Metabolomic Profiling of Fungal Pathogens Responsible for Root Rot in American Ginseng
Abstract
1. Introduction
2. Results
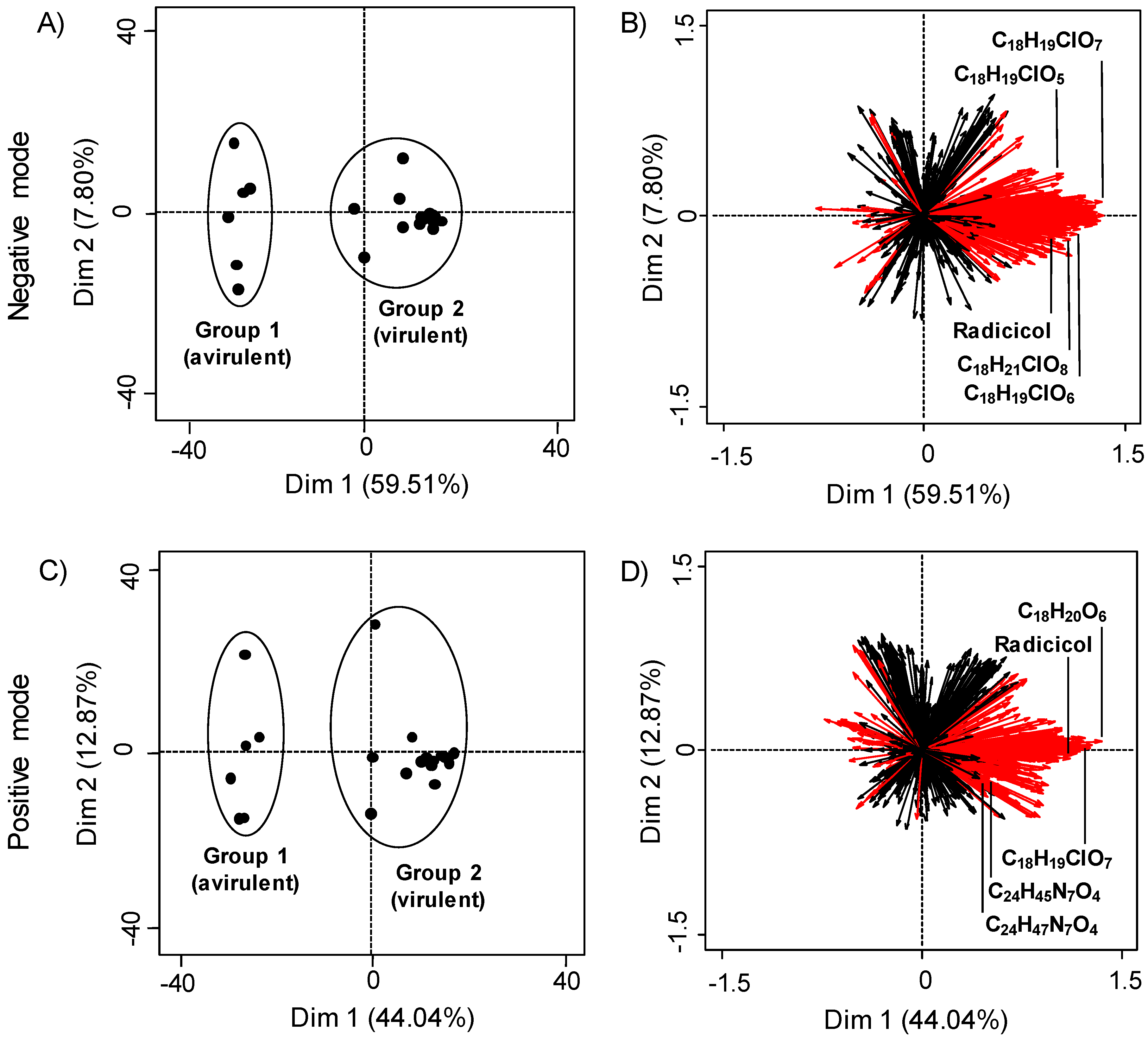
2.1. Principal Component Analysis of LC-HRMS Data
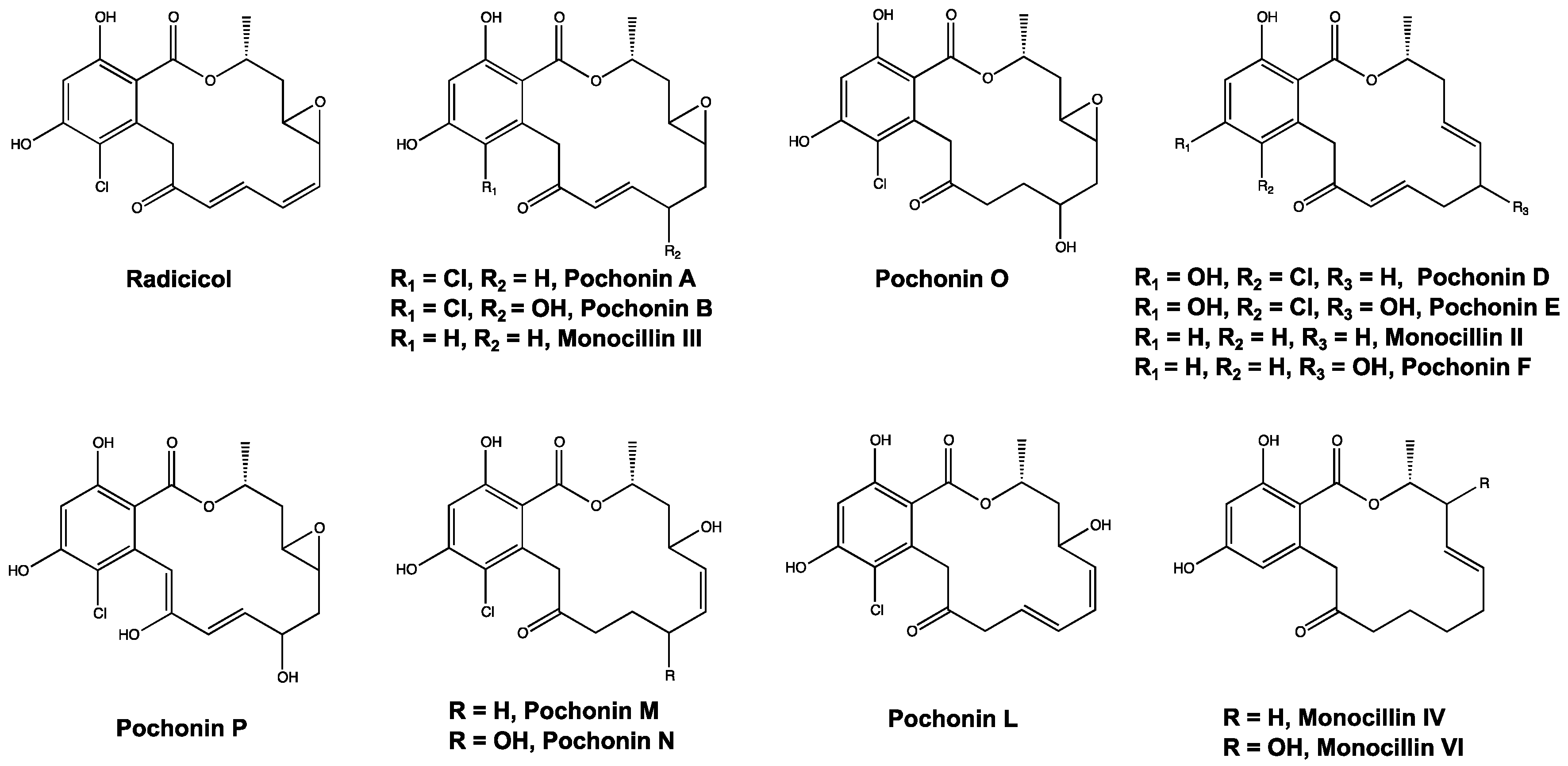
2.2. Determining LC-MS Features Significant to Virulent Strains
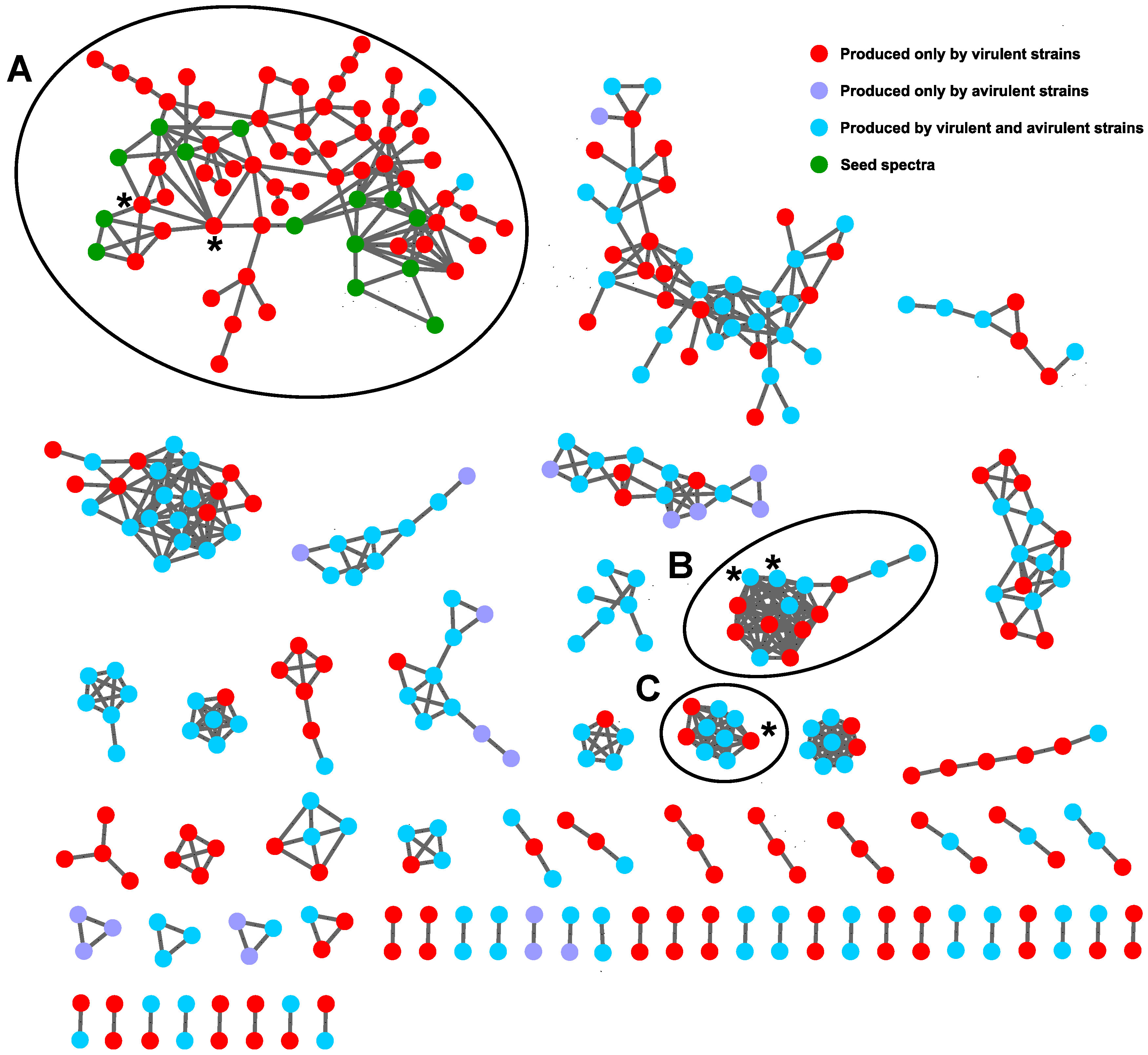
2.3. Molecular Networking with GNPS
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Secondary Metabolite Extraction
4.2. LC-MS Experiments
4.3. LC-MS Data Processing
4.4. Molecular Networking Parameters and Visualization
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Baeg, I.-H.; So, S.-H. The world ginseng market and the ginseng (Korea). J. Ginseng Res. 2013, 37, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Westerveld, S. Ginseng Production in Ontario; Queen’s Printer for Ontario: Toronto, ON, Canada, 2010. [Google Scholar]
- Westerveld, S. Cost of Production of Ginseng in Ontario. Available online: http://www.omafra.gov.on.ca/english/crops/facts/gincop.htm (accessed on 27 October 2019).
- Rahman, M.; Punja, Z.K. Factors influencing development of root rot on ginseng caused by Cylindrocarpon destructans. Phytopathology 2005, 95, 1381–1390. [Google Scholar] [CrossRef] [PubMed]
- Cabral, A.; Groenewald, J.Z.; Rego, C.; Oliveira, H.; Crous, P.W. Cylindrocarpon root rot: Multi-gene analysis reveals novel species within the Ilyonectria radicicola species complex. Mycol. Prog. 2012, 11, 655–688. [Google Scholar] [CrossRef]
- Seifert, K.; McMullen, C.; Yee, D.; Reeleder, R.; Dobinson, K. Molecular differentiation and detection of ginseng-adapted isolates of the root rot fungus Cylindrocarpon destructans. Phytopathology 2003, 93, 1533–1542. [Google Scholar] [CrossRef] [PubMed]
- Farh, M.E.-A.; Kim, Y.-J.; Kim, Y.-J.; Yang, D.-C. Cylindrocarpon destructans/Ilyonectria radicicola-species complex: Causative agent of ginseng root-rot disease and rusty symptoms. J. Ginseng Res. 2018, 42, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Reeleder, R.; Roy, R.; Capell, B. Seed and root rots of ginseng (Panax quinquefolius L.) caused by Cylindrocarpon destructans and Fusarium spp. J. Ginseng Res. 2002, 26, 151–158. [Google Scholar]
- Reeleder, R.; Brammall, R. Pathogenicity of Pythium species, Cylindrocarpon destructans, and Rhizoctonia solani to ginseng seedlings in Ontario. Can. J. Plant Pathol. 1994, 16, 311–316. [Google Scholar] [CrossRef]
- Shin, J.-H.; Fu, T.; Park, K.H.; Kim, K.S. The effect of fungicides on mycelial growth and conidial germination of the ginseng root rot fungus, Cylindrocarpon destructans. Mycobiology 2017, 45, 220–225. [Google Scholar] [CrossRef]
- Kang, Y.; Lee, S.-H.; Lee, J. Development of a selective medium for the fungal pathogen Cylindrocarpon destructans using radicicol. Plant Pathol. J. 2014, 30, 432–436. [Google Scholar] [CrossRef]
- Walsh, J.P.; DesRochers, N.; Renaud, J.B.; Seifert, K.A.; Yeung, K.K.C.; Sumarah, M.W. Identification of N,N′,N″-triacetylfusarinine C as a key metabolite for root rot disease virulence in American ginseng. J. Ginseng Res. 2019, in press. [Google Scholar] [CrossRef]
- Kragl, C.; Schrettl, M.; Abt, B.; Sarg, B.; Lindner, H.H.; Haas, H. EstB-mediated hydrolysis of the siderophore triacetylfusarinine C optimizes iron uptake of Aspergillus fumigatus. Eukaryot. Cell 2007, 6, 1278–1285. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Sanchez, L.M.; Rath, C.M.; Liu, X.; Boudreau, P.D.; Bruns, N.; Glukhov, E.; Wodtke, A.; De Felicio, R.; Fenner, A. Molecular networking as a dereplication strategy. J. Nat. Prod. 2013, 76, 1686–1699. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Maille, G.O.; Want, E.J.; Qin, C.; Trauger, S.A.; Brandon, T.R.; Custodio, D.E.; Abagyan, R.; Siuzdak, G. METLIN: A Metabolite Mass Spectral Database. Ther. Drug Monit. 2005, 27, 747–751. [Google Scholar] [CrossRef]
- Evans, G.; White, N. Radicicolin and radicicol, two new antibiotics produced by Cylindrocarpon radicicola. Trans. Br. Mycol. Soc. 1966, 49, 563–567. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Watrous, J.; Roach, P.; Alexandrov, T.; Heath, B.S.; Yang, J.Y.; Kersten, R.D.; van der Voort, M.; Pogliano, K.; Gross, H.; Raaijmakers, J.M.; et al. Mass spectral molecular networking of living microbial colonies. Proc. Natl. Acad. Sci. USA 2012, 109, E1743–E1752. [Google Scholar] [CrossRef]
- Evans, G.; Cartwright, J.B.; White, N.H. The production of a phytotoxin, nectrolide, by some root-surface isolates of Cylindrocarpon radicicola, Wr. Plant Soil 1967, 26, 253–260. [Google Scholar] [CrossRef]
- Hellwig, V.; Mayer-Bartschmid, A.; Müller, H.; Greif, G.; Kleymann, G.; Zitzmann, W.; Tichy, H.-V.; Stadler, M. Pochonins A−F, new antiviral and antiparasitic resorcylic acid lactones from Pochonia chlamydosporia var. catenulata. J. Nat. Prod. 2003, 66, 829–837. [Google Scholar] [CrossRef]
- Shinonaga, H.; Kawamura, Y.; Ikeda, A.; Aoki, M.; Sakai, N.; Fujimoto, N.; Kawashima, A. The search for a hair-growth stimulant: New radicicol analogues as WNT-5A expression inhibitors from Pochonia chlamydosporia var. chlamydosporia. J. Tetrahedron Lett. 2009, 50, 108–110. [Google Scholar] [CrossRef]
- Aver, W.A.; Peña-Rodriguez, L. Minor metabolites of Monocillium nordinii. Phytochemistry 1987, 26, 1353–1355. [Google Scholar] [CrossRef]
- Smedsgaard, J. Micro-scale extraction procedure for standardized screening of fungal metabolite production in cultures. J. Chromatogr. A 1997, 760, 264–270. [Google Scholar] [CrossRef]
- Dunn, W.B.; Wilson, I.D.; Nicholls, A.W.; Broadhurst, D. The importance of experimental design and QC samples in large-scale and MS-driven untargeted metabolomic studies of humans. Bioanalysis 2012, 4, 2249–2264. [Google Scholar] [CrossRef] [PubMed]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918. [Google Scholar] [CrossRef]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- Benton, H.P.; Want, E.J.; Ebbels, T.M. Correction of mass calibration gaps in liquid chromatography–mass spectrometry metabolomics data. Bioinformatics 2010, 26, 2488–2489. [Google Scholar] [CrossRef]
- Tautenhahn, R.; Boettcher, C.; Neumann, S. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinform. 2008, 9, 504. [Google Scholar] [CrossRef]
- McMillan, A.; Rulisa, S.; Sumarah, M.; Macklaim, J.M.; Renaud, J.; Bisanz, J.E.; Gloor, G.B.; Reid, G. A multi-platform metabolomics approach identifies highly specific biomarkers of bacterial diversity in the vagina of pregnant and non-pregnant women. Sci. Rep. 2015, 5, 14174. [Google Scholar] [CrossRef]
- van den Berg, R.A.; Hoefsloot, H.C.J.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R package for multivariate analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Nyamundanda, G.; Brennan, L.; Gormley, I.C. Probabilistic principal component analysis for metabolomic data. BMC Bioinform. 2010, 11, 571. [Google Scholar] [CrossRef] [PubMed]
- Scrucca, L.; Fop, M.; Murphy, T.B.; Raftery, A.E. Mclust 5: Clustering, classification and density estimation using Gaussian finite mixture models. R J. 2016, 8, 289. [Google Scholar] [CrossRef] [PubMed]
- The Metabolomics Workbench. Available online: https://www.metabolomicsworkbench.org/ (accessed on 16 December 2019).

| Positive | Negative | ||
|---|---|---|---|
| m/z | Formula | m/z | Formula |
| 355.115 [M + Na]+ | C18H20O6 | 349.0847 [M − H]− | C18H19ClO5 |
| * 365.0785 [M + H]+ | C18H17ClO6 | * 363.0641 [M − H]− | C18H17ClO6 |
| 383.0890 [M + H]+ | C18H19ClO7 | 365.0795 [M − H]− | C18H19ClO6 |
| 496.3631 [M + H]+ | C28H49NO6 | 381.0746 [M − H]− | C18H19ClO7 |
| 498.3787 [M + H]+ | C28H51NO6 | 399.085 [M − H]− | C18H21ClO8 |
| ID | Species | Host | Origin |
|---|---|---|---|
| 139398 | Ilyonectria robusta | Prunus cerasus (Sour cherry) | Canada, ON |
| 144524 | I. torresensis | Vitis vinifera (Grape) | ON |
| 220159 | I. mors-panacis | Panax quinquefolius (American ginseng) | ON |
| 226721 | I. rufa | Pseudotsuga menziesii (Douglas fir) | Canada, BC |
| 226727 | I. mors-panacis | P. quinquefolius | ON |
| 226729 | I. robusta | P. quinquefolius | ON |
| 226730 | I. estremocensis | Picea glauca (White spruce) | Canada, QC |
| 230337 | I. mors-panacis | Panax ginseng (Asian ginseng) | Japan |
| 230338 | I. mors-panacis | P. ginseng | Japan |
| 234582 | I. mors-panacis | P. quinquefolius | ON |
| 251601 | I. mors-panacis | P. quinquefolius | ON |
| 251602 | I. mors-panacis | P. quinquefolius | ON |
| 251603 | I. mors-panacis | P. quinquefolius | ON |
| 251604 | I. mors-panacis | P. quinquefolius | ON |
| 251605 | I. mors-panacis | P. quinquefolius | ON |
| 251606 | I. mors-panacis | P. quinquefolius | ON |
| 251607 | I. mors-panacis | P. quinquefolius | ON |
| 251608 | I. rufa | P. menziesii | BC |
| 251609 | I. rufa | P. glauca | QC |
| 251610 | I. mors-panacis | P. quinquefolius | ON |
| 251611 | I. mors-panacis | P. quinquefolius | ON |
| 94-1356 * | Neonectria obtusisporum | Picea mariana (Black spruce) | QC |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
DesRochers, N.; Walsh, J.P.; Renaud, J.B.; Seifert, K.A.; Yeung, K.K.-C.; Sumarah, M.W. Metabolomic Profiling of Fungal Pathogens Responsible for Root Rot in American Ginseng. Metabolites 2020, 10, 35. https://doi.org/10.3390/metabo10010035
DesRochers N, Walsh JP, Renaud JB, Seifert KA, Yeung KK-C, Sumarah MW. Metabolomic Profiling of Fungal Pathogens Responsible for Root Rot in American Ginseng. Metabolites. 2020; 10(1):35. https://doi.org/10.3390/metabo10010035
Chicago/Turabian StyleDesRochers, Natasha, Jacob P. Walsh, Justin B. Renaud, Keith A. Seifert, Ken K.-C. Yeung, and Mark W. Sumarah. 2020. "Metabolomic Profiling of Fungal Pathogens Responsible for Root Rot in American Ginseng" Metabolites 10, no. 1: 35. https://doi.org/10.3390/metabo10010035
APA StyleDesRochers, N., Walsh, J. P., Renaud, J. B., Seifert, K. A., Yeung, K. K.-C., & Sumarah, M. W. (2020). Metabolomic Profiling of Fungal Pathogens Responsible for Root Rot in American Ginseng. Metabolites, 10(1), 35. https://doi.org/10.3390/metabo10010035






