A Score-Based Rapid Screening and Network Visualization Method Based on Bioactive Ingredient-Induced Variations in Skin Cell Gene Expression
Abstract
1. Introduction
2. Materials and Methods
2.1. List of Bioactive Ingredients Used
2.2. Epidermal 3D Model
2.3. RNA-Seq Protocol
2.4. Preprocessing of Data
2.5. Determination of Differentially Expressed Genes (DEGs)
2.6. Calculation of Relationship Score
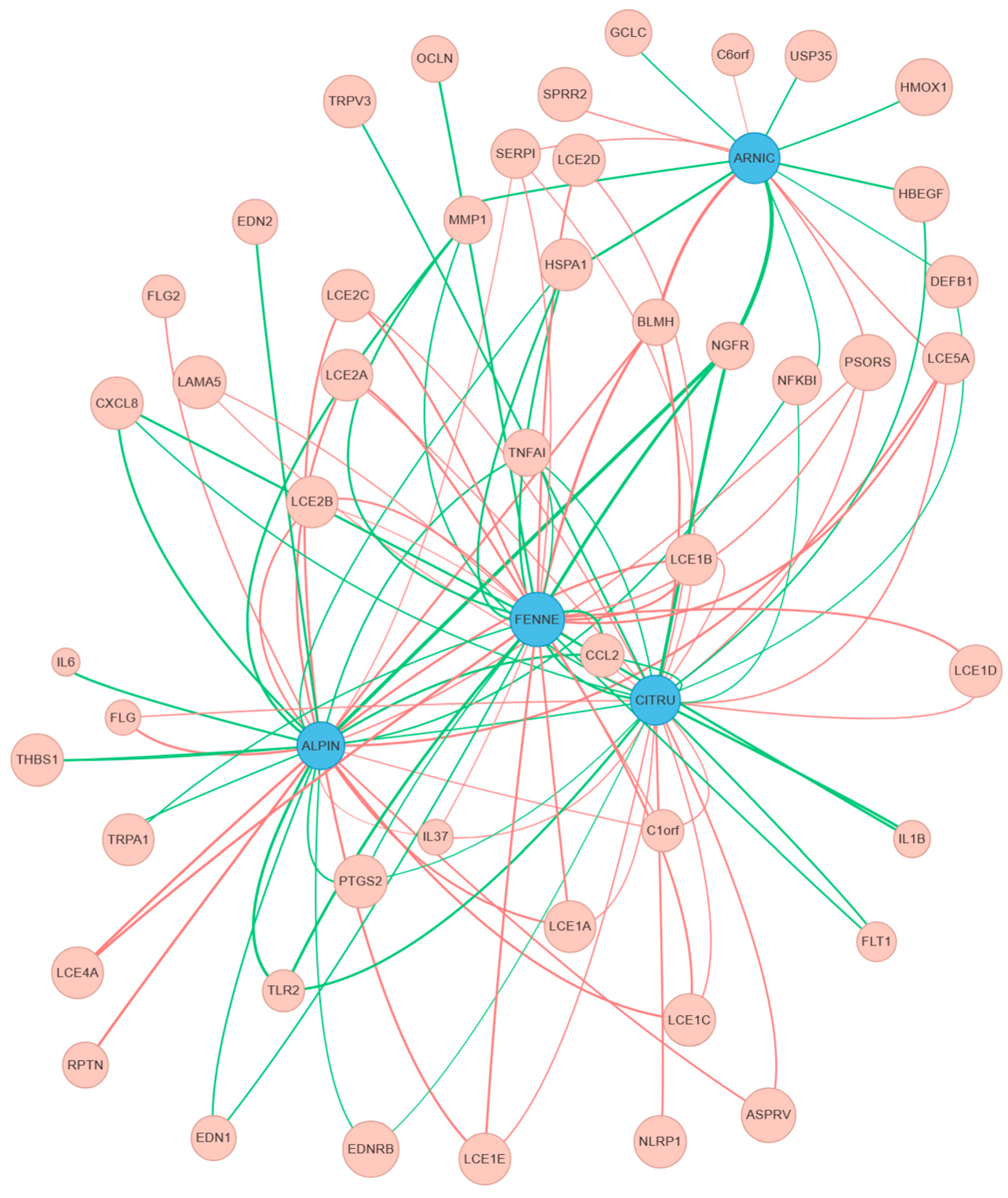
2.7. Generation of Network Graphs
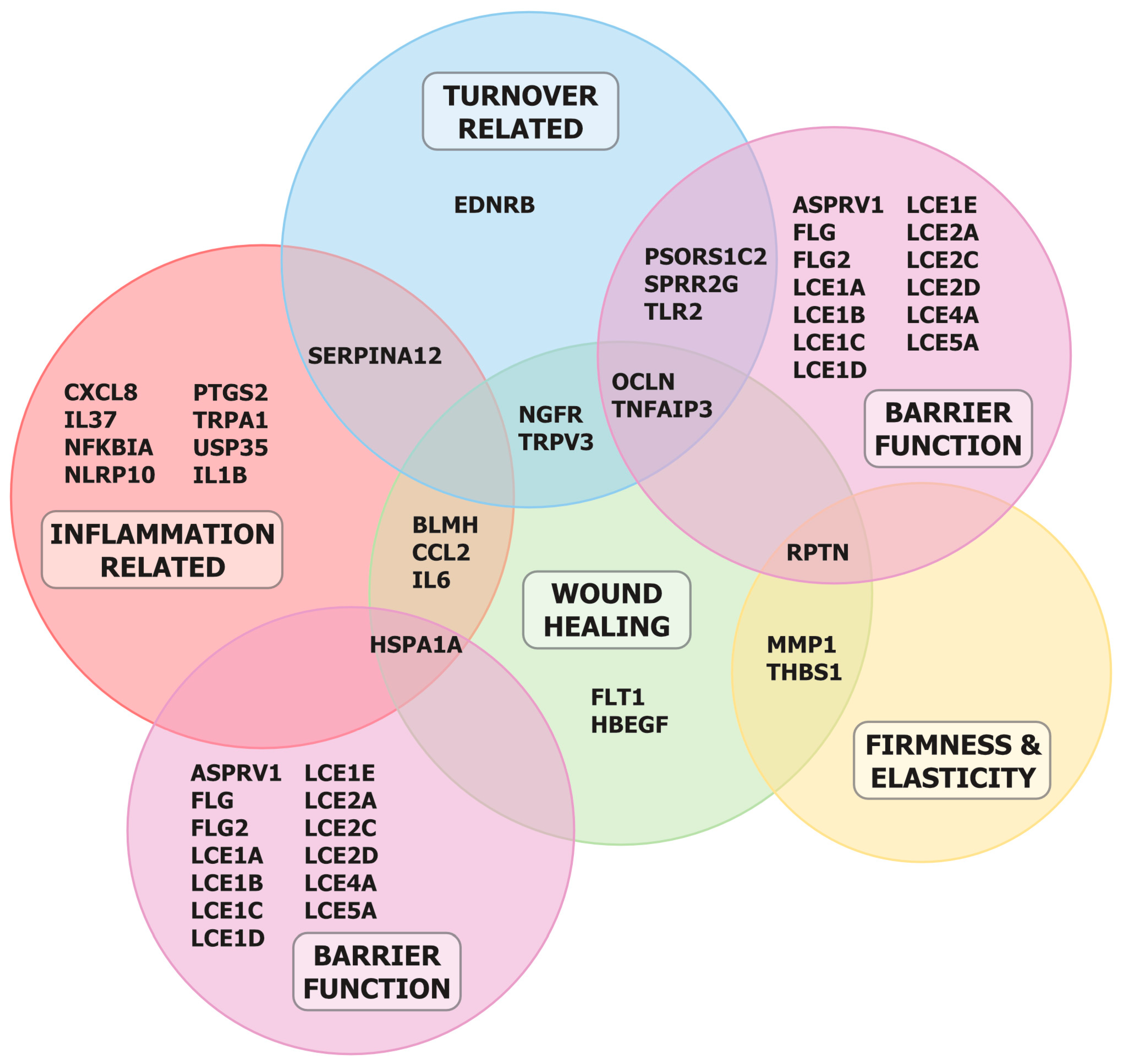
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- National Cancer Institute. NCI Dictionary of Cancer Terms. Available online: https://www.cancer.gov/publications/dictionaries/cancer-terms/def/bioactive-compound (accessed on 1 September 2025).
- Weaver, C.M. Bioactive Foods and Ingredients for Health. Adv. Nutr. 2014, 5, 306S–311S. [Google Scholar] [CrossRef]
- Samtiya, M.; Aluko, R.E.; Dhewa, T.; Moreno-Reyes, J.M. Potential Health Benefits of Plant Food-Derived Bioactive Components: An Overview. Foods 2021, 10, 839. [Google Scholar] [CrossRef]
- Donn, P.; Prieto, M.A.; Mejuto, J.C.; Cao, H.; Simal-Gandara, J. Functional foods based on the recovery of bioactive ingredients from food and algae by-products by emerging extraction technologies and 3D printing. Food Biosci. 2022, 49, 101853. [Google Scholar] [CrossRef]
- Kussmann, M.; Abe Cunha, D.H.; Berciano, S. Bioactive compounds for human and planetary health. Front. Nutr. 2023, 10, 1193848. [Google Scholar] [CrossRef]
- Farhud, D.D.; Yeganeh, M.Z. Nutrigenomics and Nutrigenetics. Iran. J. Public Health 2010, 39, 1. [Google Scholar] [PubMed]
- Liu, J.; Tuvblad, C.; Raine, A.; Baker, L. Genetic and environmental influences on nutrient intake. Genes Nutr. 2012, 8, 241–242. [Google Scholar] [CrossRef] [PubMed]
- Pokimica, B.; Garcia-Conesa, M.-T. Critical Evaluation of Gene Expression Changes in Human Tissues in Response to Supplementation with Dietary Bioactive Compounds: Moving Towards Better-Quality Studies. Nutrients 2018, 10, 807. [Google Scholar] [CrossRef]
- Niforou, A.; Konstantinidou, V.; Naska, A. Genetic Variants Shaping Inter-individual Differences in Response to Dietary Intakes—A Narrative Review of the Case of Vitamins. Front. Nutr. 2020, 7, 558598. [Google Scholar] [CrossRef]
- Mierziak, J.; Kostyn, K.; Boba, A.; Czemplik, M.; Kulma, A.; Wojtasik, W. Influence of the Bioactive Diet Components on the Gene Expression Regulation. Nutrients 2021, 13, 3673. [Google Scholar] [CrossRef] [PubMed]
- Chilton, F.H.; Manichaikul, A.; Yang, C.; O’COnnor, T.D.; Johnstone, L.M.; Blomquist, S.; Schembre, S.M.; Sergeant, S.; Zec, M.; Tsai, M.Y.; et al. Interpreting Clinical Trials With Omega-3 Supplements in the Context of Ancestry and FADS Genetic Variation. Front. Nutr. 2021, 8, 808504. [Google Scholar] [CrossRef]
- National Institute of Health; National Human Genome Research Institute. Talking Glossary of Genomic and Genetic Terms. Available online: https://www.genome.gov/genetics-glossary/Personalized-Medicine (accessed on 1 September 2025).
- Johnson, K.B.; Wei, W.; Weeraratne, D.; Frisse, M.E.; Misulis, K.; Rhee, K.; Zhao, J.; Snowdon, J.L. Precision Medicine, AI, and the Future of Personalized Health Care. Clin. Transl. Sci. 2021, 14, 86–93. [Google Scholar] [CrossRef]
- Chen, O.Y.; Roberts, B. Personalized Healthcare and Public Health in the Digital Age. Front. Digit. Health 2021, 3, 595704. [Google Scholar] [CrossRef]
- Blanco-Gonzalez, A.; Cabezon, A.; Seco-Gonzalez, A.; Conde-Torres, D.; Antelo-Riveiro, P.; Pineiro, A.; Garcia-Fandino, R. The Role of AI in Drug Discovery: Challenges, Opportunities, and Strategies. Pharmaceuticals 2023, 16, 891. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Rusnac, D.-V.; Park, H.; Canzani, D.; Nguyen, H.M.; Stewart, L.; Bush, M.F.; Nguyen, P.T.; Wulff, H.; Yarov-Yarovoy, V.; et al. An artificial intelligence accelerated virtual screening platform for drug discovery. Nat. Commun. 2024, 15, 7761. [Google Scholar] [CrossRef] [PubMed]
- Druedahl, L.C.; Price, W.N.; Minssen, T.; Sarpatwari, A. Use of Artificial Intelligence in Drug Development. JAMA Netw. Open. 2024, 7, e2414139. [Google Scholar] [CrossRef]
- Pitt, W.R.; Bentley, J.; Boldron, C.; Colliandre, L.; Esposito, C.; Frush, E.H.; Kopec, J.; Labouille, S.; Meneyrol, J.; Pardoe, D.A.; et al. Real-World Applications and Experiences of AI/ML Deployment for Drug Discovery. J. Med. Chem. 2025, 68, 851–859. [Google Scholar] [CrossRef]
- Zhang, K.; Yang, X.; Wang, Y.; Yu, Y.; Huang, N.; Li, G.; Li, X.; Wu, J.C.; Yang, S. Artificial intelligence in drug development. Nat. Med. 2025, 31, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Meijer, D.; Beniddir, M.A.; Coley, C.W.; Mejri, Y.M.; Öztürk, M.; van der Hooft, J.J.J.; Medema, M.H.; Skiredj, A. Empowering natural product science with AI: Leveraging multimodal data and knowledge graphs. Nat. Prod. Rep. 2024, 42, 654–662. [Google Scholar] [CrossRef]
- Chen, H.; King, F.J.; Zhou, B.; Wang, Y.; Canedy, C.J.; Hayashi, J.; Zhong, Y.; Chang, M.W.; Pache, L.; Wong, J.L.; et al. Drug target prediction through deep learning functional representation of gene signatures. Nat. Commun. 2024, 15, 1853. [Google Scholar] [CrossRef]
- Singh, P.; Bhat, S.S.; Singh, N.; Venkanna, B.U.; Mohamed, R.; Rao, R.P. Cell-Based Model Systems for Validation of Various Efficacy-Based Claims for Cosmetic Ingredients. Cosmetics 2022, 9, 107. [Google Scholar] [CrossRef]
- Markiewicz, E.; Idowu, O.C. Evaluation of Personalized Skincare Through in-silico Gene Interactive Networks and Cellular Responses to UVR and Oxidative Stress. Clin. Cosmet. Investig. Dermatol. 2022, 15, 2221–2243. [Google Scholar] [CrossRef]
- Singh, N.; Vayer, P.; Tanwar, S.; Poyet, J.-L.; Tsaioun, K.; Villoutreix, B.O. Drug discovery and development: Introduction to the general public and patient groups. Front. Drug Discov. 2023, 3, 1201419. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.A.; Wang, Z. Next-generation transcriptome assembly. Nat. Rev. Genet. 2011, 12, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Khatoon, Z.; Figler, B.; Zhang, H.; Cheng, F. Introduction to RNA-Seq and its Applications to Drug Discovery and Development. Drug Dev. Res. 2014, 75, 324–330. [Google Scholar] [CrossRef]
- Ye, C.; Ho, D.J.; Neri, M.; Yang, C.; Kulkarni, T.; Randhawa, R.; Henault, M.; Mostacci, N.; Farmer, P.; Renner, S.; et al. DRUG-seq for miniaturized high-throughput transcriptome profiling in drug discovery. Nat. Commun. 2018, 9, 4307. [Google Scholar] [CrossRef]
- Van de Sande, B.; Lee, J.S.; Mutasa-Gottgens, E.; Naughton, B.; Bacon, W.; Manning, J.; Wang, Y.; Pollard, J.; Mendez, M.; Hill, J.; et al. Applications of single-cell RNA sequencing in drug discovery and development. Nat. Rev. Drug Discov. 2023, 22, 496–520. [Google Scholar] [CrossRef] [PubMed]
- Aja, P.M.; Agu, P.C.; Ogbu, C.; Alum, E.U.; Fasogbon, I.V.; Musyoka, A.M.; Ngwueche, W.; Egwu, C.O.; Tusubira, D.; Ross, K. RNA research for drug discovery: Recent advances and critical insight. Gene 2025, 947, 149342. [Google Scholar] [CrossRef]
- Jobanputra, V.; Wrzeszczynski, K.O.; Buttner, R.; Caldas, C.; Cuppen, E.; Grimmond, S.; Haferlach, T.; Mullighan, C.; Schuh, A.; Elemento, O. Clinical interpretation of whole-genome and whole-transcriptome sequencing for precision oncology. Semin. Cancer Biol. 2022, 84, 23–31. [Google Scholar] [CrossRef]
- Fu, X.; Fu, N.; Guo, S.; Yan, Z.; Xu, Y.; Hu, H.; Menzel, C.; Chen, W.; Li, Y.; Zeng, R.; et al. Estimating accuracy of RNA-Seq and microarrays with proteomics. BMC Genom. 2009, 10, 161. [Google Scholar] [CrossRef]
- Ruan, M.; Liu, J.; Ren, X.; Li, C.; Zhao, A.Z.; Li, L.; Yang, H.; Dai, Y.; Wang, Y. Whole transcriptome sequencing analyses of DHA treated glioblastoma cells. J. Neurol. Sci. 2019, 396, 247–253. [Google Scholar] [CrossRef]
- Hernandez, D.F.; Cervantes, E.L.; Luna-Vital, D.A.; Mojica, L. Food-derived bioactive compounds with anti-aging potential for nutricosmetic and cosmeceutical products. Crit. Rev. Food Sci. Nutr. 2020, 61, 3740–3755. [Google Scholar] [CrossRef]
- Michalak, M.; Błońska-Sikora, E.; Dobros, N.; Spałek, O.; Zielińska, A.; Paradowska, K. Bioactive Compounds, Antioxidant Properties, and Cosmetic Applications of Selected Cold-Pressed Plant Oils from Seeds. Cosmetics 2024, 11, 153. [Google Scholar] [CrossRef]
- Goyal, A.; Sharma, A.; Kaur, J.; Kumari, S.; Garg, M.; Sindhu, R.K.; Rahman, H.; Akhtar, M.F.; Tagde, P.; Najda, A.; et al. Bioactive-Based Cosmeceuticals: An Update on Emerging Trends. Molecules 2022, 27, 828. [Google Scholar] [CrossRef] [PubMed]
- Gimenez Martinez, R.J.; García, F.R.; Cerdá, J.C.M.; Hernández-Ruíz, Á.; Castro, M.I.G.; Valverde-Merino, M.-I.; Camarasa, F.J.H.; Meseguer, F.L.; Gallardo, M.L.-V. Bioactive Substance and Skin Health: An Integrative Review from a Pharmacy and Nutrition Perspective. Pharmaceuticals 2025, 18, 373. [Google Scholar] [CrossRef] [PubMed]
- Youn, I.; Han, A.-R.; Piao, D.; Lee, H.; Kwak, H.; Lee, Y.; Nam, J.-W.; Seo, E.K. Phytochemical and pharmacological properties of the genus Alpinia from 2016 to 2023. Nat. Prod. Rep. 2024, 21, 1346–1367. [Google Scholar] [CrossRef] [PubMed]
- Min, K.Y.; Lee, K.A.; Kim, H.J.; Kim, K.-T.; Chung, M.-S.; Chang, P.-S.; Park, H.; Paik, H.-D. Antioxidative and anti-inflammatory activities of Citrus unshiu peel extracts using a combined process of subcritical water extraction and acid hydrolysis. Food Sci. Biotechnol. 2014, 23, 1441–1446. [Google Scholar] [CrossRef]
- Badgujar, S.B.; Patel, V.V.; Bandivdekar, A.H. Foeniculum vulgare Mill: A Review of Its Botany, Phytochemistry, Pharmacology, Contemporary Application, and Toxicology. Biomed Res. Int. 2014, 2014, 842674. [Google Scholar] [CrossRef]
- Kriplani, P.; Guarve, K.; Baghael, U.S. Arnica montana L.—A plant of healing: Review. J. Pharm. Pharmacol. 2017, 69, 925–945. [Google Scholar] [CrossRef]
- Hoang, H.T.; Moon, J.-Y.; Lee, Y.-C. Natural Antioxidants from Plant Extracts in Skincare Cosmetics: Recent Applications, Challenges and Perspectives. Cosmetics 2021, 8, 106. [Google Scholar] [CrossRef]
- Lin, T.-K.; Zong, L.; Santiago, J.L. Anti-Inflammatory and Skin Barrier Repair Effects of Topical Application of Some Plant Oils. Int. J. Mol. Sci. 2018, 19, 70. [Google Scholar] [CrossRef]
- Reddy, B.H.V.; Hussain, S.M.S.; Hussain, M.S.H.; Kumar, R.N.; Gupta, J. Essential oils in cosmetics: Antioxidant properties and advancements through nanoformulations. PRENAP 2025, 6, 100192. [Google Scholar] [CrossRef]
- Joyce-Brady, M.; Hiratake, J. Inhibiting Glutathione Metabolism in Lung Lining Fluid as a Strategy to Augment Antioxidant Defense. Curr. Enzym. Inhib. 2011, 7, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Aiuchi, T.; Nakajo, S.; Nakaya, K. Reducing Activity of Colloidal Platinum Nanoparticles for Hydrogen Peroxide, 2,2-Diphenyl-1-picrylhydrazyl Radical and 2,6-Dichlorophenol Indophenol. Biol. Pharm. Bull. 2004, 27, 736–738. [Google Scholar] [CrossRef] [PubMed]
- Benech, P.D.; Patatian, A. From experimental design to functional gene networks: DNA microarray contribution to skin ageing research. Int. J. Cosmet. Sci. 2014, 36, 516–526. [Google Scholar] [CrossRef][Green Version]
- Rao, M.S.; Van Vleet, T.R.; Ciurlionis, R.; Buck, W.R.; Mittelstadt, S.W.; Blomme, E.A.; Liguori, M.J. Comparison of RNA-Seq and Microarray Gene Expression Platforms for the Toxicogenomic Evaluation of Liver From Short-Term Rat Toxicity Studies. Front. Genet. 2018, 9, 636. [Google Scholar] [CrossRef] [PubMed]
- Anaconda, Inc. Conda. Version 3.27. [Computer Software]. Anaconda, Inc.: Austin, TX, USA, 2023. Available online: https://www.anaconda.com/ (accessed on 1 September 2025).
- Ritchie, M.E.; Phipson, B.; Wu, D.I.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Chen, Y.; Lun, A.T.L.; Smyth, G.K. From reads to genes to pathways: Differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Research 2016, 5, 1438. [Google Scholar]
- Law, C.W.; Alhamdoosh, M.; Su, S.; Dong, X.; Tian, L.; Smyth, G.K.; Ritchie, M.E. RNA-seq analysis is easy as 1-2-3 with limma, Glimma and edgeR. F1000Research 2016, 5, 1408. [Google Scholar] [CrossRef]
- Genesis Healthcare, Co. GenesisGaia [Computer Software]. 2024. Available online: https://www.adam-innovations.com/genesisgaia (accessed on 9 November 2025).
- Martin, F.J.; Amode, M.R.; Aneja, A.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Becker, A.; Bennett, R.; Berry, A.; Bhai, J.; et al. Ensembl 2023. Nucleic Acids Res. 2023, 51, D933–D941. [Google Scholar] [CrossRef]
- Edqvist, P.-H.D.; Fagerberg, L.; Hallström, B.M.; Danielsson, A.; Edlund, K.; Uhlén, M.; Pontén, F. Expression of Human Skin-Specific Genes Defined by Transcriptomics and Antibody-Based Profiling. J. Histochem. Cytochem. 2014, 63, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Gerber, P.A.; Hevezi, P.; Buhren, B.A.; Martinez, C.; Schrumpf, H.; Gasis, M.; Grether-Beck, S.; Krutmann, J.; Homey, B.; Zlotnik, A. Systematic Identification and Characterization of Novel Human Skin-Associated Genes Encoding Membrane and Secreted Proteins. PLoS ONE 2013, 8, e63949. [Google Scholar] [CrossRef] [PubMed]

| Bioactive Ingredient Name | Concentration |
|---|---|
| Alpinia speciosa Leaf Extract | 0.5% |
| Arnica montana Flower Extract | 1.0% |
| Citrus unshiu Peel Extract | 0.3% |
| Colloidal Platinum | 1.0% |
| Foeniculum vulgare Fruit Extract | 2.0% |
| Nahlsgen | 1.0% |
| Ingredient Name | Gene Name | Accession Number | Regulation | Log2 Fold Change | Adj. p. Val. |
|---|---|---|---|---|---|
| Alpinia speciosa | ASPRV1 | NM_152792 | DOWN | −1.12 | 8.73 × 10−4 |
| BLMH | NM_000386 | DOWN | −1.45 | 3.61 × 10−4 | |
| C1orf68 | NM_001024679 | DOWN | −1.31 | 1.05 × 10−3 | |
| CCL2 | NM_002982 | UP | 1.24 | 3.10 × 10−2 | |
| CXCL8 | NM_000584 | UP | 1.67 | 4.26 × 10−3 | |
| EDN1 | NM_001955 | UP | 1.12 | 4.48 × 10−2 | |
| EDN2 | NM_001956 | UP | 2.61 | 1.85 × 10−2 | |
| EDNRB | NM_001122659 | UP | 1.18 | 1.24 × 10−2 | |
| FLG | NM_002016 | DOWN | −1.32 | 4.33 × 10−3 | |
| FLG2 | NM_001014342 | DOWN | −1.07 | 6.71 × 10−3 | |
| HSPA1 | NM_005345 | UP | 1.01 | 4.27 × 10−3 | |
| IL37 | NM_014439 | DOWN | −1.30 | 2.25 × 10−3 | |
| IL6 | NM_000600 | UP | 1.23 | 1.58 × 10−3 | |
| LCE1A | NM_178348 | DOWN | −1.08 | 7.94 × 10−4 | |
| LCE1B | NM_178349 | DOWN | −1.12 | 1.50 × 10−3 | |
| LCE1C | NM_178351 | DOWN | −1.15 | 8.73 × 10−4 | |
| LCE1E | NM_178353 | DOWN | −1.32 | 8.88 × 10−4 | |
| LCE2A | NM_178428 | DOWN | −1.04 | 1.07 × 10−3 | |
| LCE2B | NM_014357 | DOWN | −1.24 | 6.73 × 10−4 | |
| LCE2C | NM_178429 | DOWN | −1.08 | 1.07 × 10−3 | |
| LCE4A | NM_001387222 | DOWN | −2.35 | 2.40 × 10−2 | |
| LCE5A | NM_178438 | DOWN | −1.26 | 3.93 × 10−3 | |
| MMP1 | NM_002421 | UP | 1.27 | 1.27 × 10−2 | |
| NFKBIA | NM_020529 | UP | 1.14 | 1.30 × 10−3 | |
| NGFR | NM_002507 | UP | 3.83 | 1.29 × 10−2 | |
| PSORS1C2 | NM_014069 | DOWN | −1.58 | 1.00 × 10−3 | |
| PTGS2 | NM_000963 | UP | 1.29 | 1.81 × 10−3 | |
| RPTN | NM_001122965 | DOWN | −1.29 | 1.44 × 10−2 | |
| SERPINA12 | NM_001382267 | DOWN | −1.77 | 3.61 × 10−4 | |
| THBS1 | NM_003246 | UP | 1.17 | 8.57 × 10−3 | |
| TLR2 | NM_001318789 | UP | 1.12 | 1.07 × 10−3 | |
| TNFAIP3 | NM_001270508 | UP | 1.21 | 1.32 × 10−3 | |
| TRPA1 | NM_007332 | UP | 1.13 | 4.06 × 10−2 | |
| Arnica montana | BLMH | NM_000386 | DOWN | −1.44 | 3.29 × 10−4 |
| C6orf15 | NM_014070 | DOWN | −1.09 | 7.89 × 10−4 | |
| DEFB103A | NM_001081551 | UP | 1.57 | 6.55 × 10−4 | |
| GCLC | NM_001498 | UP | 1.43 | 7.50 × 10−3 | |
| HBEGF | NM_001945 | UP | 1.03 | 4.85 × 10−3 | |
| HMOX1 | NM_002133 | UP | 1.81 | 2.37 × 10−3 | |
| HSPA1 | NM_005345 | UP | 1.06 | 1.12 × 10−2 | |
| LCE5A | NM_178438 | DOWN | −2.05 | 5.71 × 10−3 | |
| MMP1 | NM_002421 | UP | 1.33 | 1.78 × 10−2 | |
| NFKBIA | NM_020529 | UP | 1.08 | 9.59 × 10−3 | |
| NGFR | NM_002507 | UP | 4.80 | 1.38 × 10−3 | |
| PSORS1C2 | NM_014069 | DOWN | −1.79 | 4.52 × 10−3 | |
| SERPINA12 | NM_001382267 | DOWN | −2.11 | 6.07 × 10−3 | |
| SPRR2G | NM_001014291 | DOWN | −1.11 | 9.44 × 10−3 | |
| USP35 | NM_020798 | UP | 1.24 | 7.34 × 10−4 | |
| Citrus unshiu Peel | ASPRV1 | NM_152792 | DOWN | −1.36 | 3.29 × 10−4 |
| BLMH | NM_000386 | DOWN | −1.46 | 7.89 × 10−4 | |
| C1orf68 | NM_001024679 | DOWN | −1.82 | 6.55 × 10−4 | |
| CCL2 | NM_002982 | UP | 1.10 | 7.50 × 10−3 | |
| CXCL8 | NM_000584 | UP | 1.33 | 4.85 × 10−3 | |
| DEFB103A | NM_001081551 | UP | 1.82 | 2.37 × 10−3 | |
| EDNRB | NM_001122659 | UP | 1.22 | 1.12 × 10−2 | |
| FLG | NM_002016 | DOWN | −1.19 | 5.71 × 10−3 | |
| FLT1 | NM_002019 | UP | 1.16 | 1.78 × 10−2 | |
| HBEGF | NM_001945 | UP | 1.10 | 9.59 × 10−3 | |
| HSPA1A | NM_005345 | UP | 1.22 | 1.38 × 10−3 | |
| IL1B | NM_000576 | UP | 3.08 | 4.52 × 10−3 | |
| IL37 | NM_014439 | DOWN | −1.44 | 6.07 × 10−3 | |
| LAMA5 | NM_005560 | DOWN | −1.16 | 9.44 × 10−3 | |
| LCE1A | NM_178348 | DOWN | −1.07 | 7.34 × 10−4 | |
| LCE1B | NM_178349 | DOWN | −1.18 | 1.15 × 10−3 | |
| LCE1C | NM_178351 | DOWN | −1.44 | 4.29 × 10−4 | |
| LCE1D | NM_178352 | DOWN | −1.01 | 2.65 × 10−3 | |
| LCE1E | NM_178353 | DOWN | −1.34 | 4.44 × 10−3 | |
| LCE2A | NM_178428 | DOWN | −1.30 | 5.16 × 10−4 | |
| LCE2B | NM_014357 | DOWN | −1.42 | 6.32 × 10−4 | |
| LCE2C | NM_178429 | DOWN | −1.28 | 6.43 × 10−4 | |
| LCE2D | NM_178430 | DOWN | −1.25 | 1.15 × 10−3 | |
| LCE5A | NM_178438 | DOWN | −1.91 | 5.45 × 10−3 | |
| MMP1 | NM_002421 | UP | 1.04 | 7.90 × 10−4 | |
| NFKBIA | NM_020529 | UP | 1.29 | 3.99 × 10−4 | |
| NGFR | NM_002507 | UP | 3.84 | 1.31 × 10−2 | |
| NLRP10 | NM_001391958 | DOWN | −1.23 | 1.89 × 10−2 | |
| PSORS1C2 | NM_014069 | DOWN | −1.62 | 6.49 × 10−4 | |
| PTGS2 | NM_000963 | UP | 1.20 | 6.49 × 10−4 | |
| SERPINA12 | NM_001382267 | DOWN | −1.75 | 4.45 × 10−4 | |
| THBS1 | NM_003246 | UP | 1.07 | 3.53 × 10−3 | |
| TLR2 | NM_001318789 | UP | 1.08 | 3.41 × 10−3 | |
| TNFAIP3 | NM_001270508 | UP | 1.55 | 7.59 × 10−4 | |
| TRPV3 | NM_145068 | UP | 1.03 | 1.79 × 10−2 | |
| Foeniculum vulgare (Fennel) Fruit | BLMH | NM_000386 | DOWN | −1.17 | 1.46 × 10−3 |
| C1orf68 | NM_001024679 | DOWN | −1.57 | 2.88 × 10−3 | |
| CCL2 | NM_002982 | UP | 1.48 | 3.08 × 10−3 | |
| CXCL8 | NM_000584 | UP | 1.45 | 1.15 × 10−2 | |
| EDN1 | NM_001955 | UP | 1.13 | 2.83 × 10−2 | |
| FLT1 | NM_002019 | UP | 1.21 | 1.61 × 10−2 | |
| HSPA1 | NM_005345 | UP | 1.00 | 1.88 × 10−3 | |
| IL1B | NM_000576 | UP | 2.55 | 1.69 × 10−2 | |
| IL37 | NM_014439 | DOWN | −1.26 | 2.40 × 10−3 | |
| LAMA5 | NM_005560 | DOWN | −1.04 | 2.56 × 10−2 | |
| LCE1A | NM_178348 | DOWN | −1.04 | 1.79 × 10−2 | |
| LCE1B | NM_178349 | DOWN | −1.21 | 3.41 × 10−2 | |
| LCE1C | NM_178351 | DOWN | −1.18 | 1.70 × 10−2 | |
| LCE1D | NM_178352 | DOWN | −1.10 | 3.04 × 10−2 | |
| LCE1E | NM_178353 | DOWN | −1.50 | 1.59 × 10−2 | |
| LCE2A | NM_178428 | DOWN | −1.23 | 1.42 × 10−2 | |
| LCE2B | NM_014357 | DOWN | −1.20 | 8.86 × 10−3 | |
| LCE2C | NM_178429 | DOWN | −1.16 | 1.23 × 10−2 | |
| LCE2D | NM_178430 | DOWN | −1.19 | 3.13 × 10−2 | |
| LCE4A | NM_001387222 | DOWN | −2.18 | 3.33 × 10−2 | |
| LCE5A | NM_178438 | DOWN | −1.57 | 5.08 × 10−3 | |
| MMP1 | NM_002421 | UP | 1.17 | 3.41 × 10−3 | |
| NGFR | NM_002507 | UP | 3.43 | 3.54 × 10−2 | |
| OCLN | NM_001205254 | UP | 1.00 | 1.32 × 10−2 | |
| PSORS1C2 | NM_014069 | DOWN | −1.31 | 1.46 × 10−3 | |
| SERPINA12 | NM_001382267 | DOWN | 1.08 | 1.57 × 10−2 | |
| TLR2 | NM_001318789 | UP | −1.83 | 1.46 × 10−3 | |
| TNFAIP3 | NM_001270508 | UP | 1.01 | 2.67 × 10−3 | |
| TRPA1 | NM_007332 | UP | 1.01 | 5.26 × 10−3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Published by MDPI on behalf of the Österreichische Pharmazeutische Gesellschaft. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ogawa, M.; Crawford, C.W.; Ishigaki, A.; Sato-Baran, I.; Ordinario, D.D.; Miyashita, T. A Score-Based Rapid Screening and Network Visualization Method Based on Bioactive Ingredient-Induced Variations in Skin Cell Gene Expression. Sci. Pharm. 2025, 93, 56. https://doi.org/10.3390/scipharm93040056
Ogawa M, Crawford CW, Ishigaki A, Sato-Baran I, Ordinario DD, Miyashita T. A Score-Based Rapid Screening and Network Visualization Method Based on Bioactive Ingredient-Induced Variations in Skin Cell Gene Expression. Scientia Pharmaceutica. 2025; 93(4):56. https://doi.org/10.3390/scipharm93040056
Chicago/Turabian StyleOgawa, Mio, Charles W. Crawford, Ayumu Ishigaki, Iri Sato-Baran, David D. Ordinario, and Tadayoshi Miyashita. 2025. "A Score-Based Rapid Screening and Network Visualization Method Based on Bioactive Ingredient-Induced Variations in Skin Cell Gene Expression" Scientia Pharmaceutica 93, no. 4: 56. https://doi.org/10.3390/scipharm93040056
APA StyleOgawa, M., Crawford, C. W., Ishigaki, A., Sato-Baran, I., Ordinario, D. D., & Miyashita, T. (2025). A Score-Based Rapid Screening and Network Visualization Method Based on Bioactive Ingredient-Induced Variations in Skin Cell Gene Expression. Scientia Pharmaceutica, 93(4), 56. https://doi.org/10.3390/scipharm93040056








