 | 5.92 | 10 eV—255.0428 (100), 175.0871 (7), 254.9162 (6), 219.8448 (5) |
| 4-Hydroxy-3-methoxyphenylacetic acid sulfate (33) | C9H9O7S− | 261.0074 | 7.46 | 10 eV—261.0052 (100), 181.0484 (32), 217.0153 (9) 30 eV—137.0602 (100), 122.0366 (80), 181.0496 (23), 79.9563 (10), 105.0337 (6) |
| 3-Hydroxyhippuric acid sulfate (34) | C9H8NO7S− | 274.0027 | 6.73 | 10 eV—274.0007 (100), 194.0435 (17), 150.0539 (7) 30 eV—150.0551 (100), 93.0335 (38), 194.0448 (16) |
| 4-Hydroxyhippuric acid sulfate (35) | C9H8NO7S− | 274.0027 | 6.43 | 10 eV—274.0005 (100), 194.0435 (68), 100.0017 (9) 30 eV—93.0335 (100), 100.0029 (97), 74.0233 (39), 150.0550 (19), 194.0448 (10) |
| Syringic acid sulfate (36) | C9H9O8S− | 277.0024 | 6.99 | 10 eV—197.0447 (100), 277.0015 (47), 182.0210 (17), 233.0114 (8), 96.9591 (5) 30 eV—123.0079 (100), 182.0211 (79), 95.0131 (43), 166.9975 (41), 197.0445 (14) |
| Sinapic acid sulfate (37) | C11H11O8S− | 303.0180 | 8.11 | 10 eV—223.0601 (100), 303.0169 (39), 208.0366 (12), 259.0269 (5) 30 eV—193.0134 (100), 121.0287 (71), 149.0236 (64), 208.0368 (61), 164.0471 (51) |
| 3-Hydroxyflavone sulfate (38) | C15H9O6S− | 317.0125 | 12.8 | 10 eV—237.0574 (100), 317.0130 (58) 30 eV—237.0554 (100), 181.0655 (83), 180.0574 (71), 209.060 (39), 153.0701 (17) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Correia, M.S.P.; Lin, W.; Aria, A.J.; Jain, A.; Globisch, D. Rapid Preparation of a Large Sulfated Metabolite Library for Structure Validation in Human Samples. Metabolites 2020, 10, 415. https://doi.org/10.3390/metabo10100415
Correia MSP, Lin W, Aria AJ, Jain A, Globisch D. Rapid Preparation of a Large Sulfated Metabolite Library for Structure Validation in Human Samples. Metabolites. 2020; 10(10):415. https://doi.org/10.3390/metabo10100415
Chicago/Turabian StyleCorreia, Mario S. P., Weifeng Lin, Arash J. Aria, Abhishek Jain, and Daniel Globisch. 2020. "Rapid Preparation of a Large Sulfated Metabolite Library for Structure Validation in Human Samples" Metabolites 10, no. 10: 415. https://doi.org/10.3390/metabo10100415
APA StyleCorreia, M. S. P., Lin, W., Aria, A. J., Jain, A., & Globisch, D. (2020). Rapid Preparation of a Large Sulfated Metabolite Library for Structure Validation in Human Samples. Metabolites, 10(10), 415. https://doi.org/10.3390/metabo10100415