Comprehensive Circulatory Metabolomics in ME/CFS Reveals Disrupted Metabolism of Acyl Lipids and Steroids
Abstract
1. Introduction
2. Results
2.1. Population Statistics and Dataset Handling
2.1.1. Cohort Statistics
2.1.2. Global Metabolomics Panel
2.1.3. Complex Lipid PanelTM
2.2. Statistical Analysis of the Global Metabolomics Dataset
2.2.1. Whole Dataset Analysis
- Non-parametric tests
- Fold change analysis
- Volcano plot analysis
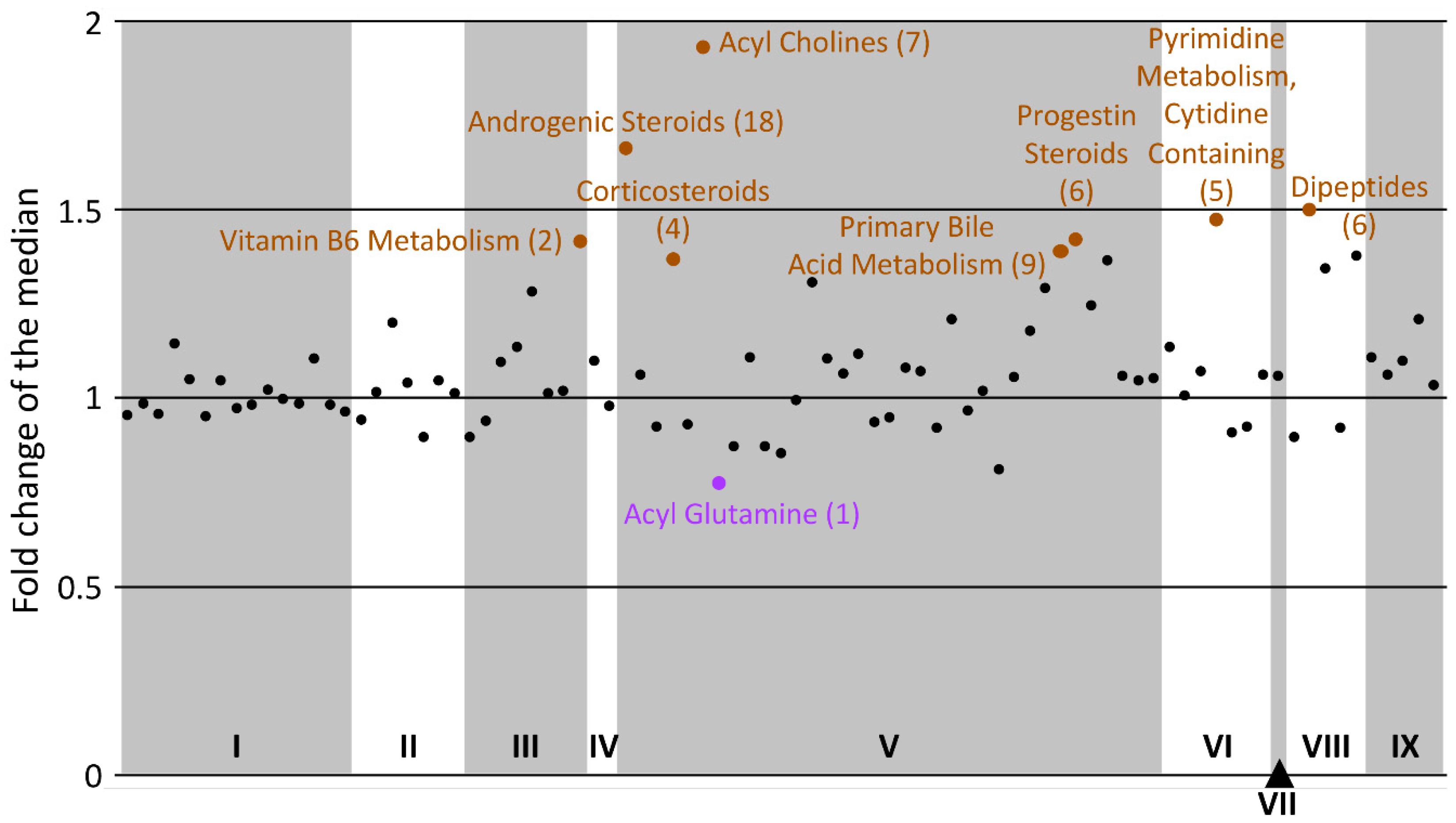
2.2.2. Super-Pathway Dichotomized Analysis
2.3. Statistical Analysis of the Lipidomics Dataset
2.4. Metabolomics Data Insight
2.4.1. Pathway Enrichment
- Global metabolomics panel
- Complex lipid panelTM
2.4.2. Statistical Enrichment
- Global metabolomics panel
- Complex lipid panelTM
- Sub-pathway-based enrichment analysis
2.4.3. Inclusive Observation of the Metabolomics Data
3. Discussion
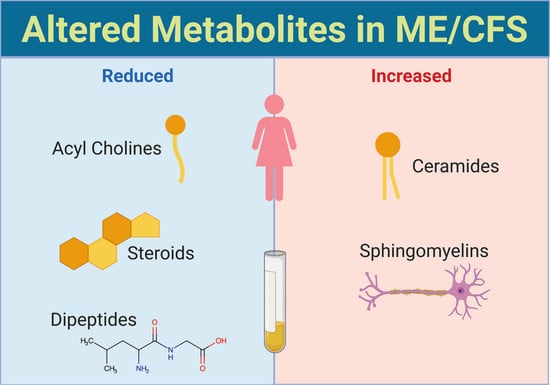
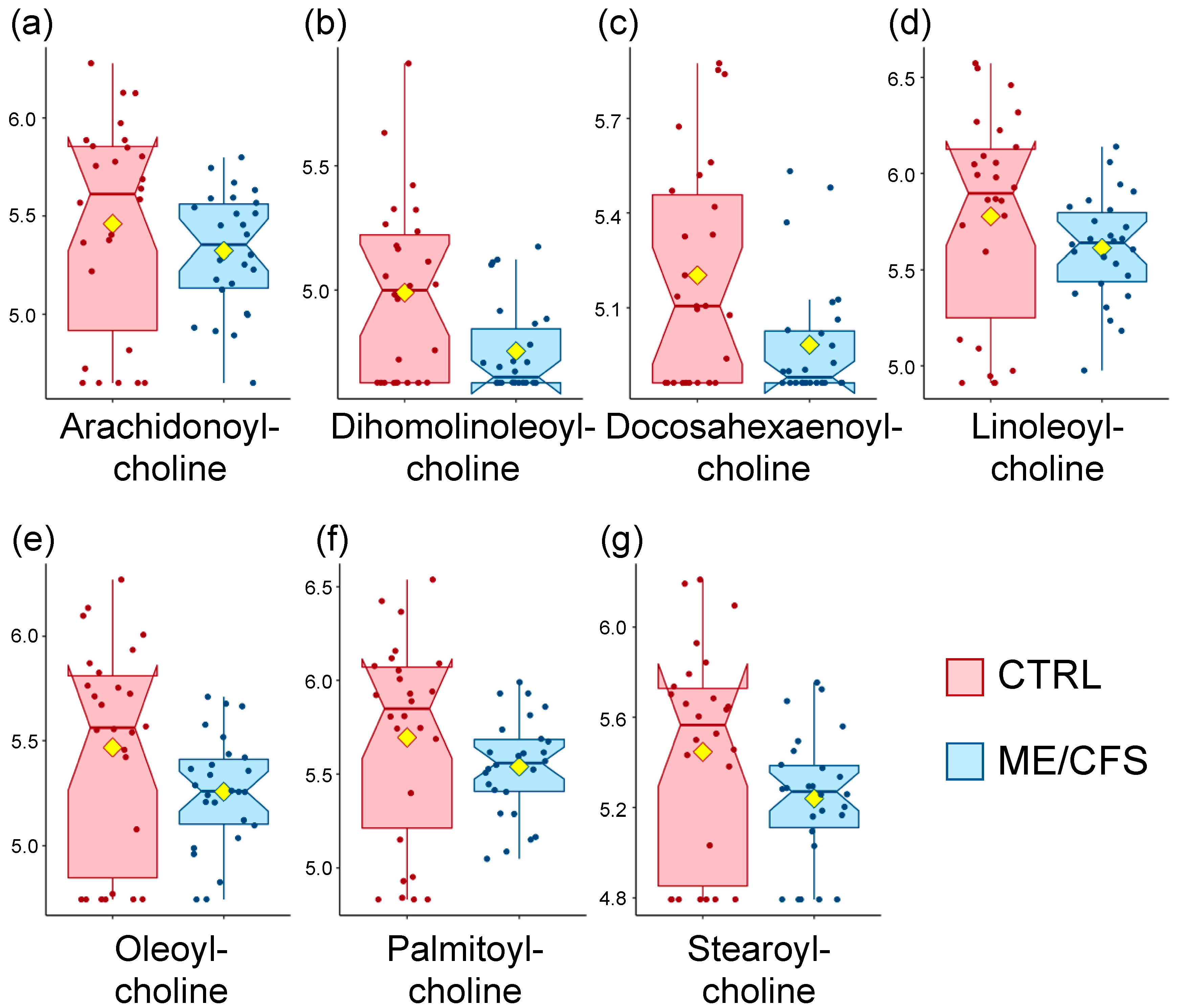
3.1. Acyl Cholines Are Decreased
3.2. Dipeptides Are Decreased
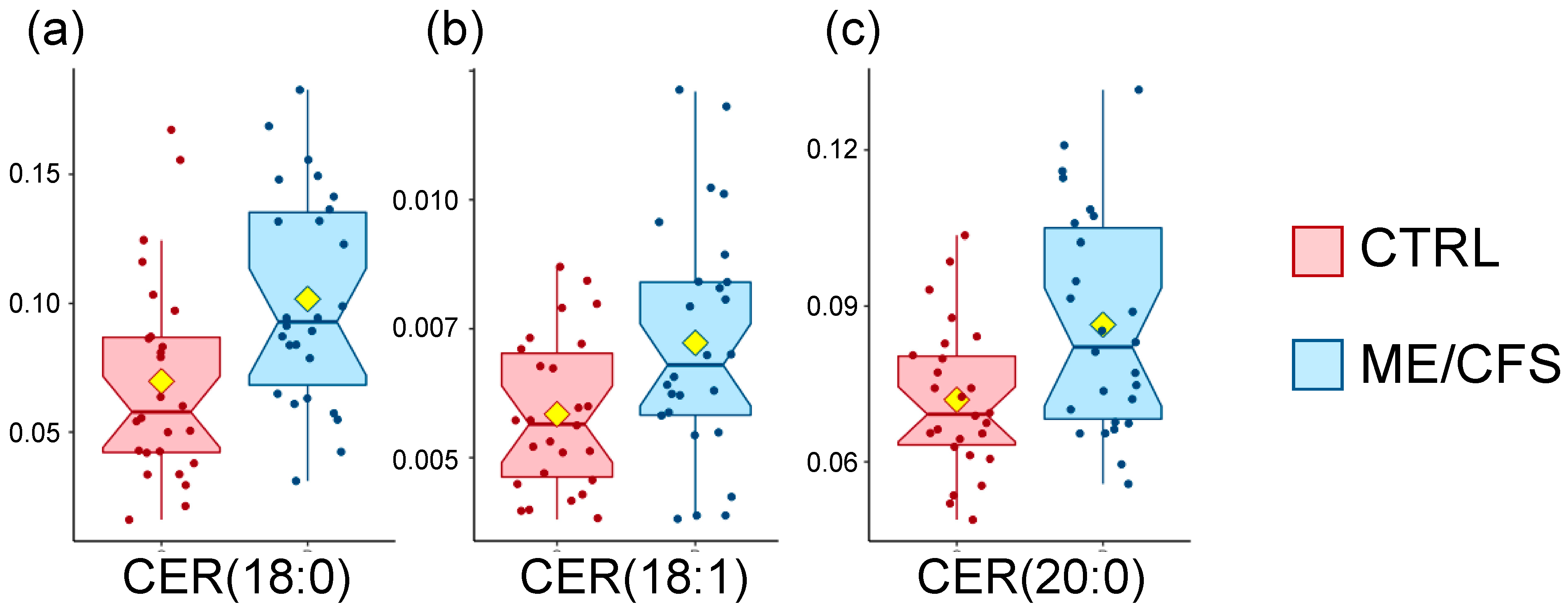
3.3. Sphingolipids Are Increased
3.4. Three Classes of Steroids Are Decreased
3.5. Acyls and ME/CFS
3.6. Reproducibility
4. Materials and Methods
4.1. Cohort and Blood Sampling
4.2. Metabolomics Panels
4.3. Data Analysis
4.4. Data Availability
4.5. Study Approval
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Valdez, A.R.; Hancock, E.E.; Adebayo, S.; Kiernicki, D.J.; Proskauer, D.; Attewell, J.R.; Bateman, L.; DeMaria, A.; Lapp, C.W.; Rowe, P.C.; et al. Estimating prevalence, demographics, and costs of ME/CFS using large scale medical claims data and machine learning. Front. Pediatr. 2019, 6, 412. [Google Scholar] [CrossRef]
- Cliff, J.M.; King, E.C.; Lee, J.S.; Sepulveda, N.; Wolf, A.S.; Kingdon, C.; Bowman, E.; Dockrell, H.M.; Nacul, L.; Lacerda, E.; et al. Cellular immune function in myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS). Front. Immunol. 2019, 10, 796. [Google Scholar] [CrossRef]
- Rivas, J.L.; Palencia, T.; Fernandez, G.; Garcia, M. Association of T and NK cell phenotype with the diagnosis of myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS). Front. Immunol. 2018, 9, 1028. [Google Scholar] [CrossRef]
- Maes, M.; Bosmans, E.; Kubera, M. Increased expression of activation antigens on CD8+T lymphocytes in Myalgic Encephalomyelitis/chronic fatigue syndrome: Inverse associations with lowered CD19+expression and CD4+/CD8+ratio, but no associations with (auto)immune, leaky gut, oxidative and nitrosative stress biomarkers. Neuroendocrinol. Lett. 2015, 36, 439–446. [Google Scholar]
- Klimas, N.G.; Salvato, F.R.; Morgan, R.; Fletcher, M.A. Immunological abnormalities in chronic fatigue syndrome. J. Clin. Microbiol. 1990, 28, 1403–1410. [Google Scholar] [CrossRef]
- Hornig, M.; Montoya, J.G.; Klimas, N.G.; Levine, S.; Felsenstein, D.; Bateman, L.; Peterson, D.L.; Gottschalk, C.G.; Schultz, A.F.; Che, X.; et al. Distinct plasma immune signatures in ME/CFS are present early in the course of illness. Sci. Adv. 2015, 1, e1400121. [Google Scholar] [CrossRef]
- Montoya, J.G.; Holmes, T.H.; Anderson, J.N.; Maecker, H.T.; Rosenberg-Hasson, Y.; Valencia, I.J.; Chu, L.; Younger, J.W.; Tato, C.M.; Davis, M.M. Cytokine signature associated with disease severity in chronic fatigue syndrome patients. Proc. Natl. Acad. Sci. USA 2017, 114, E7150–E7158. [Google Scholar] [CrossRef]
- Boissoneault, J.; Letzen, J.; Lai, S.; O’Shea, A.; Craggs, J.; Robinson, M.E.; Staud, R. Abnormal resting state functional connectivity in patients with chronic fatigue syndrome: An arterial spin-labeling fMRI study. Magn. Reson. Imaging 2016, 34, 603–608. [Google Scholar] [CrossRef]
- Gay, C.W.; Robinson, M.E.; Lai, S.; O’Shea, A.; Craggs, J.G.; Price, D.D.; Staud, R. Abnormal resting-state functional connectivity in patients with chronic fatigue syndrome: Results of seed and data-driven analyses. Brain Connect 2016, 6, 48–56. [Google Scholar] [CrossRef]
- Boissoneault, J.; Letzen, J.; Robinson, M.; Staud, R. Cerebral blood flow and heart rate variability predict fatigue severity in patients with chronic fatigue syndrome. Brain Imaging Behav. 2019, 13, 789–797. [Google Scholar] [CrossRef]
- Aaron, L.A.; Herrell, R.; Ashton, S.; Belcourt, M.; Schmaling, K.; Goldberg, J.; Buchwald, D. Comorbid clinical conditions in chronic fatigue—A co-twin control study. J. Gen. Intern. Med. 2001, 16, 24–31. [Google Scholar] [CrossRef][Green Version]
- Nagy-Szakal, D.; Williams, B.L.; Mishra, N.; Che, X.; Lee, B.; Bateman, L.; Klimas, N.G.; Komaroff, A.L.; Levine, S.; Montoya, J.G.; et al. Fecal metagenomic profiles in subgroups of patients with myalgic encephalomyelitis/chronic fatigue syndrome. Microbiome 2017, 5, 44. [Google Scholar] [CrossRef]
- Armstrong, C.W.; McGregor, N.R.; Lewis, D.P.; Butt, H.L.; Gooley, P.R. Metabolic profiling reveals anomalous energy metabolism and oxidative stress pathways in chronic fatigue syndrome patients. Metabolomics 2015, 11, 1626–1639. [Google Scholar] [CrossRef]
- Armstrong, C.W.; McGregor, N.R.; Lewis, D.P.; Butt, H.L.; Gooley, P.R. The association of fecal microbiota and fecal, blood serum and urine metabolites in myalgic encephalomyelitis/chronic fatigue syndrome. Metabolomics 2017, 13, 8. [Google Scholar] [CrossRef]
- Fluge, O.; Mella, O.; Bruland, O.; Risa, K.; Dyrstad, S.E.; Alme, K.; Rekeland, I.G.; Sapkota, D.; Rosland, G.V.; Fossa, A.; et al. Metabolic profiling indicates impaired pyruvate dehydrogenase function in myalgic encephalopathy/chronic fatigue syndrome. JCI Insight 2016, 1, e89376. [Google Scholar] [CrossRef]
- Naviaux, R.K.; Naviaux, J.C.; Li, K.F.; Bright, A.T.; Alaynick, W.A.; Wang, L.; Baxter, A.; Nathan, N.; Anderson, W.; Gordon, E. Metabolic features of chronic fatigue syndrome. Proc. Natl. Acad. Sci. USA 2016, 113, E5472–E5480. [Google Scholar] [CrossRef]
- Yamano, E.; Sugimoto, M.; Hirayama, A.; Kume, S.; Yamato, M.; Jin, G.H.; Tajima, S.; Goda, N.; Iwai, K.; Fukuda, S.; et al. Index markers of chronic fatigue syndrome with dysfunction of TCA and urea cycles. Sci. Rep. 2016, 6, 34990. [Google Scholar] [CrossRef]
- Nagy-Szakal, D.; Barupal, D.K.; Lee, B.; Che, X.; Williams, B.L.; Kahn, E.J.R.; Ukaigwe, J.E.; Bateman, L.; Klimas, N.G.; Komaroff, A.L.; et al. Insights into myalgic encephalomyelitis/chronic fatigue syndrome phenotypes through comprehensive metabolomics. Sci. Rep. 2018, 8, 10056. [Google Scholar] [CrossRef]
- Germain, A.; Ruppert, D.; Levine, S.M.; Hanson, M.R. Metabolic profiling of a myalgic encephalomyelitis/chronic fatigue syndrome discovery cohort reveals disturbances in fatty acid and lipid metabolism. Mol. Biosyst. 2017, 13, 371–379. [Google Scholar] [CrossRef]
- Germain, A.; Ruppert, D.; Levine, S.M.; Hanson, M.R. Prospective biomarkers from plasma metabolomics of myalgic encephalomyelitis/chronic fatigue syndrome implicate redox imbalance in disease symptomatology. Metabolites 2018, 8, 90. [Google Scholar] [CrossRef]
- McGregor, N.R.; Armstrong, C.W.; Lewis, D.P.; Gooley, P.R. Post-exertional malaise is associated with hypermetabolism, hypoacetylation and purine metabolism deregulation in ME/CFS cases. Diagnostics 2019, 9, 70. [Google Scholar] [CrossRef] [PubMed]
- Ware, J.E.; Kosinski, M. Interpreting SF-36 summary health measures: A response. Qual. Life Res. 2001, 10, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Moghimipour, E.; Ameri, A.; Handali, S. Absorption-enhancing effects of bile salts. Molecules 2015, 20, 14451–14473. [Google Scholar] [CrossRef]
- Hanson, M.R.; Giloteaux, L. The gut microbiome in myalgic encephalomyelitis. Biochemist 2017, 39, 10–13. [Google Scholar] [CrossRef]
- Sperringer, J.E.; Addington, A.; Hutson, S.M. Branched-chain amino acids and brain metabolism. Neurochem. Res. 2017, 42, 1697–1709. [Google Scholar] [CrossRef]
- Cruzat, V.F.; Bittencourt, A.; Scomazzon, S.P.; Leite, J.S.M.; de Bittencourt, P.I.H.; Tirapegui, J. Oral free and dipeptide forms of glutamine supplementation attenuate oxidative stress and inflammation induced by endotoxemia. Nutrition 2014, 30, 602–611. [Google Scholar] [CrossRef]
- Ano, Y.; Kita, M.; Kitaoka, S.; Furuyashiki, T. Leucine-histidine dipeptide attenuates microglial activation and emotional disturbances induced by brain inflammation and repeated social defeat stress. Nutrients 2019, 11, 2161. [Google Scholar] [CrossRef]
- Summers, S.A.; Chaurasia, B.; Holland, W.L. Metabolic messengers: Ceramides. Nat. Metab. 2019. [Google Scholar] [CrossRef]
- Mitsnefes, M.M.; Fitzpatrick, J.; Sozio, S.M.; Jaar, B.G.; Estrella, M.M.; Monroy-Trujillo, J.M.; Zhang, W.J.; Setchell, K.; Parekh, R.S. Plasma glucosylceramides and cardiovascular risk in incident hemodialysis patients. J. Clin. Lipidol. 2018, 12, 1513–1522. [Google Scholar] [CrossRef]
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 175–191. [Google Scholar] [CrossRef]
- Prough, R.A.; Clark, B.J.; Klinge, C.M. Novel mechanisms for DHEA action. J. Mol. Endocrinol. 2016, 56, R139–R155. [Google Scholar] [CrossRef] [PubMed]
- de Kloet, E.R.; Meijer, O.C.; de Nicola, A.F.; de Rijk, R.H.; Joels, M. Importance of the brain corticosteroid receptor balance in metaplasticity, cognitive performance and neuro-inflammation. Front. Neuroendocr. 2018, 49, 124–145. [Google Scholar] [CrossRef]
- Shao, Y.P.; Le, W.D. Recent advances and perspectives of metabolomics-based investigations in Parkinson’s disease. Mol. Neurodegener 2019, 14, 3. [Google Scholar] [CrossRef]
- Hunt, R.; de Mortemer Taveau, R. The Effects of a Number of Derivatives of Choline and Analogous Compounds of the Blood-Pressure; US Government Printing Office: Washington, DC, USA, 1911; p. 73.
- Schneider, R.; Timms, A.R.; Kyi, Z.Y.; Wilson, W. Some aspects of the pharmacology of an homologous series of choline esters of fatty acids. Br. J. Pharmacol. Chemother. 1956, 12, 30–38. [Google Scholar] [CrossRef] [PubMed]
- IOM. Beyond Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: Redefining an Illness; The National Academies Press: Washington, DC, USA, 2015. [Google Scholar]
- Cornelis, M.C.; Erlund, I.; Michelotti, G.A.; Herder, C.; Westerhuis, J.A.; Tuomilehto, J. Metabolomic response to coffee consumption: Application to a three-stage clinical trial. J. Intern. Med. 2018, 283, 544–557. [Google Scholar] [CrossRef] [PubMed]
- Kuang, A.; Erlund, I.; Herder, C.; Westerhuis, J.A.; Tuomilehto, J.; Cornelis, M.C. Lipidomic response to coffee consumption. Nutrients 2018, 10, 1851. [Google Scholar] [CrossRef]
- Li, K.J.; Borresen, E.C.; Jenkins-Puccetti, N.; Luckasen, G.; Ryan, E.P. Navy bean and rice bran intake alters the plasma metabolome of children at risk for cardiovascular disease. Front. Nutr. 2018, 4, 71. [Google Scholar] [CrossRef]
- Zarei, I.; Oppel, R.C.; Borresen, E.C.; Brown, R.J.; Ryan, E.P. Modulation of plasma and urine metabolome in colorectal cancer survivors consuming rice bran. Integr. Food Nutr. Metab. 2019, 6. [Google Scholar] [CrossRef]
- Lujuan, X.; MacKenzie, E.C.; Hua, Z.; Wangang, Z.; Yoshinori, M. Carnosine—A natural bioactive dipeptide: Bioaccessibility, bioavailability and health benefits. J. Food Bioact. 2019, 5. [Google Scholar] [CrossRef]
- Raizel, R.; Tirapegui, J. Role of glutamine, as free or dipeptide form, on muscle recovery from resistance training: A review study. Nutrire 2018, 43, 28. [Google Scholar] [CrossRef]
- Ano, Y.; Yoshino, Y.; Uchida, K.; Nakayama, H. Preventive effects of tryptophan-methionine dipeptide on neural inflammation and alzheimer’s pathology. Int. J. Mol. Sci. 2019, 20, 3206. [Google Scholar] [CrossRef] [PubMed]
- Cole, T.J.; Short, K.L.; Hooper, S.B. The science of steroids. Semin. Fetal Neonatl Med. 2019, 24, 170–175. [Google Scholar] [CrossRef]
- Baulieu, E.-E.; Robel, P. Dehydroepiandrosterone (DHEA) and dehydroepiandrosterone sulfate (DHEAS) as neuroactive neurosteroids. Proc. Natl. Acad. Sci. USA 1998, 95, 4089–4091. [Google Scholar] [CrossRef] [PubMed]
- Castro-Marrero, J.; Sáez-Francàs, N.; Santillo, D.; Alegre, J. Treatment and management of chronic fatigue syndrome/myalgic encephalomyelitis: All roads lead to Rome. Br. J. Pharmacol. 2017, 174, 345–369. [Google Scholar] [CrossRef] [PubMed]
- de Vega, W.C.; Herrera, S.; Vernon, S.D.; McGowan, P.O. Epigenetic modifications and glucocorticoid sensitivity in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS). BMC Med. Genom. 2017, 10, 11. [Google Scholar] [CrossRef]
- Fukuda, K.; Straus, S.E.; Hickie, I.; Sharpe, M.C.; Dobbins, J.G.; Komaroff, A. The chronic fatigue syndrome: A comprehensive approach to its definition and study. International Chronic Fatigue Syndrome Study Group. Ann. Intern. Med. 1994, 121, 953–959. [Google Scholar] [CrossRef]
- Ware, J.E.; Sherbourne, C.D. The Mos 36-Item Short-Form Health Survey (Sf-36). 1. Conceptual-Framework and Item Selection. Med. Care 1992, 30, 473–483. [Google Scholar] [CrossRef]
- Ware, J.E., Jr.; Kosinski, M.; Bayliss, M.S.; McHorney, C.A.; Rogers, W.H.; Raczek, A. Comparison of methods for the scoring and statistical analysis of SF-36 health profile and summary measures: Summary of results from the Medical Outcomes Study. Med. Care 1995, 33, AS264–AS279. [Google Scholar]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef]
- Barupal, D.K.; Fiehn, O. Chemical Similarity Enrichment Analysis (ChemRICH) as alternative to biochemical pathway mapping for metabolomic datasets. Sci. Rep. 2017, 7, 14567. [Google Scholar] [CrossRef]
| Controls | ME/CFS | Mann-Whitney U Test | ||
|---|---|---|---|---|
| Gender (n) | Female | 26 | 26 | ND |
| Age | Mean +/− SD | 41.5 +/− 15 | 49.7 +/− 13.7 | p = 0.05 |
| Median +/− SD | 43 (22–66) | 52 (22–72) | ||
| BMI | Mean +/− SD | 21.9 +/− 3.2 | 24.6 +/− 5.6 | p = 0.8 |
| Median +/− SD | 21.4 (16.3–28.9) | 23 (16.8–40.7) | ||
| Type of onset | Gradual | ND | 42% | ND |
| Sudden | ND | 58% | ND | |
| Gut symptoms * | 4% | 50% | ND | |
| Positive tilt table test ** (n = 16) | ND | 69% | ND | |
| Bell’s disability scale *** | 10–20 | 0 | 11 | p < 0.001 |
| 30–40 | 0 | 11 | ||
| 50–60 | 1 | 4 | ||
| 90–100 | 25 | 0 | ||
| SF-36 *** | Physical Component Summary (PCS) | 55.5 +/− 5.3 | 25.7 +/− 7.8 | p < 0.001 |
| Mental Component Summary (MCS) | 55.1 +/− 6 | 40.6 +/− 10.9 | p < 0.001 |
| Super-Pathway | Sub-Pathway | Metabolite | HMDB ID | Fold Change | p-Value |
|---|---|---|---|---|---|
| Amino-Acids | Glutamate Metabolism | 4-hydroxyglutamate | HMDB01344 | 0.5 | 0.005 |
| Lipids | Acyl Cholines | Dihomo-linolenoyl-choline | NA | 2.3 | 0.02 |
| Linoleoylcholine | NA | 2.2 | 0.07 | ||
| Oleoylcholine | NA | 2.3 | 0.04 | ||
| Palmitoylcholine | NA | 2.1 | 0.07 | ||
| Stearoylcholine | NA | 2.2 | 0.04 | ||
| Xenobiotics | Chemical | Dimethyl Sulfone | HMDB04983 | 2.7 | 0.03 |
| Food Component/Plant | Erythritol | HMDB02994 | 6 | 0.09 | |
| Piperine | HMDB29377 | 3 | 0.03 | ||
| Sulfate of piperine metabolite C16H19NO3 (3) | NA | 2.5 | 0.03 |
| Super-Pathway | Sub-Pathway | Metabolite | HMDB ID | p-Value | Fold Change |
|---|---|---|---|---|---|
| Sphingolipids | Ceramides | CER(18:0) | HMDB04950 | 0.01 | 0.7 |
| CER(18:1) | HMDB04948 | 0.03 | 0.8 | ||
| CER(20:0) | HMDB04951 | 0.004 | 0.8 |
| Metabolite Class | KEGG ID | Importance | p-Value |
|---|---|---|---|
| Sphingomyelins | C00550 | 0.01 | 0.007 |
| Ceramides | C00195 | 0.29 | 0.02 |
| Glucosylceramides | C01190 | 0.03 | 0.5 |
| Lactosylceramides | C01290 | 0 | 0.2 |
| Super-Pathway | Sub-Pathway | Metabolite | HMDB ID | Fold Change | p-Value |
|---|---|---|---|---|---|
| Sphingolipids | Ceramides | CER (18:0) | HMDB04950 | 0.7 | 0.01 |
| CER (18:1) | HMDB04948 | 0.8 | 0.03 | ||
| CER (24:1) | HMDB06728 | 0.9 | 0.02 | ||
| Sphingomyelins | SM (18:0) | HMDB01348 | 0.8 | 0.008 | |
| SM (18:1) | HMDB12101 | 0.8 | 0.003 |
| Sub-Pathway | Cluster Size | p-Value | q-Value | Altered | Increased | Decreased |
|---|---|---|---|---|---|---|
| Androgenic Steroids | 18 | 1.9 × 10−8 | 2.1 × 10−6 | 11 | 0 | 11 |
| Analgesics, Anesthetics | 20 | 5.5 × 10−7 | 0.00003 | 10 | 2 | 8 |
| Acyl Cholines | 7 | 0.00002 | 0.0007 | 6 | 0 | 6 |
| Ceramides | 12 | 0.00004 | 0.001 | 7 | 7 | 0 |
| Dipeptides | 6 | 0.005 | 0.1 | 4 | 0 | 4 |
| Acyl Carnitines | 39 | 0.008 | 0.1 | 8 | 3 | 5 |
| Sphingomyelins | 12 | 0.03 | 0.5 | 4 | 4 | 0 |
| Super-Pathway | Sub-Pathway | Metabolite | HMDB ID | Fold Change | p-Value |
|---|---|---|---|---|---|
| Lipids | Acyl Cholines | Arachidonoylcholine | NA | 1.7 | 0.03 |
| Linoleoylcholine | NA | 1.7 | 0.02 | ||
| Oleoylcholine | NA | 1.6 | 0.06 | ||
| Palmitoylcholine | NA | 1.6 | 0.01 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Germain, A.; Barupal, D.K.; Levine, S.M.; Hanson, M.R. Comprehensive Circulatory Metabolomics in ME/CFS Reveals Disrupted Metabolism of Acyl Lipids and Steroids. Metabolites 2020, 10, 34. https://doi.org/10.3390/metabo10010034
Germain A, Barupal DK, Levine SM, Hanson MR. Comprehensive Circulatory Metabolomics in ME/CFS Reveals Disrupted Metabolism of Acyl Lipids and Steroids. Metabolites. 2020; 10(1):34. https://doi.org/10.3390/metabo10010034
Chicago/Turabian StyleGermain, Arnaud, Dinesh K. Barupal, Susan M. Levine, and Maureen R. Hanson. 2020. "Comprehensive Circulatory Metabolomics in ME/CFS Reveals Disrupted Metabolism of Acyl Lipids and Steroids" Metabolites 10, no. 1: 34. https://doi.org/10.3390/metabo10010034
APA StyleGermain, A., Barupal, D. K., Levine, S. M., & Hanson, M. R. (2020). Comprehensive Circulatory Metabolomics in ME/CFS Reveals Disrupted Metabolism of Acyl Lipids and Steroids. Metabolites, 10(1), 34. https://doi.org/10.3390/metabo10010034