Abstract
This study focuses on the preparation and optimization of caffeic acid (CA)-loaded casein nanoparticles (CS NPs) via the Box–Behnken design (BBD) for potential applications in cancer treatment. CS NPs were loaded with CA as a promising anti-cancer molecule. Non-hazardous green materials were exploited for nanoparticle fabrication. The BBD was used, followed by a desirability function to select the optimum formulation. The BBD was adopted as it avoids the runs implemented in extreme conditions, hence making it suitable for proteins. CS NPs were characterized regarding particle size (PS), size distribution (PDI), zeta potential (ZP), drug entrapment, morphology using TEM, differential scanning calorimetry, molecular docking, in vitro release, and cytotoxicity studies. PS, PDI, and ZP had significant responses, while EE% was insignificant. The suggested models were quadratic with high fitting. Optimized NPs showed PS = 110.31 ± 1.02 nm, PDI = 0.331 ± 0.029, ZP = −23.94 ± 1.64 mV, and EE% = 95.4 ± 2.56%. Molecular modeling indicated hydrophobic and electrostatic interactions between CA and CS, accounting for the high EE%. Almost spherical particles were realized with a sustained CA release pattern. Optimized NPs effectively suppressed the growth of MCF-7 cell lines by scoring the lowest IC50 = 78.45 ± 1.7 µg/mL. A novel combination of bioinspired-derived materials was developed for use in breast cancer treatment.
1. Introduction
Breast cancer (BC) is one of the most common lethal malignancies that cause death in women [1,2]. Its pathology is characterized by the overproduction of Rho proteins, resulting in cancer development, proliferation, breast cancer invasion, oncogenic transformation, and metastasis [3,4,5,6]. There are several tactics for BC management, such as surgery, chemotherapy, and radiotherapy. Surgery is commonly followed by the use of chemotherapeutic agents, also known as cytotoxic drugs, that interfere with cell division and growth. Although chemotherapy and radiotherapy showed great effectiveness in curbing tumor growth, acute and long-term adverse effects on patients’ healthy organs were observed, affecting patients’ survival rates and their quality of life [7,8]. Effects such as fatigue, weight loss, peripheral neuropathy, and nausea were seen and may extend to severe ones such as heart problems, osteoporosis, lymphedema, and concerns about cognitive functions [9,10,11]. Improving patients’ quality of life was our motivation for the nomination of safe green materials.
The main aim of the current research is to implement safe materials in nano-formulations in order to exploit the benefits of loaded drug while minimizing their undesirable effects. Accordingly, protein-based nanocarriers acquired researchers’ attention. They are biocompatible, biodegradable, cheap, and have several functional groups, allowing for their interaction with different compounds. They are GRAS (generally recognized as safe) compounds and can be easily manipulated into micro- and nanocarriers [12]. Animal and plant proteins can be used as drug carriers. In the former group, gelatin, collagen, albumin, and casein were extensively used as drug delivery systems, while later plant proteins such as zein, gliadin, soy protein, and lectins were also reported to be used for this task [13]. In our current research, we focus on casein as a nanocarrier system.
Casein (CS) is the major component of milk and belongs to the family of phosphorylated proteins (αS1-, αS2-, β, and k-caseins) [14,15]. It has a flexible structure composed of unfolded or randomly coiled peptides, rendering feasible intermolecular interactions as well as a paucity of disulfide bonds [16,17]. Consequently, CS is heat-stable and thus durable during pasteurization [18]. Moreover, CS has a low price and is easily produced, so its implementation in the pharmaceutical industry is recommended [19]. CS nanoparticles were easily prepared via three approaches. The first is copolymerization with synthetic polymers such as acrylic acid derivatives [20,21] or dextrans [22]. The second tactic is electrostatic complexation with polysaccharides [23,24]. The last one is based on simple coacervation by adding CaCl2 [25], enzymatic treatment with genipen [26], or chemical cross-linkage with glutaraldehyde [16].
Since we adopted the idea of gaining benefits from natural materials, the implemented drug chosen was caffeic acid (CA). CA originates from many fruits, vegetables, beverages (wine and coffee), and olive oil [27]. It exhibits anti-cancer effects due to its antioxidant effects by scavenging oxygen free radicals and chelating pro-oxidant metal ions [28]. CA exerts a significant role in reducing monocyte chemotactic protein 1 (MCP-1), inhibiting epithelial–mesenchymal transition (EMT), and downregulating E-cadherin expression accordingly, inhibiting BC metastasis. To sum up, CA possesses a significant influence in curbing cancer metastasis [29,30,31,32].
Nowadays, developing nanocarriers by utilizing quality in design is a main component in the pharmaceutical field. Mathematical models using statistics are strongly recommended in the pharmaceutical field. Response surface methodology (RSM) is a smart tool for both assessing and optimizing prepared nano-formulations. It gives a detailed statistical analysis of the factors implemented in the design and the required responses [33,34]. One of the main forms of the RSM is the Box–Behnken design (BBD). This is a very effective model for surveying the interactions between different involved factors and their impact on responses. Using the BBD is a significant tactic for investigating the maximum number of variables at different levels with a reduced number of experimental runs [35]. The BBD exhibits great merit in that it avoids combinations of extreme conditions that hinder the performance of experiments under extreme conditions that might affect the protein structure, resulting in potentially unsatisfactory conditions such as coagulation [36,37].
A great challenge in the present era of nanomedicine is the implementation and preparation of NPs using safe bioinspired materials free from any organic solvents. Accordingly, the entrapment of CA within CS NPs using a non-toxic cross-linking agent such as CaCl2 is considered a safe tactic. There is a scarcity of the literature regarding the use of CA as a loaded anti-cancer drug in nanoparticulate systems. CA and its derivative caffeic acid phenethyl ester (CAPE) exhibited antitumor and anti-metastatic effects against liver, breast, cervical, ovarian, lung, and colon cancer [38,39,40,41,42]. CAPE was synthesized in 1988 and extracted from propolis plants [43]. Several studies showed that CAPE is more cytotoxic than CA in cancerous cells [44]. Although several studies have encountered its efficacy as an anti-cancer agent, the chemical synthesis of CAPE involves the use of toxic chemicals and difficulties in its plant extraction and purification. A low-yield product is also realized. Moreover, if it is extracted from honey bee propolis, the process will be time-consuming in addition to the presence of many impurities [45]. On the other hand, Mücahit Secme and his team proved the safety of CA in human embryonic kidney (HEK293) cells used as normal cells. They observed that CA did not affect the cell viability or proliferation of HEK293 cells [44]. CA exhibits the characteristic merit that it is well tolerated by cancer patients and directly targets cancer stem cells (CSCs), thus preventing cancer recurrence, as CSCs are responsible for the resistance to chemotherapy and radiotherapy [39,46,47]. To the best of our knowledge, no research has focused on the encapsulation of CA as an anti-cancer active moiety in protein-based nanoparticulate systems.
The aim of our work was to formulate CA-loaded CSNPs via the simple coacervation method. The loaded CS NPs were then examined for their particle size (PS), polydispersity index (PDI), zeta potential (ZP), and entrapment efficiency (EE). The practical results were determined and then emulated by the response surface methodology using the Box–Behnken design (BBD) where the responses were evaluated statistically. Model validation was then performed; hence, successful navigation through the model space was endorsed. After that, the optimized formulation was further evaluated by applying in vitro characterization tests using transmission electron microscopy (HR-TEM), differential scanning calorimetry (DSC), in vitro drug release, and cytotoxicity studies.
2. Materials and Methods
2.1. Materials
Caffeic acid (CA) (HPLC grade) and casein (CS) from bovine milk of technical grade off-white powder for R&D use were obtained from Sigma-Aldrich Chemical Co. (Steinheim, Germany). Tween 80®, calcium chloride (CaCl2), sodium chloride, potassium chloride, disodium hydrogen phosphate, and potassium dihydrogen phosphate utilized as salts for phosphate buffer saline ‘PBS’ were purchased from Adwic, El-Nasr Pharmaceutical Co. (Cairo, Egypt). A Spectra/Por dialysis membrane having a 12,000–14,000 molecular weight cut off was obtained from Spectrum Laboratories Inc. (Rancho Dominguez, Compton, CA, USA). Deionized water was obtained from the MilliQ® purification system (Millipore, Bedford, MA, USA). RPMI-1640 medium, inactivated fetal calf serum, glutamine, and gentamycin were provided by BioWhittaker (Lonza, Belgium). Human breast carcinoma (MCF-7) cells were obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA).
2.2. Methodology
2.2.1. Preparation of CA-Loaded CS NPs
CA-loaded CS NPs were prepared by the simple coacervation method as previously reported [12,48]. A specific amount of CS was dissolved in deionized water adjusted at pH 8 using 0.1 N NaOH in order to increase the aqueous solubility of CS. Then, a weighed amount of CA was added, followed by stirring for 30 min to enhance drug–protein interaction and hence the entrapment of CA into CS NPs. The cross-linking agent CaCl2 (1 M) was added to the pre-mentioned solution, followed by stirring for 15 min. to allow for the interaction between Ca2+ ions and the formed NPs; hence, CA-loaded CS NPs would be precipitated. Centrifugation was then adopted for 2 min. at a low speed of 2000 rpm to remove the aggregated undesired microparticles [12]. The supernatant comprising the recommended particles was subjected to further characterization.
2.2.2. Computational Optimization and Modeling of CA-Loaded CS NPs
CA-loaded CS NPs were optimized using the response surface methodology where a relationship was constructed between the design factors (independent variables) and the experimental data responses (dependent variables). The Box–Behnken design (BBD) was selected to optimize the best design for drug-loaded CS NPs. The three independent factors were (A) CS concentration, (B) CaCl2 volume, and (C) CA amount, while the dependent variables were (Y1) PS, (Y2) PDI, and (Y3) ZP. According to the BBD, a total of 17 formulae of CA-loaded CS NPs were prepared; their compositions are presented in Table 1.

Table 1.
The composition of the prepared caffeic acid-loaded casein nanoparticles according to the Box–Behnken design.
2.2.3. Characterization of CA-Loaded CS NPs
PS, PDI and ZP Analysis
The PS and PDI values of all prepared CS NPs were determined through the dynamic light scattering (DLS) technique using a Zetasizer Nano ZS (Malvern instruments, Malvern, UK) at 25 °C using disposable polystyrene cells, while the ZP of the prepared NPs was measured via the laser doppler anemometry (LDA) technique using disposable, plain, folded capillary zeta cells. The samples were diluted with deionized water before measurements. The measurements were conducted in triplicate.
2.2.4. Determination of CA Entrapment Efficiency Percentage (EE %)
The EE% of CA-loaded CS NPs was determined by measuring the concentration of free CA in the formed dispersions. A total of 500 µL of each formula was diluted with 1.5 mL deionized water. The total volume (2 mL) was then placed in the Eppendorf® and subjected to centrifugation using a cooling centrifuge (Hermlez216MK, Hermle, Germany) at 9000 rpm and 4 °C for 1.5 h. The amount of unloaded CA in the filtrate was quantitatively determined using an HPLC assay, and the EE% was calculated as follows:
where Wt is the total amount of drug used in the formulation, and Wf is the amount of free CA remaining in the supernatant.
2.2.5. Molecular Docking Experiments
In order to interpret the entrapment efficiency of caffeic acid on casein nanoparticles, molecular docking experiments were performed. The crystal structure of casein (κ-casein) was obtained from the EMBL-EBI AlphaFold protein Structure Database (https://alphafold.ebi.ac.uk/entry/Q5ZGI1 accessed on 30 May 2024). The structure of the k-casein chain was encoded as UniProt Q5ZGI1. The IUPAC name corresponding to the chemical structure of the investigated molecule, caffeic acid, was obtained through PubChem®. The 3D configuration of the molecule was generated using the ChemDraw Ultra version 10 (Cambridgesoft, Waltham, MA, USA). Additionally, the energy minimization was attained using the MM2 forcefield of the same program [49]. The docking experiment was accomplished using ArgusLab v.4.50 (Mark Thompson and Planaria Software LLC, Seattle, WA, USA). The sybyl2 file of casein was imported to ArgusLab where the identification of the binding sites was performed [50] to be ready for docking using the ASscore of the program [51]. The binding site box size was 15 × 15 × 15 angstroms. Additionally, the Dock and Flexible calculations were used at the maximum number of 150 poses.
2.2.6. Transmission Electron Microscope Imaging
The shape and morphology of the optimized CS NP formulae were examined using an HR-TEM instrument (Jeol Electron Microscope, JEM-1010, Tokyo, Japan). A droplet of the nanosuspension was placed on a carbon film-covered copper grid (200-mesh) and stained using 2% phosphotungstic acid. A filter paper placed at the edge of the copper grid was used to remove any excess liquid. Subsequently, the sample was air-dried for about 5 min. before observation by TEM [52,53,54].
2.2.7. Differential Scanning Calorimetry
The thermal properties of CA, CS, unloaded CS NPs, and the optimized CA-loaded CS NPs were investigated using DSC (Shimadzu DC-60, Kyoto, Japan). Samples were sealed in aluminum pans with lids and heated at a rate of 10 °C/min. to a temperature of 300 °C, using dry nitrogen as the carrier gas with a flow rate of 25 mL/min. Indium was used as the standard reference material to calibrate the energy and the scale temperature of the instrument.
2.2.8. In Vitro Release Study of CA from Optimized CS NPs
The in vitro drug release of CA from the optimized formulae was performed by the dialysis bag diffusion technique using a shaking water bath (GFL1083 Gesellschaft für Labortechnik, Burgwedel, Germany). An accurately weighed amount of the CA-loaded CS NP formula equivalent to 8.9 mg CA was filled in a dialysis bag and soaked in a bottle filled with 100 mL PBS, pH 7.4, containing 0.2% w/v Tween 80 [26,55], to provide medium preservation and ensure sink conditions during the whole duration of the release experiment. The bottle was tightly closed and then placed in a shaking water bath maintained at a temperature of 37 ± 0.5 °C. Two-milliliter samples were withdrawn at different time intervals, 0.5, 1, 2, 4, 6, 8, 24, 48, 72, and 96 h, replaced with a fresh volume of dissolution medium, diluted appropriately, and drug concentrations were then measured by HPLC. The experiments were performed in triplicate.
2.2.9. IC50 Determination by Cell Culture Studies
Human Breast Carcinoma Cell Line
Human breast carcinoma (MCF-7) cells were grown on RPMI-1640 medium supplemented with 10% inactivated fetal calf serum and 50 µg/mL gentamycin. The cells were maintained at 37 °C in a humidified atmosphere with 5% CO2 and were sub-cultured two to three times a week.
2.2.10. Evaluation of In Vitro Antitumor Activity of Optimized CA-Loaded CS NPs
The optimized loaded CS NPs and the drug solution were assessed in terms of their antitumor activities against MCF-7 cell lines. Moreover, the plain formula was implemented in this study for proving the safety of the bioinspired NPs. The RPMI-1640 medium was supplemented with 10% inactivated fetal calf serum and 50 µg/mL gentamycin. The MCF-7 cells were grown as monolayers of 10,000 cells in the previously mentioned medium. The monolayers were adhered at the bottom of the wells in a 96-well microliter plate and incubated in a humidified incubator (Shellab water jacketed CO2 Incubators, Inc., American Laboratory Trading, San Diego, CA, USA) for 24 h at 37 °C with 5% CO2. They were washed with sterile PBS (0.01 M, pH 7.2). A total of 100 µL from different dilutions of the tested sample was then added to the cells while being incubated at 37 °C. Each concentration of the examined sample was examined using six wells. Untreated cells served as the control ones. After 24 h, the cells were stained with crystal violet (Sigma-Aldrich Chemical Co., Steinheim, Germany) for determining the number of surviving cells. Cell lysis was caused by 33% glacial acetic acid, and the absorbance of the stained cells using crystal violet was estimated at 590 nm using an ELISA reader (SunRise, TECAN, Inc., San Jose, CA, USA) after well mixing. The number of viable cells was determined by the ELISA reader. The absorbance of the control cells being untreated was determined as 100% proliferation.
The viability percentage was estimated using the following equation
where ODt is the mean optical density of wells treated with the examined sample, and ODc is the mean optical density of untreated cells. The graphical plots determined the 50% inhibitory concentration (IC50), which is the concentration required to cause toxic effects in 50% of intact cells.
(ODt/ODc) × 100
2.2.11. Statistical Analysis
Each experiment was conducted in triplicate, and the mean values ± SD were calculated. Designing the experiments using the Box–Behnken design and the generation of models and 3D response surface and contour plots were performed using Design Expert v. 7.0. (Design Expert® Software, Stat-Ease Inc., Minneapolis, MN, USA). The obtained data were statistically analyzed by applying a one-way ANOVA test. Differences with p-values < 0.05 were considered significant.
The validity of the generated models was assessed by calculating the percentage bias using the following equation for three randomly chosen formulae other than those prepared in the statistical design:
Numerical optimization using the desirability function (D) was conducted by minimizing the values for the PS, PDI, and ZP responses. The optimized formulation was picked up based on the formulation scoring the highest D value.
The desirability function “D” is a functional tool used to optimize the studied responses. It aims to reach the most appropriate and compromising point in the design space that accomplishes the set goal for the dependent variables [56]. It involves the transformation of each estimated response variable to a desirability value di, where 0 ≤ di ≤ 1. The higher the desirability of the corresponding responses, the higher the D value that would be attained.
The desirability value for the response to be minimized can be calculated from the following equation:
where di,min is the individual desirability of the response to be minimized, Ymin is the lowest acceptable value, Ymax is the highest acceptable value, and Yi indicates the experimental value. If Yi is greater than Ymax, then di = 0, and if Yi is less than Ymin, then di = 1 [56].
To obtain the D value by combining individual desirabilities, the geometric mean is utilized as follows:
D = (d1x d2x d3x…….x dk)1/k
The D value becomes higher as the properties of the responses become more favorable. In other words, if any response has di = 0, meaning that one of the response variables is unacceptable, then D = 0, meaning that the overall product value is unacceptable, which is the underlying reason for using the geometric mean rather than the arithmetic mean [57].
3. Results and Discussion
3.1. Preparation and Optimization of CA-Loaded CS NPs
CA-loaded CS NPs were prepared by the simple coacervation method as reported by Sona Gandhi, Indrajit Roy, 2019 and Rebeca Penalva, et al., 2014 [12,48]. The selected parameters adopted in this study were based on previous studies where CS concentration and CaCl2 volume were used at 2% w/v and 50 µL, respectively [12]. As for CS concentration the ranges from 1 to 3% w/v were selected while CaCl2 volume ranged from 25 to 75 µL in addition to the CA amount ranging from 10 to 30 mg. By raising the CS concentration or CA amount, a significant increase in PS was realized, reaching 1 µm along with observed precipitation. The selected CA amount range was related to its reported cytotoxic effect [58].
The BBD was adopted for the optimization of the prepared CS NPs rather than other response surface models, i.e., central composite and full factorial designs as the BBD does not govern points in which all factors are simultaneously at their highest or lowest levels. Therefore, the BBD was used in the safe operating zone of the experimental space [59]. Accordingly, the BBD is beneficial for delicate structures such as proteins and for the avoidance of experiments performed under extreme conditions for which unsatisfactory results might occur [60,61]. The BBD implements fewer runs [62]. The number of runs is usually calculated as follows, where the design points lie on the center or in the middle of the edges of the design cubic space for a three-factor experiment: [59]
where N = the number of experiments (runs) performed according to the BBD, k = the number of the investigated factors, and C0 represents the central points. It is worthy to note that additional central points or replicates were added for further confirmation.
N = (2k(k − 1)) + C0
CS concentration (A), CaCl2 volume (B), and CA amount (C) were the three independent variables; hence, a set of 17 formulae were prepared in addition to three formulae that were used as check points to validate the model.
Table 2 demonstrates that the PS of the prepared CA-loaded CS NPs is in the range from 108.93 ± 2.25 to 268.63 ± 3.88 nm with PDI values varying from 0.301 ± 0.133 to 0.499 ± 0.018. CA-loaded CS NPs manifested negative surface charges within the range from 10.45 ± 0.37 up to −27.44 ± 0.17 mV.

Table 2.
The response data of CA-loaded CS NP formulae prepared according to the BBD.
CA is highly encapsulated within the matrix of CS NPs exhibiting an EE% range from 93.52 ± 4.98 to 97.26 ± 1.98%, as depicted in Table 2. This could be from the high binding affinity between the CS protein and the drug [63] ascribed to the electrostatic interaction between these two entities that enhanced the CA entrapment within the CS matrix, thus lowering the presence of free molecules [64].
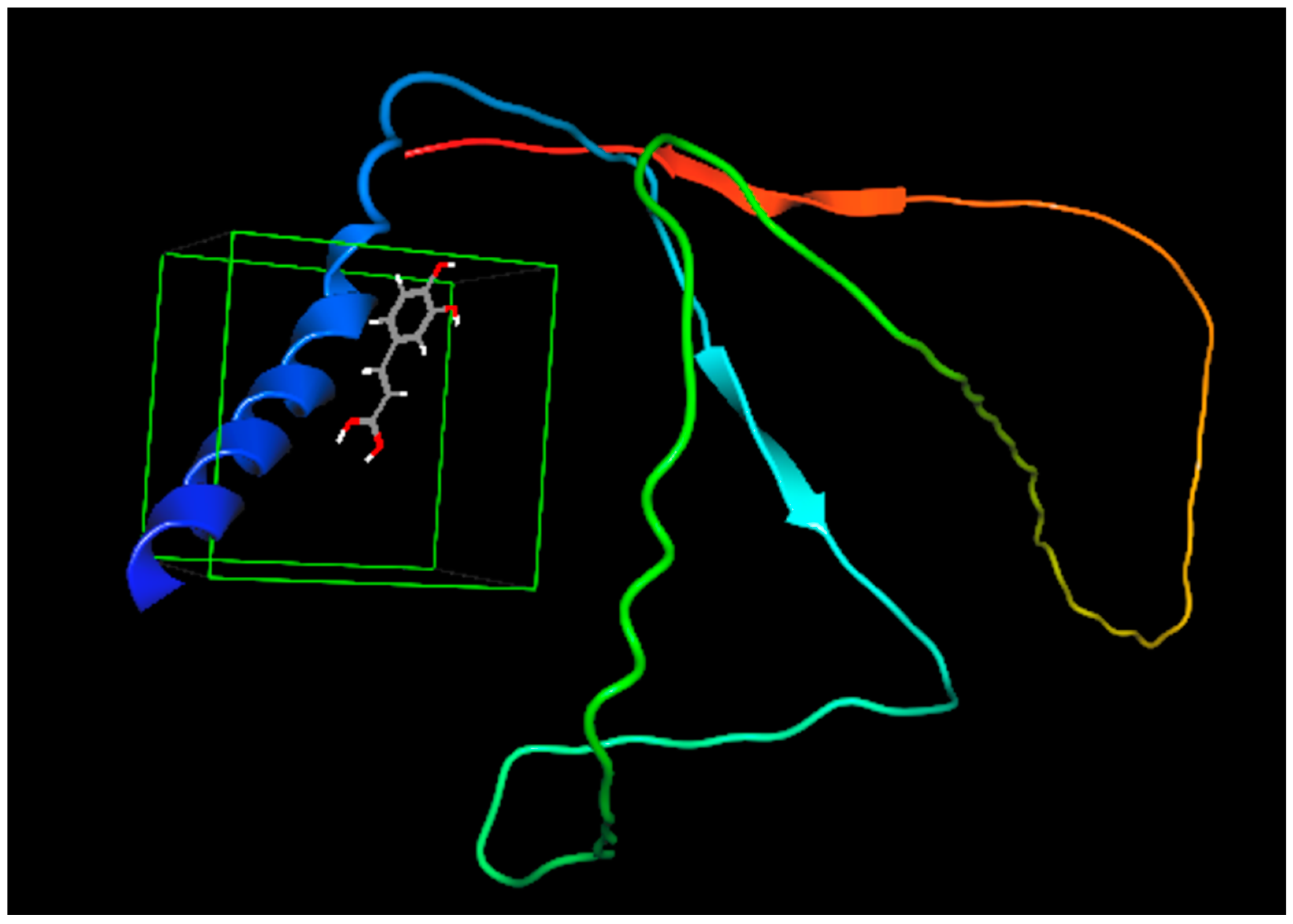
CA possesses a double bond between the carboxylic group and the aromatic ring. This double bond represents a large electron-less conjugated system. Moreover, the CS protein includes aromatic amino acids, e.g., tryptophan, tyrosine, and phenylalanine groups, in its binding cavity of the protein. These amino acids encompass conjugated π-electrons, giving them the chance to form charge-transfer complexes with other functional groups lacking electrons or with π-electrons [65]. Therefore, the double bond in the CA molecule would prolong the contact with the binding pockets in the protein. These could account for the greater affinity (ΔG = −6.021 kCal/mole) and thus the enhanced binding mode between CA and the CS protein [66]. The interaction between caffeic acid (log P = 1.35) [67] and casein is demonstrated in Figure 1. The interaction of caffeic acid with similar hydrophobic amino acids such as leucine, isoleucine, valine, and proline is postulated, where hydrophobic forces prevail.
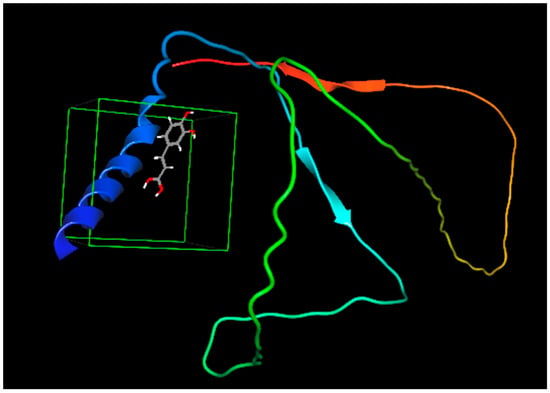
Figure 1.
Binding affinity between caffeic acid and casein. The colored structures are the protein chains (red and green), the α-helices (blue coiled ribbon), β-sheets (the red and light blue colored ar-rows) are of the protein while the grey structure is caffeic acid.
Furthermore, based on the chemical structure of CA, the functional groups para-OH, meta-OH, and COOH would act as hydrogen bond donors/acceptors for different side chain groups of the protein; hence, a further stabilization of the CS-CA interaction would be implemented by the electrostatic interaction [64].
3.2. Model Analysis
After an analysis of the results using an ANOVA, the suggested models for the studied responses, PS, PDI, and ZP, are quadratic ones, as shown in Table 3. A p-value less than 0.5 indicates that the model is significant. As revealed in Table 3, the suggested models for the responses demonstrate a high level of significance as the p-value is <0.0001 for the PS, PDI, and ZP responses.

Table 3.
Model analysis results of investigated responses according to BBD.
Furthermore, these three responses are highly fitted to the suggested models, with an R2 of 0.9996, 0.8760, and 0.8835 for the PS, PDI, and ZP, respectively, as shown in Table 3.
Moreover, the suggested models were tremendously successful where navigating the design space was possible. This was confirmed by the term ‘adequate precision’ as it measures the range of a predicted response relative to its associated error, or with further elucidation, it measures the signal-to-noise ratio. A ratio greater than 4 is usually desirable for navigating any design space [68]. This was accomplished for the three responses where these ratios attained 119.867, 14.311, and 14.366 for the PS, PDI, and ZP models, respectively.
‘PRESS’ is the Predicted Residual Sum of Squares for the model, and it measures how well a particular model fits each point in the design. As clearly shown in Table 2, the three models are fitting each point in the design where the PS, PDI, and ZP data are greatly fitted to the model with PRESS values equal to 266.66, 0.017, and 125.06, respectively. Similar values of the PRESS were obtained by Garg and Prasad, showing the high fitting of the model [69,70].
Table 3 shows low values for C.V.% which are found to be equal to 1.08, 7.06, and 10.12 for the three responses: PS, PDI, and ZP, respectively. The low value for C.V.% means a lower deviation of the data around the mean; hence, the suggested models were successful, and navigation through the design space was credible. A C.V.% less than 10% indicates lower variation in the actual and model-predicted data, and this is desirable [69]. Furthermore, a non-significant lack of fit was obtained for all the models, confirming the fitting of the data on the investigated models [59].
3.3. Model Diagnostics
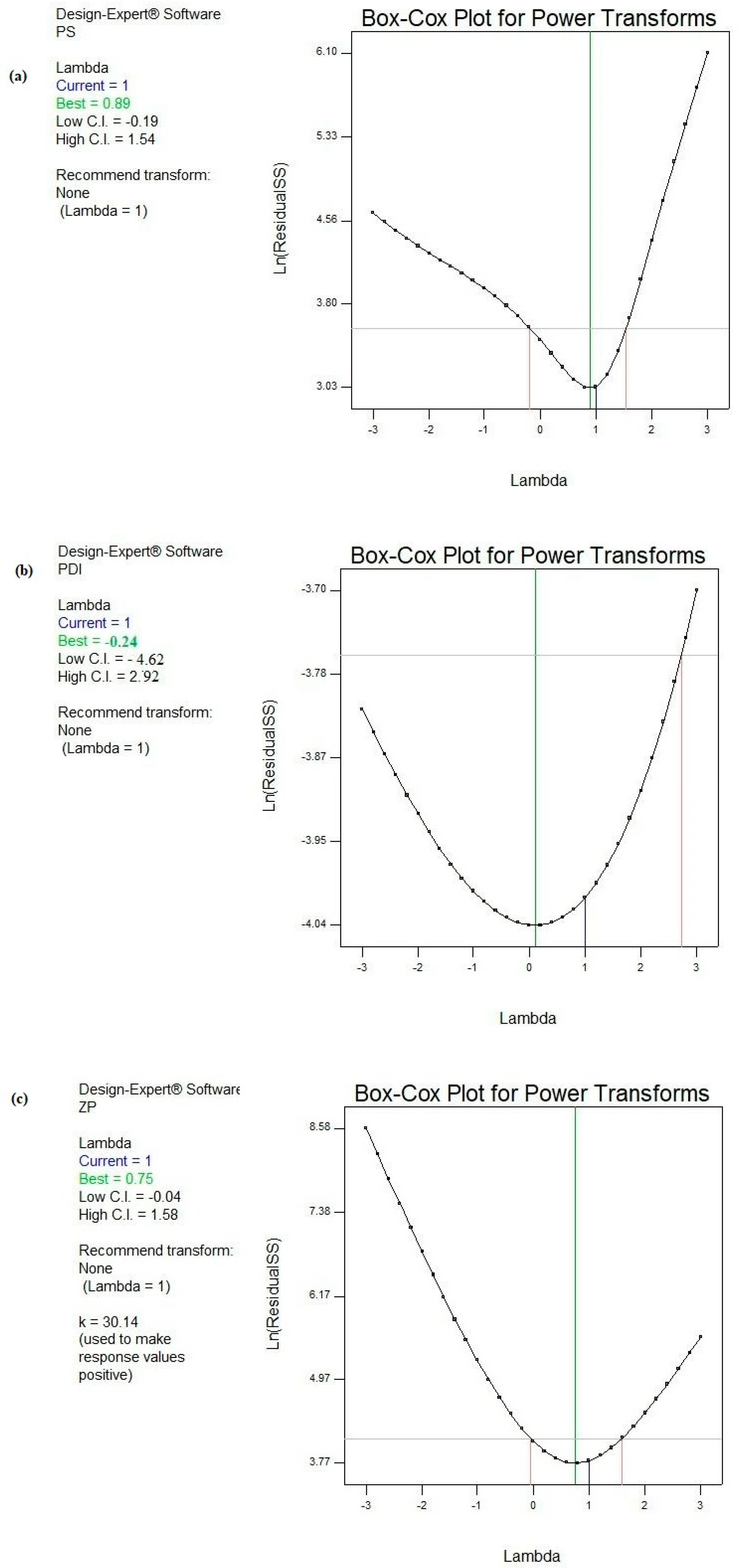
Figure 2 demonstrates the Box-Cox power transformation that usually suggests the best power (lambda) that all the response data should be raised to in order to obtain the best-fitting models.
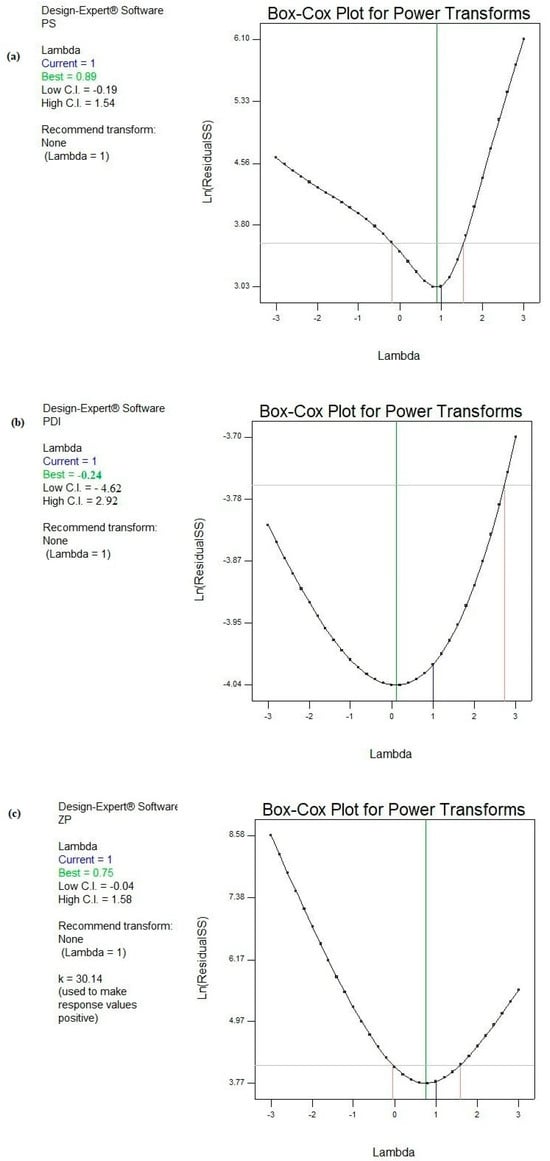
Figure 2.
Box–Cox diagnostic test for power transformations for responses (a) PS, (b) PDI, and (c) ZP.
Consequently, Box–Cox plots did not suggest any recommendations for the responses PS, PDI, and ZP. In other words, the current powers (lambda) were found to precisely coincide with the best lambdas and were located between the low and high confidence intervals for the pre-mentioned models [71,72,73].
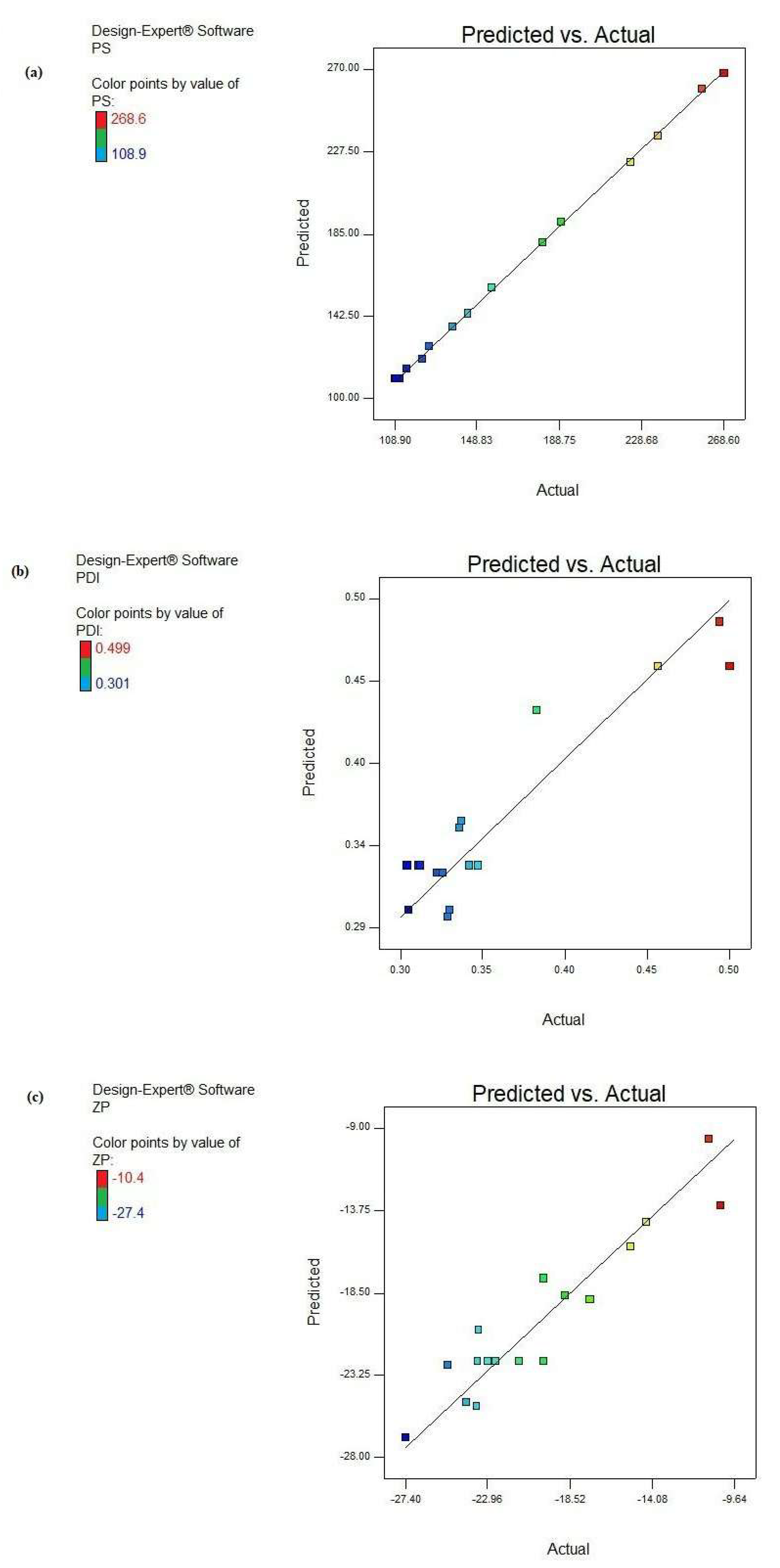
Figure 3 shows a graphical representation of the predicted data versus the actual ones.
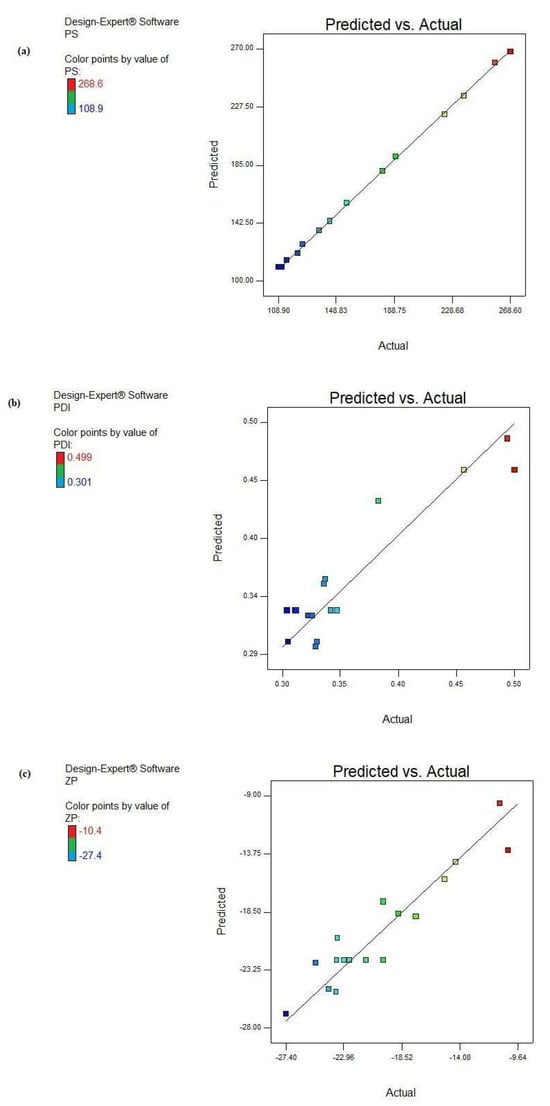
Figure 3.
Predicted versus actual result plots of the responses: (a) PS, (b) PDI, and (c) ZP.
It is clear from this figure that almost all the points were in close vicinity to the line 45°, elucidating a higher correlation between the experimental and predicted results for the three examined models. These graphs were in great harmony with the high obtained R2 values.
3.4. The Interpretation of the Models’ Results
3.4.1. The PS Response
CA-loaded CS NPs recorded PS in a range from 108.93 ± 2.25 to 268.63 ± 3.88 nm, as shown in Table 2. The adopted method for the preparation of CS NPs was the simple coacervation technique where CaCl2 served as a cross-linking agent. These data were consistent with those obtained by Sona Gandhi, Indrajit Roy, 2019 and Rebeca Penalva, et al., 2014 [12,48].
The three independent variables studied, CS concentration (A), CaCl2 volume (B), and CA amount (C), were found to be highly significant terms (p < 0.0001) affecting the PS response, as depicted from the ANOVA results in Table 4. Moreover, all the possible 2-FI interactions are highly significant terms scoring p < 0.0001, as observed in Table 4.

Table 4.
ANOVA test results of all responses studied according to BBD.
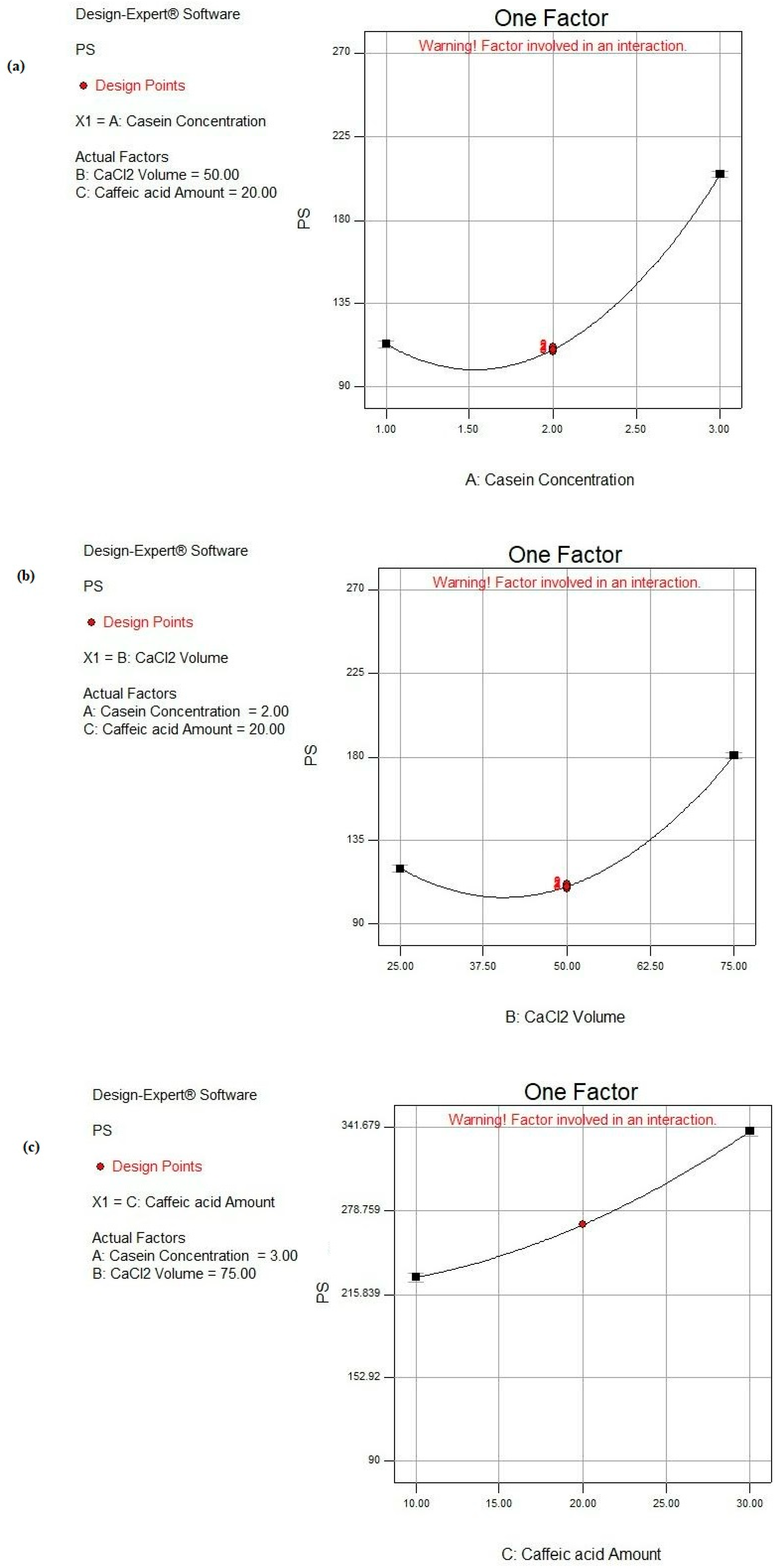
As shown in Figure 4, by increasing the CS concentration from 1 to 3%, the PS increased significantly (p < 0.05) from 136.81 ± 0.33 to 257.81 ± 3.14 nm, as attained in F2 and F16, from 189.74 ± 0.95 to 268.63 ± 3.88 nm in F1 and F14, from 122.11 ± 3.52 to 180.83 ± 2.89 nm in F3 and F17, and similarly in F4 and F15, the PS increased from 114.52 ± 5.13 to 223.34 ± 5.76 nm, respectively, while maintaining other parameters constant. When the proportion of the CS protein in the colloidal phase increased, denser self-assembled CS NPs were attained, so a greater PS of the CS NPs together with more dispersed and more negatively charged NPs were fulfilled [74].
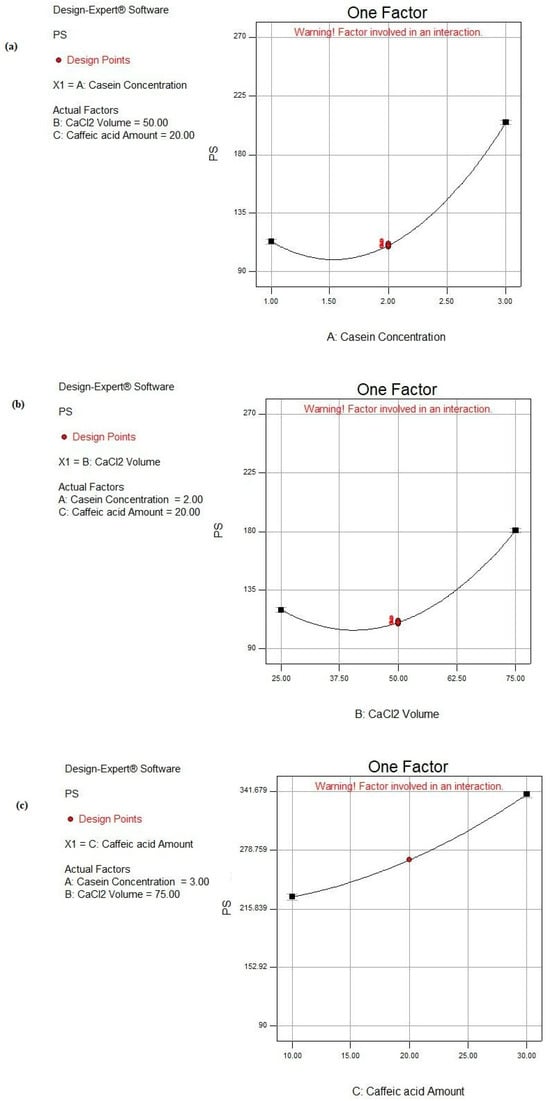
Figure 4.
Main effect plots of individual factors: (a) A: CS concentration, (b) B: CaCl2 volume, and (c) C: CA amount on PS response.
CaCl2 was the source of the Ca2+ pool that induced the aggregation of CS micelles. The Ca2+ ions performed bridging between the CS molecules. Accordingly, a reduction in the electrostatic repulsion of the CS molecules was attained owing to ion binding and the electrostatic screening effect [75]. Protein aggregation was executed under certain conditions where the net attractive forces (i.e., van der Waals and hydrophobic) surpassed the net repulsive forces (i.e., electrostatic and steric), and this occurred at high ionic strength [76]. Accordingly, the addition of Ca2+ to CS micelles lessened the strength of the electrostatic repulsive forces, causing the self-aggregation and more packing of free CS molecules. Therefore, the subsequent inevitable precipitation of nanomicelles into dense NPs was achieved [12,76]. As observed in Table 2 and Figure 4, the PS significantly increased (p < 0.05) by increasing the volume of CaCl2 from 25 to 75 μL under similar formulation conditions, as in the cases of F4 and F1, F12 and F13, and F15 and F14, where the PS raised from 114.52 ± 5.23 to 189.74 ± 0.95 nm, 125.44 ± 0.96 to 156.53 ± 0.28 nm, and 223.34 ± 5.76 to 268.63 ± 3.88 nm, respectively.
As shown in Figure 4, increasing the CA amount led to a significant increase in PS (p < 0.05), as demonstrated in the cases of F3 and F2 where PS increased from 122.11 ± 3.52 to 136.81 ± 0.33 nm when the CA amount increased from 10 to 30 mg. Similar observations were observed where PS raised from 125.44 ± 0.96 to 144.34 ± 2.81 nm, from 156.53 ± 0.28 to 236.83 ± 1.34 nm, and from 180.83 ± 2.89 to 257.81 ± 3.14 nm when elevating the CA amount from 10 to 30 mg in the cases of F12 and F7, F13 and F8, and F17 and F16, respectively. These observations were also documented by Görner et al. [77]. This could be due to the fact that more CA was entrapped within the core of the CS NPs. The later observation was based on the high hydrophobic interaction between the phenolic rings of both CA and the CS protein [78].
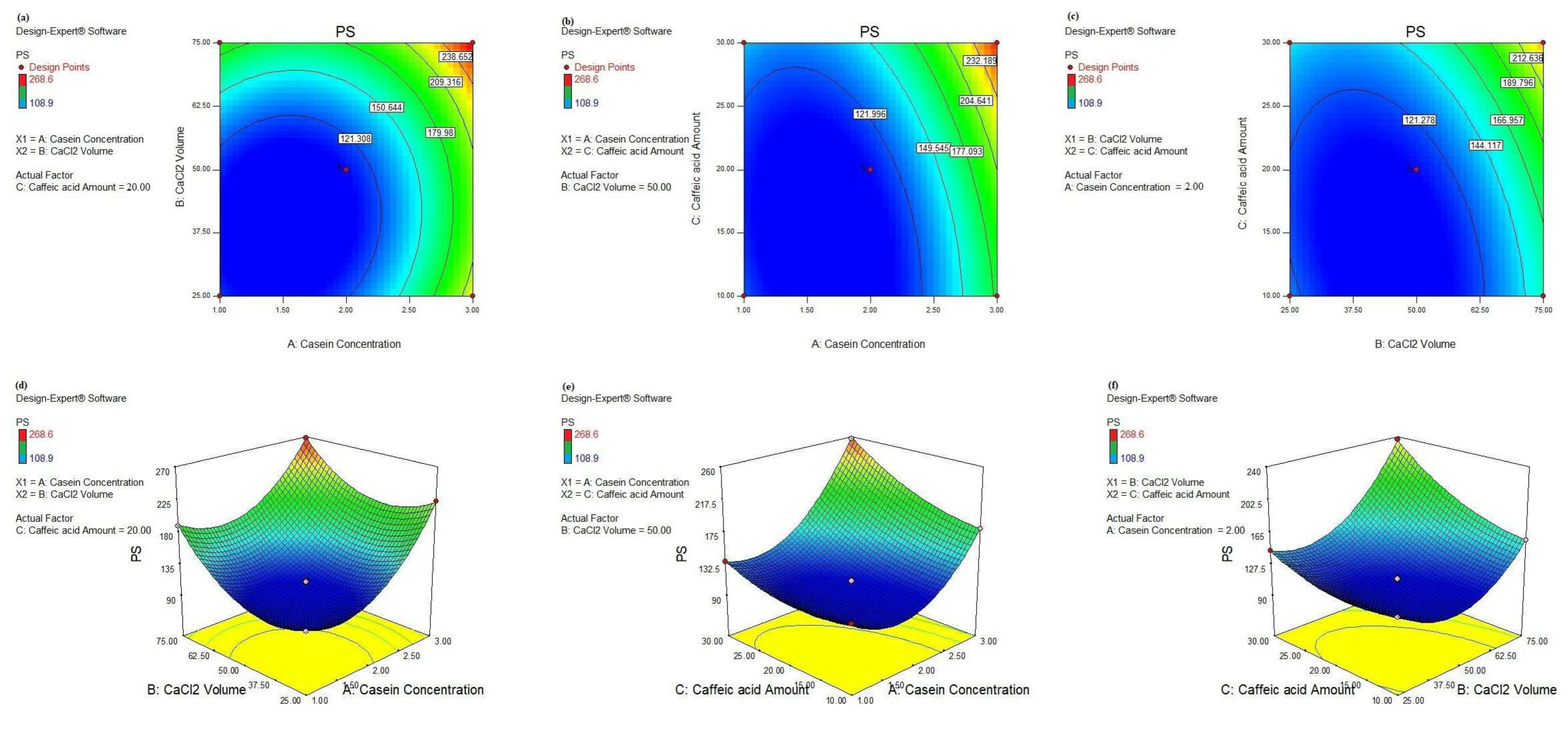
Further navigating through the parameters affecting the PS response, we investigated the effect of two-way interactions as well as their magnitudes, as depicted in Table 4. The significant 2-FI AB interaction scored the lowest F-value of 75.68, confirming its smaller effect on the PS response than the other interactions. As observed in Figure 5a,d, the minimal PS (108.9 nm) was attained when the CS concentration was less than 2 g% and the CaCl2 volume was less than 62.5 µL. Also, the 2-FI AC interaction realized the highest F-value = 328.56, affirming its significant effect on the PS where the smallest PS was achieved by lowering the CS concentration and CA amount to less than 2.5 g% and 25 mg, respectively, as shown in Figure 5b,e. A similar pattern was achieved in the case of the significant 2-FI BC interaction where the lowest PS was scored when the CaCl2 volume and the CA amount were less than 62.5 µL and 25 mg, respectively, as illustrated in Figure 5c,f.
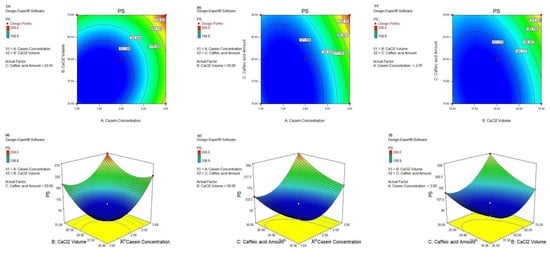
Figure 5.
Contour plots (a–c): AB, AC, and BC and 3D response surface plots (d–f): AB, AC, and BC, respectively, of significant 2-FI interactions on PS response, respectively.
3.4.2. The PDI Response
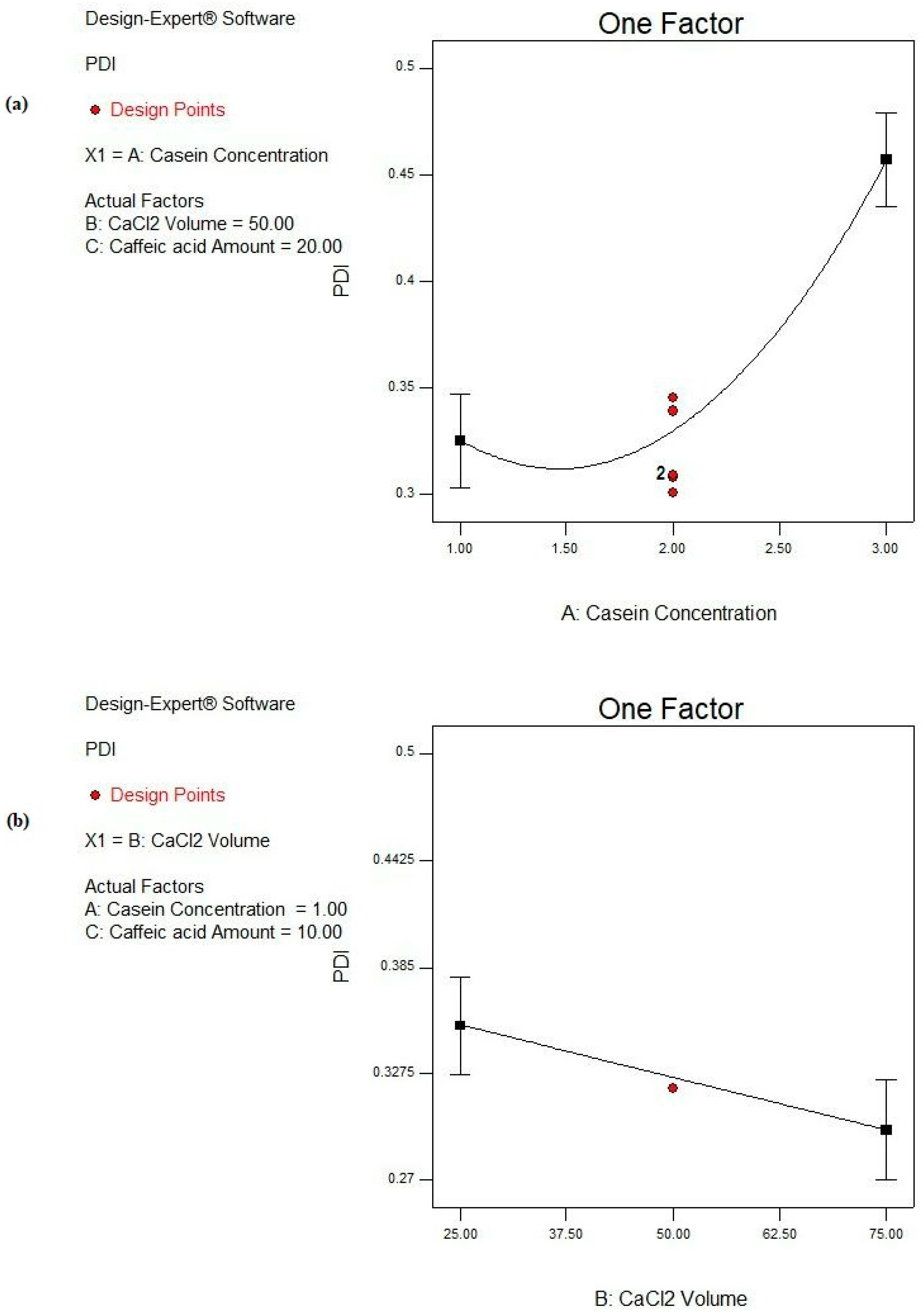
Based on the ANOVA results shown in Table 4, the CS concentration (A) is a highly significant factor (p ˂ 0.0001) affecting the PDI response. Moreover, the CaCl2 volume (B) reveals a significant effect on the PDI response (p ˂ 0.05), while the CA amount (C) is a non-significant parameter (p > 0.05), as depicted in Table 4 and Figure 6. Moreover, no significant interactions between the studied factors are deduced from the design, as observed in Table 4.
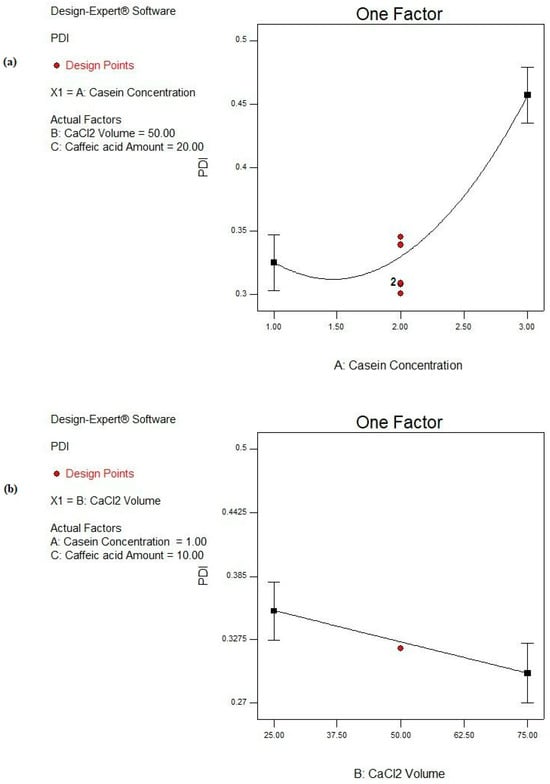
Figure 6.
Main effect plots of significant individual factors; (a) A: CS concentration and (b) B: CaCl2 volume on PDI response.
By increasing the CS concentration from 1 to 3%, the PDI is significantly increased (p < 0.05), as illustrated in Figure 6a, and this is revealed in the cases of F2 and F16, F3 and F17, and F4 and F15 from 0.323 ± 0.041 to 0.455 ± 0.076, from 0.326 ± 0.092 to 0.499 ± 0.018, and from 0.333 ± 0.128 to 0.493 ± 0.004, respectively, as presented in Table 2. Soleimanifar et al. and Amighi et al. showed similar observations where the increase in the protein concentration resulted in a heterogeneous distribution of the particles [79,80]. This could be due to the fact that the elevation in the protein concentration would result in a higher viscosity of the resulting solution, as well as enhancing molecular crowding. The latter rendered higher PDI values [81]. Furthermore, the increase in the protein concentration would evolve a higher protein aggregation with more diverse nanoparticles [82].
Moreover, CaCl2 volume shows a great impact on the degree of heterogeneity of the formed CS NPs as increasing the amount of Ca2+ significantly reduces the PDI (p < 0.05), as shown in Figure 6b. This observation is fulfilled in the case of F15 and F14 where the values of the PDI are minimized from 0.493 ± 0.004 to 0.381 ± 0.012 upon raising the volume of CaCl2 from 25 to 75 µL, as depicted from Table 2. This could be interpreted on the basis of the structure of CS molecules. More packing of free CS molecules together boosted their self-assembly and resulted in the precipitation of CS NPs in a more homogenous, less dispersed pattern [83,84].
3.4.3. The ZP Response
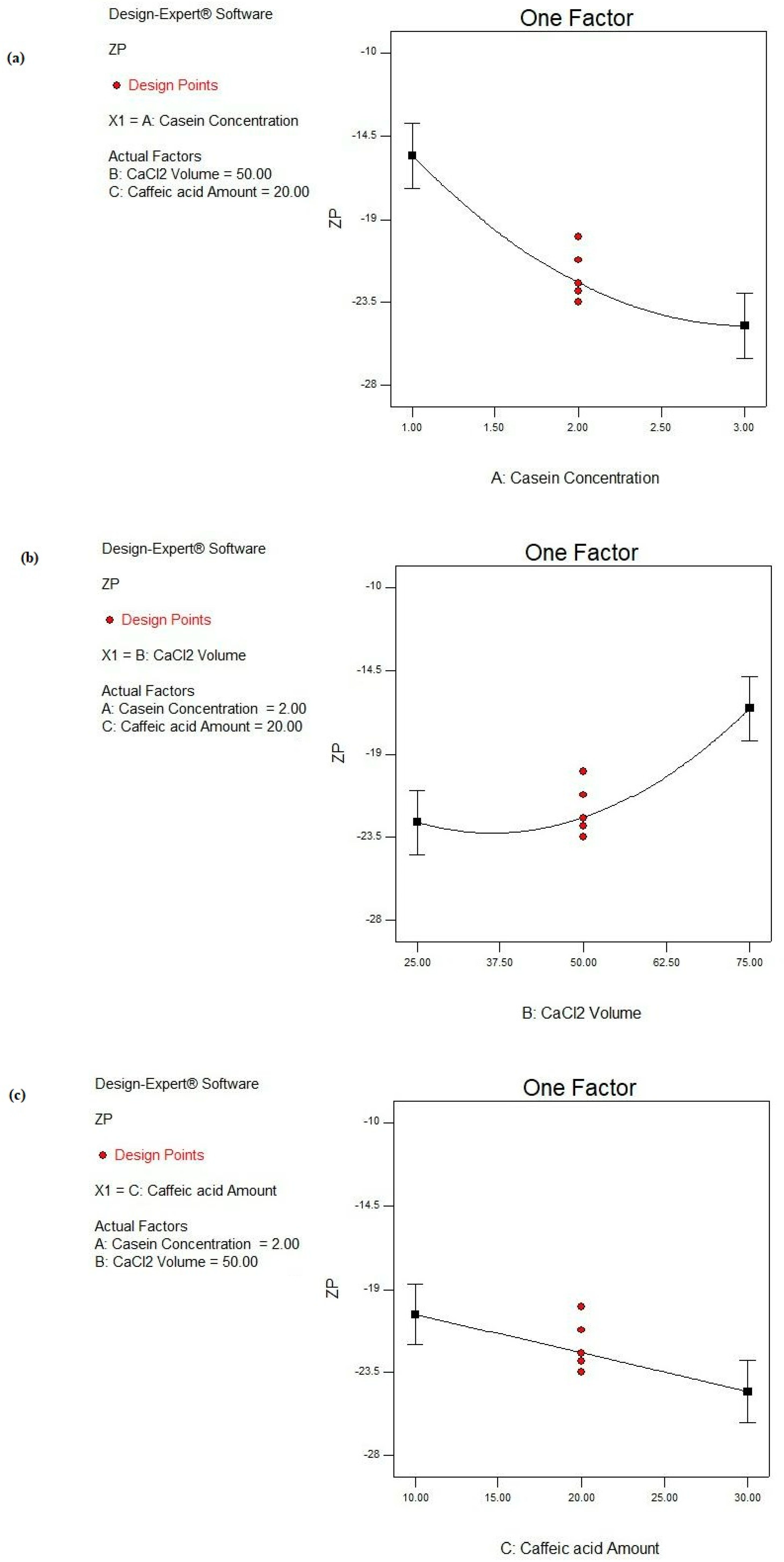
According to the ANOVA results (Table 4), the CS concentration (A) is a highly significant variable (p ˂ 0.0001) affecting the ZP response. Also, both the CaCl2 volume (B) and CA amount (C) demonstrate significant impacts on the ZP response (p ˂ 0.05), as shown in Figure 7. However, no significant interactions between the studied factors are deduced (p > 0.05), as observed in Table 4.
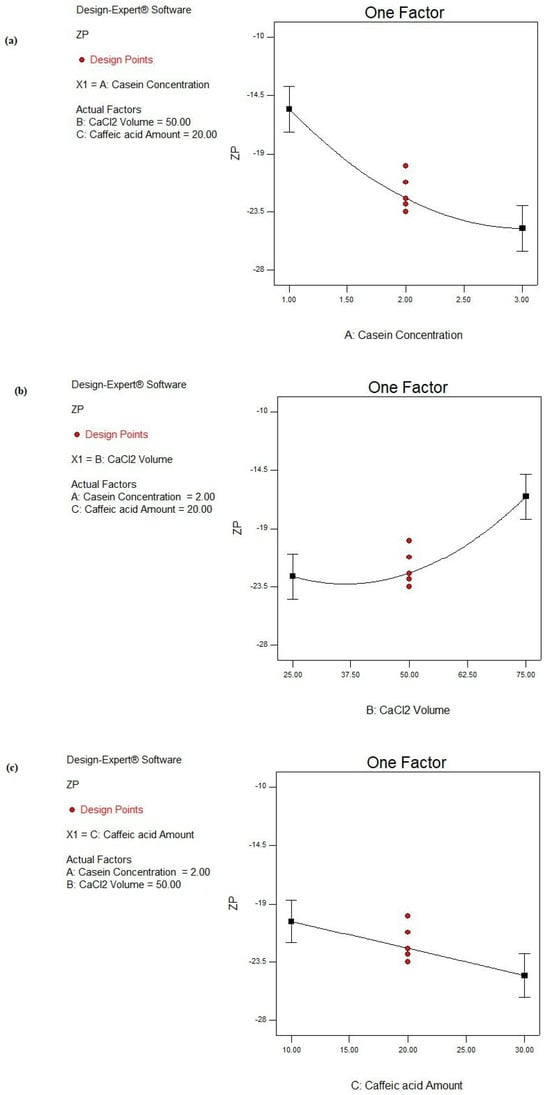
Figure 7.
Main effect plots of studied factors: (a) A: CS concentration, (b) B: CaCl2 volume, and (c) C: CA amount on ZP response.
As shown in Figure 7a, by increasing the CS concentration from 1 to 3%, the negative ZP value is increased from −11.01 ± 1.45 to −17.42 ± 0.23 mV as in the case of F1 and F14, respectively, under similar conditions. Similarly, the negativities of ZP values were increased from −15.22 ± 0.75 to −23.53 ± 1.24 mV, from −19.93 ± 0.92 mV to −27.44 ± 0.17 mV, and from −10.45 ± 0.37 to −25.12 ± 1.99 mV in F4 and F15, F2 and F16, and F3 and F17, respectively (Table 2). This could be interpreted by the presence of the negatively charged K layer of CS covering the CS NPs due to ester phosphate and the carboxylate groups. Accordingly, more negative charges were introduced to the NPs associated by increasing the protein concentration [85].
Upon the addition of Ca2+, the negative ZP value of CS NPs is decreased due to the binding of Ca2+ to the phosphoserine residues of the CS protein, resulting in electrostatic screening effects [76], as shown in Figure 7b. By increasing the CaCl2 volume from 25 to 75 μL, the negative ZP values are significantly decreased, as shown in Table 2. The latter observation was also noticed in F4 vs. F1, F7 vs. F8, F12 vs. F13, and F15 vs. F14, where the ZP values decreased from −15.22 ± 0.75 to −11.01 ± 1.45 mV, from −24.11 ± 2.46 to −18.74 ± 2.52 mV, from −23.43 ± 0.23 to −14.42 ± 0.43 mV, and from −23.53 ± 1.24 to −17.42 ± 0.23 mV, respectively.
By a further analysis of the ZP response (Figure 7c), increasing the CA amount from 10 to 30 mg significantly increases the negative ZP value from −10.45 ± 0.37 to −19.93 ± 0.92 mV as in F3 and F2, respectively (p < 0.05). Also, by comparing F13 to F8, the ZP was significantly raised from −14.42 ± 0.43 to −18.74 ± 2.52 mV (p < 0.05). This could be explained based on the pH of the adjusted alkaline medium involved during the NP fabrication. The alkaline medium caused the ionization of CA. Consequently, CA carried a negative charge as the pKa of CA equaled 4.62 [86]. Moreover, the CS protein had a negative charge, so a greater amount of negative charge enclosed the NPs.
3.5. Model Validation
The validation of the models was assessed by calculating the percentage bias. This was realized through choosing three random check points other than those prepared in the BBD and were figured out using the equation stated in the Section 2.2.
Table 5 shows the bias percentages for the three inspected models, recording very low values (<10%) with an overall mean percentage bias of 1.7% for the PS response, 2.3% for the PDI response, and 3.5% for the ZP response, indicating the validity of the models and their success on navigating through the design space [87,88].

Table 5.
Model validation using the calculated percent of bias.
3.6. Optimization of Prepared CA-Loaded CS NPs Using Desirability Function (D)
From the previous investigation, the desirability function was adopted to obtain the optimized formulation based on the target goals: minimizing the PS and PDI and maximizing the ZP values. The optimized formula showing the highest D value which reached 0.884 constituted of a CS concentration of 1.84 g%, CaCl2 volume of 40.23 μL, and CA amount of 25.19 mg. The chosen formula (O1) manifested a PS of 110.31 ± 1.02 nm, PDI of 0.331 ± 0.029, ZP of −23.94 ± 1.64 mV, and EE% of 95.4 ± 2.56%.
3.7. Imaging Using High-Resolution Transmission Electron Microscopy (HR-TEM)
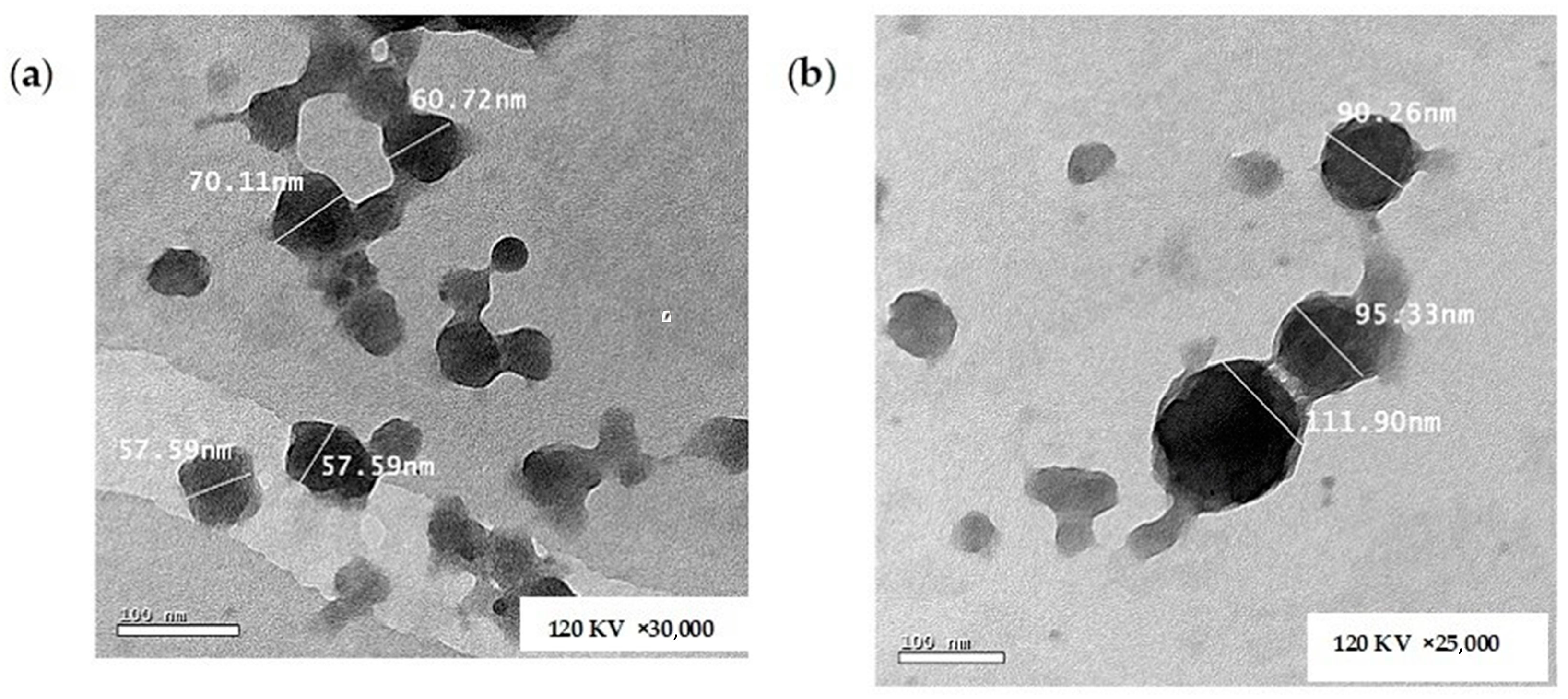
Figure 8a,b show the morphology of the unloaded CS NPs and the optimized CA-loaded CS NPs (O1), respectively. It is obvious from the TEM images that the formulations were regular in geometry, demonstrating an almost spherical structure. The measured sizes using the microscope software tool (Gatan Microscopy Suite GMS 3.X) were in good agreement with the results obtained from the DLS measurement [12,89]. The efficient loading of CA was confirmed by the TEM images where the loaded particles are greater in PS compared with the unloaded ones, as shown in the respective Figure 8a,b.
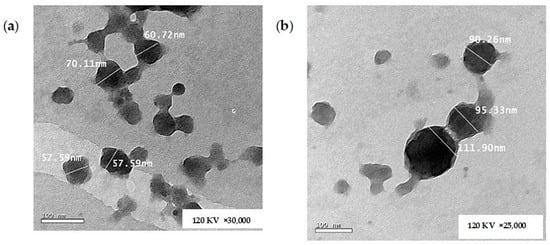
Figure 8.
TEM images of (a) unloaded CS NPs and (b) CA-loaded CS NPs.
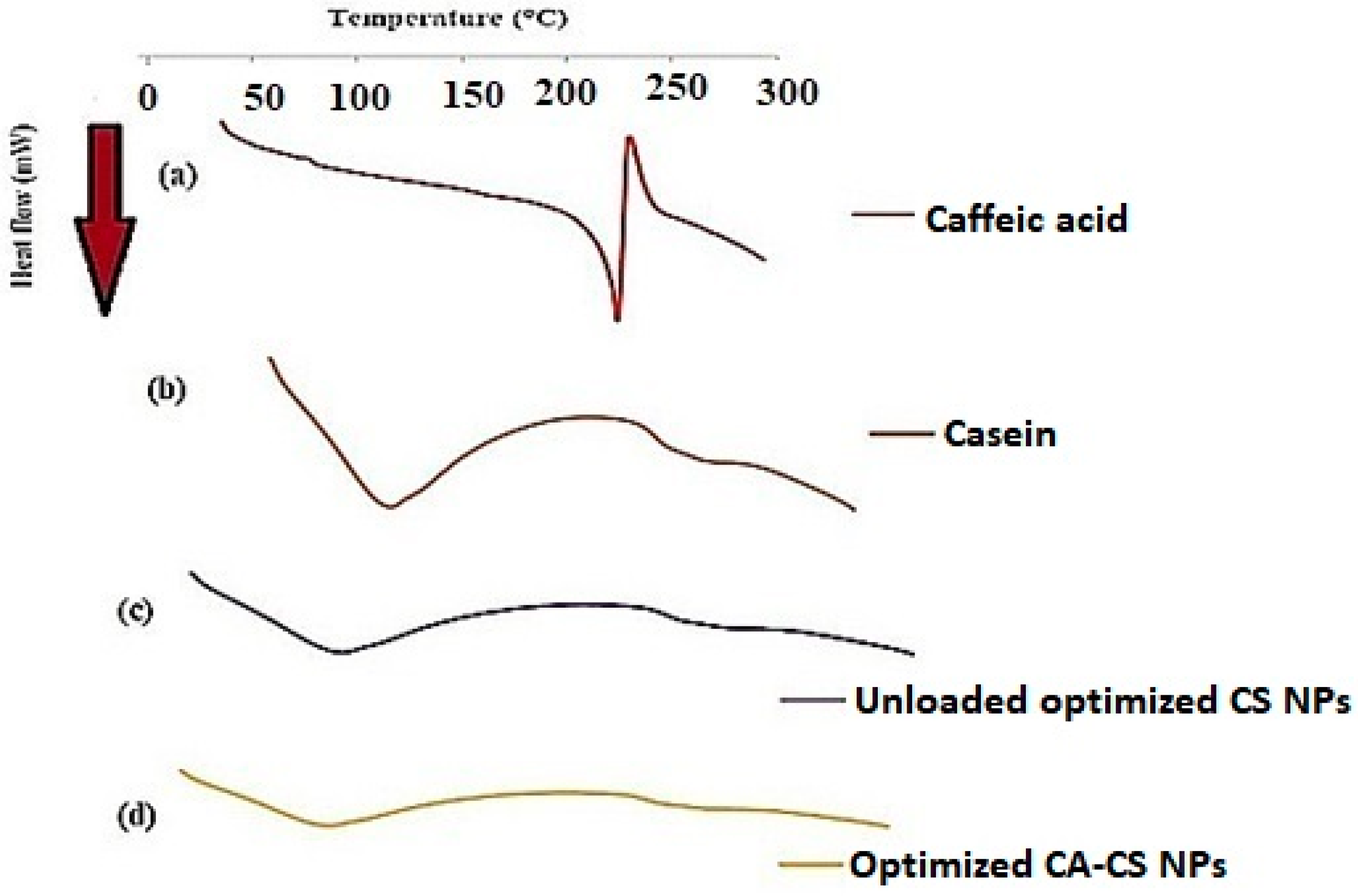
3.8. DSC
DSC was performed in order to determine the crystalline state of CA entrapped in CS NPs. As shown in Figure 9a, CA exhibits a single endothermic peak at 225 °C corresponding to its melting point [90], while the CS protein shows a broad endothermic peak in the range of 50 to 100 °C due to CS dehydration [91], as illustrated in Figure 9b. Both unloaded and loaded CS NPs (O1) display a broad endothermic peak in the range of 50 to 100 °C corresponding to that of CS, as depicted from Figure 9c and Figure 9d, respectively. The results proved that CA was completely entrapped within the CS NPs as the drug peak completely disappeared from the corresponding thermogram, as shown in Figure 9d. This emphasized the molecular dispersion of CA molecules and their efficient encapsulation within the protein matrix [26].
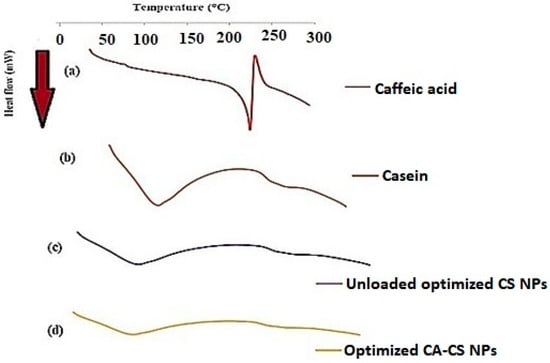
Figure 9.
DSC thermograms of (a) CA, (b) CS, (c) unloaded optimized CS NPs, and (d) optimized CA-loaded CS NPs (O1).
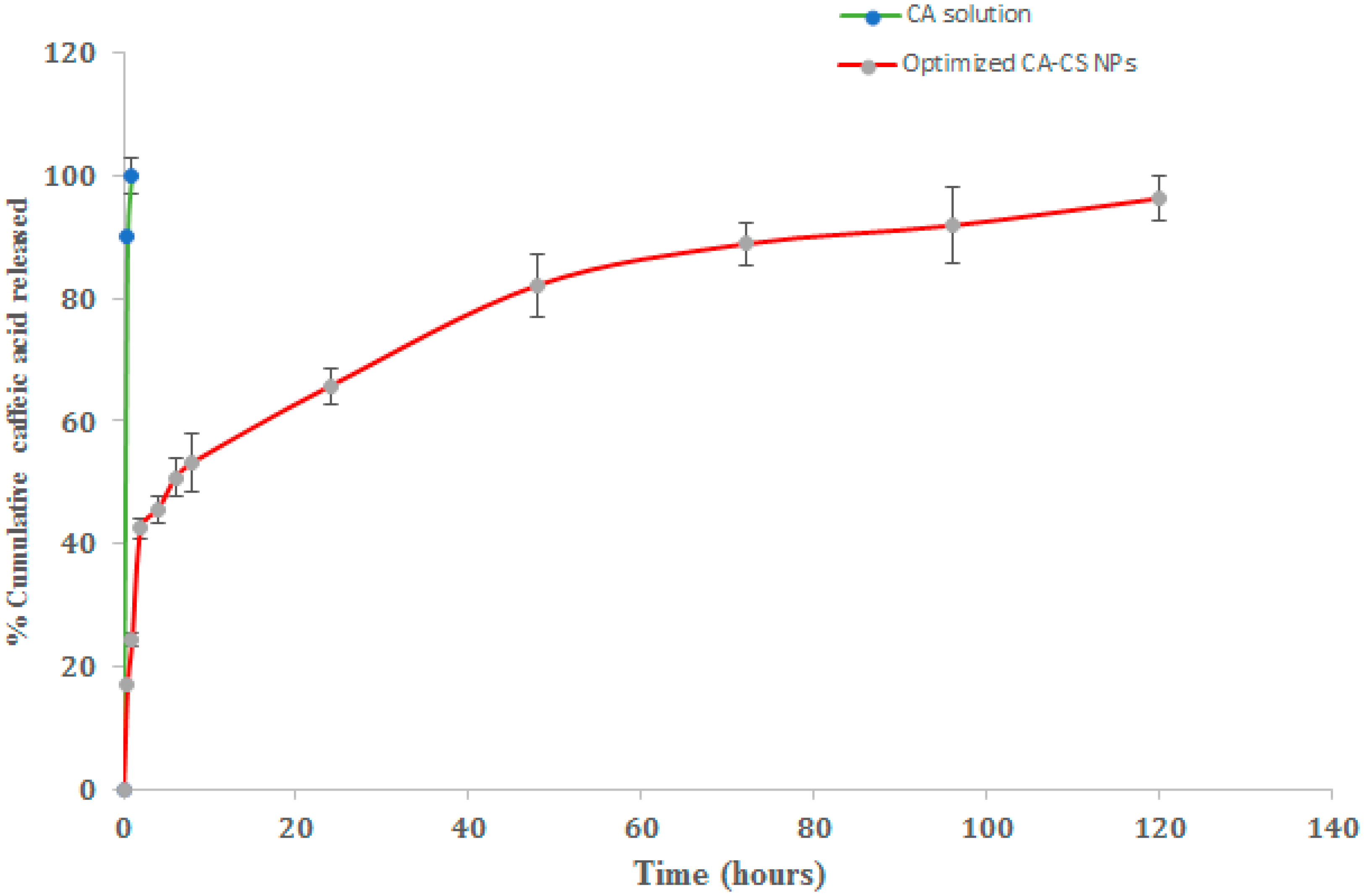
3.9. Results of the in Vitro Release Study of CA from Optimized CS NPs
The in vitro cumulative release percentages of CA from the optimized (O1) NPs were analyzed at different time intervals in PBS (pH 7.4) containing 0.2% w/v Tween 80 and at 37 ± 0.5 °C. The release profiles are illustrated in Figure 10. The CA release from the drug solution was implemented as a control.
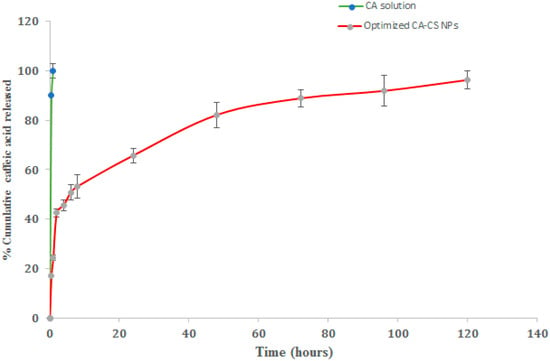
Figure 10.
In vitro release profiles of CA from optimized CA-CS NPs (O1) in PBS (pH 7.4) containing 0.2% w/v Tween 80 at 37 ± 0.5 °C.
It was clearly obvious that almost 90% of CA was released after 1 h only in the case of the CA solution. However, the release profiles of CA from the NPs were represented as biphasic patterns characterized by an initial burst release followed by a sustained rate of drug release over 120 h. The initial burst release was attained over the first 8 h where the optimized CA-CS NPs realized a 53.29 ± 2.90% release of CA. The initial burst release could be attributed to the rapid release of the CA deposited on the surface of the NPs and relatively shallow channels within the NPs [92]. The sustained drug release pattern could be due to the little pathway tackled by the drug, promoting the easy partitioning of CA between the NPs and the release medium [93].
3.10. Cell Culture Studies
The cytotoxicity study of the optimized formulae was performed on MCF-7 cell lines after 24 h incubation. The safety of the NPs’ components was proven by the IC50 of the unloaded CS NPs which scored the highest value of 537.5 ± 6.1 µg/mL, as demonstrated in Table 6. The CA-CS NPs (O1) were more cytotoxic when compared to the drug solution where CA-CS NPs attained an IC50 of 78.45 ± 1.7 µg/mL compared to 173.3 ± 3 µg/mL in the case of the drug solution, as shown in Table 6. These results conformed with the results and the conclusions driven via the meta-analysis that was conducted by Safwat and co-authors [94]. The capability of the NPs in curbing the viability of the cancerous cells was significantly greater than the drug solution since the NPs were characterized by a high surface area-to-volume ratio because of their nanometric merits augmenting their cytotoxic properties [95]. These nano-entities were able to encapsulate high drug payloads per a single nanocarrier realizing a higher cytotoxicity than the drug solution [96,97]. Another beneficial effect of these nanocarriers was the ability of CA to inhibit the efflux effect of P-gp, resulting in more NPs that were internalized inside the cancerous cells [42]. CA directly displayed an allosteric modulation on the M site of P-gp, preventing the pump-out effect of P-gp. Moreover, CA bound to the ATPase binding site. This resulted in the consumption of ATP and enhanced the repressing effect in the function of P-gp [42]. Furthermore, there was a strong correlation between oxidative stress and P-gp. CA displayed strong significant reactive oxygen species (ROS)-related antioxidant effects, resulting in the downregulation of ROS production. The insufficiency of the ROS level resulted in a higher level of ABCB1 genes that impeded the efflux mechanism by reversing the multi-drug resistance (MDR) effect [98]. From previous data, we concluded that the CA-CS NPs were highly accumulated inside MCF-7 cell lines. Once stepping inside the cancerous cells, CA triggered P53 and P21 upregulation genes in the MCF-7 cell lines [99]. P53, also known as TP53, plays a significant role in tumor suppression and the initiation of apoptosis by coding a protein that is responsible for a previous function [100,101]. This gene promotes apoptosis by target gene activation and is transactivation-independent in mitochondria [102]. The over-expression of this gene resulted in growth arrest evoking cellular differentiation, thereby inhibiting cell proliferation [101]. Furthermore, inhibiting several CDKs’ activity, such as CDK2, CDK3, and CDK4, resulted in cell cycle arrest in G1 or G2 [103]. In addition to the previous process, P21 binds to the proliferating cell nuclear antigen (PCNA) and results in the direct prevention of DNA synthesis [102,104]. From this, we came to the conclusion of the superiority of CA-CS NPs (O1) compared to the drug solution in terms of cytotoxicity on MCF-7 cancerous cells.

Table 6.
Cytotoxicity results presenting IC50 data of CA solution and optimized CA-loaded CS NPs when incubated in MCF-7 cell line after 24 h.
4. Conclusions
In our investigation, we pursued the development and optimization of a nanoparticulate system comprising bioinspired materials for potential cancer treatment. CA-loaded CS NPs were successfully prepared and optimized using the BBD. Mathematical equations were obtained, thus paving the road for the researchers to use the designated independent factors in the investigated ranges and predicting the affected responses. Accordingly, both time and cost savings can be achieved. The CS concentration, CaCl2 volume, and drug amount were significant factors affecting the PS and ZP, while the PDI was only affected by both casein concentration and CaCl2 volume. A desirability function was implemented and resulted in an optimized formulation characterized by a minimal PS and PDI and bearing great negative ZP charges. Moreover, molecular docking experiments explained the high loading capacity of casein to caffeic acid. The TEM imaging of the optimized formulae was consistent with the DLS measurements, showing almost spherical particles. The CA-loaded CS NPs showed sustained in vitro drug release. A superb reduction in the viability of the breast cancer cell lines was attained by the optimized CA-CS NPs. Consequently, prominent CA-CS NPs with splendid merits and effective anti-cancer effects were achieved. Future investigations will focus on studying the in vivo antitumor activity of caffeic acid-loaded casein nanoparticles in animal tumor models. This may include performing histopathological examinations and monitoring the different levels of related biomarkers, reflecting the effectiveness of these NPs in curbing tumor cells.
Author Contributions
Conceptualization, R.M.H.; Methodology, S.S., R.M.H. and R.A.H.I.; Software, R.M.H.; Validation, S.S., R.M.H., R.A.H.I. and N.D.M.; Formal analysis, R.M.H. and R.A.H.I.; Investigation, S.S., R.M.H., R.A.H.I. and N.D.M.; Data curation, S.S.; Writing—original draft, S.S.; Writing—review and editing, R.M.H., R.A.H.I. and N.D.M.; Visualization, R.M.H., R.A.H.I. and N.D.M.; Supervision, R.M.H., R.A.H.I. and N.D.M.; Project administration, N.D.M. All authors have read and agreed to the published version of the manuscript.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within this article.
Conflicts of Interest
The authors declare no conflicts of interests.
References
- Krishnan, P.; Yan, K.J.; Windler, D.; Tubbs, J.; Grand, R.; Li, B.D.; Aldaz, C.M.; McLarty, J.; Kleiner-Hancock, H.E. Citrus Auraptene Suppresses Cyclin D1 and Significantly Delays N-Methyl Nitrosourea Induced Mammary Carcinogenesis in Female Sprague-Dawley Rats. BMC Cancer 2009, 9, 259. [Google Scholar] [CrossRef] [PubMed]
- Weiss, R.H.; Marshall, D.; Howard, L.; Corbacho, A.M.; Cheung, A.T.; Sawai, E.T. Suppression of Breast Cancer Growth and Angiogenesis by an Antisense Oligodeoxynucleotide to P21Waf1/Cip1. Cancer Lett. 2003, 189, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Demierre, M.-F.; Higgins, P.D.R.; Gruber, S.B.; Hawk, E.; Lippman, S.M. Statins and Cancer Prevention. Nat. Rev. Cancer 2005, 5, 930–942. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, A.B.; Hall, A. Rho GTPases: Biochemistry and Biology. Annu. Rev. Cell Dev. Biol. 2005, 21, 247–269. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Olufemi, L.; Wang, M.-T.; Nie, D. Role of Rho GTPases in Breast Cancer. Front. Biosci. 2008, 13, 759–776. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sahai, E.; Marshall, C.J. RHO–GTPases and Cancer. Nat. Rev. Cancer 2002, 2, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Dao, K.-L.; Hanson, R.N. Targeting the Estrogen Receptor Using Steroid-Therapeutic Drug Conjugates (Hybrids). Bioconjug. Chem. 2012, 23, 2139–2158. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Thati, S.; Bagby, T.R.; Diab, H.-M.; Davies, N.M.; Cohen, M.S.; Forrest, M.L. Localized Doxorubicin Chemotherapy with a Biopolymeric Nanocarrier Improves Survival and Reduces Toxicity in Xenografts of Human Breast Cancer. J. Control. Release 2010, 146, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, C.L.; Recht, A. Side Effects of Adjuvant Treatment of Breast Cancer. N. Engl. J. Med. 2001, 344, 1997–2008. [Google Scholar] [CrossRef]
- Greene, L.M.N.D.; Fieler, V.K.; Dudgeon, D.; Jones, L.S. A Comparison of Patient reported Side Effects among Three Chemotherapy Regimens for Breast Cancer. Cancer Pract. 1994, 2, 57–62. [Google Scholar]
- Mao, J.J.; Chung, A.; Benton, A.; Hill, S.; Ungar, L.; Leonard, C.E.; Hennessy, S.; Holmes, J.H. Online Discussion of Drug Side Effects and Discontinuation among Breast Cancer Survivors. Pharmacoepidemiol. Drug Saf. 2013, 22, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, S.; Roy, I. Doxorubicin-Loaded Casein Nanoparticles for Drug Delivery: Preparation, Characterization and In Vitro Evaluation. Int. J. Biol. Macromol. 2019, 121, 6–12. [Google Scholar] [CrossRef] [PubMed]
- Elzoghby, A.O.; Samy, W.M.; Elgindy, N.A. Protein-Based Nanocarriers as Promising Drug and Gene Delivery Systems. J. Control. Release 2012, 161, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Semo, E.; Kesselman, E.; Danino, D.; Livney, Y.D. Casein Micelle as a Natural Nano-Capsular Vehicle for Nutraceuticals. Food Hydrocoll. 2007, 21, 936–942. [Google Scholar] [CrossRef]
- EL-Salam, M.H.A.; EL-Shibiny, S. Formation and Potential Uses of Milk Proteins as Nano Delivery Vehicles for Nutraceuticals—A Review. Int. J. Dairy Technol. 2012, 65, 13–21. [Google Scholar] [CrossRef]
- Głąb, T.K.; Boratyński, J. Potential of Casein as a Carrier for Biologically Active Agents. Top. Curr. Chem. 2017, 375, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Livney, Y.D. Milk Proteins as Vehicles for Bioactives. Curr. Opin. Colloid Interface Sci. 2010, 15, 73–83. [Google Scholar] [CrossRef]
- Sauer, A.; Moraru, C.I. Heat Stability of Micellar Casein Concentrates as Affected by Temperature and PH. J. Dairy Sci. 2012, 95, 6339–6350. [Google Scholar] [CrossRef] [PubMed]
- Broyard, C.; Gaucheron, F. Modifications of Structures and Functions of Caseins: A Scientific and Technological Challenge. Dairy Sci. Technol. 2015, 95, 831–862. [Google Scholar] [CrossRef]
- Picchio, M.L.; Minari, R.J.; Gonzalez, V.D.G.; Barandiaran, M.J.; Gugliotta, L.M. New Strategy to Improve Acrylic/Casein Compatibilization in Waterborne Hybrid Nanoparticles. J. Appl. Polym. 2015, 132, 1–9. [Google Scholar] [CrossRef]
- Zhen, X.; Wang, X.; Xie, C.; Wu, W.; Jiang, X. Cellular Uptake, Antitumor Response and Tumor Penetration of Cisplatin-Loaded Milk Protein Nanoparticles. Biomaterials 2013, 34, 1372–1382. [Google Scholar] [CrossRef]
- Pan, X.; Yao, P.; Jiang, M. Simultaneous Nanoparticle Formation and Encapsulation Driven by Hydrophobic Interaction of Casein-Graft-Dextran and Beta-Carotene. J. Colloid Interface Sci. 2007, 315, 456–463. [Google Scholar] [CrossRef]
- Nakagawa, K.; Kagemoto, M. Characterization of Casein-Based Nanoparticles Formed upon Freezing by In Situ SAXS Measurement. Colloids Surf. B Biointerfaces 2013, 103, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Ye, A.; Flanagan, J.; Singh, H. Formation of Stable Nanoparticles via Electrostatic Complexation between Sodium Caseinate and Gum Arabic. Biopolymers 2006, 82, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Malekhosseini, P.; Alami, M.; Khomeiri, M.; Esteghlal, S.; Nekoei, A.-R.; Hosseini, S.M.H. Development of Casein-Based Nanoencapsulation Systems for Delivery of Epigallocatechin Gallate and Folic Acid. Food Sci. Nutr. 2019, 7, 519–527. [Google Scholar] [CrossRef]
- Elzoghby, A.O.; Samy, W.M.; Elgindy, N.A. Novel Spray-Dried Genipin-Crosslinked Casein Nanoparticles for Prolonged Release of Alfuzosin Hydrochloride. Pharm. Res. 2013, 30, 512–522. [Google Scholar] [CrossRef] [PubMed]
- Saija, A.; Tomaino, A.; Trombetta, D.; De Pasquale, A.; Uccella, N.; Barbuzzi, T.; Paolino, D.; Bonina, F. In Vitro and in Vivo Evaluation of Caffeic and Ferulic Acids as Topical Photoprotective Agents. Int. J. Pharm. 2000, 199, 39. [Google Scholar] [CrossRef]
- Hassan, E.A.; Hathout, R.M.; Gad, H.A.; Sammour, O.A. Multi-Purpose Zein Nanoparticles for Battling Hepatocellular Carcinoma: A Green Approach. Eur. Polym. J. 2022, 176, 111396. [Google Scholar] [CrossRef]
- Wang, Q.S.; Kong, P.Z.; Li, X.Q.; Yang, F.; Feng, Y.M. FOXF2 Deficiency Promotes Epithelial-Mesenchymal Transition and Metastasis of Basal-like Breast Cancer. Breast Cancer Res. 2015, 17, 30. [Google Scholar] [CrossRef]
- Zhang, X.; Ma, G.; Liu, J.; Zhang, Y. MicroRNA-182 Promotes Proliferation and Metastasis by Targeting FOXF2 in Triplenegative Breast Cancer. Oncol. Lett. 2017, 14, 4805–4811. [Google Scholar] [CrossRef]
- Li, S.; Lu, J.; Chen, Y.; Xiong, N.; Li, L.; Zhang, J.; Yang, H.; Wu, C.; Zeng, H.; Liu, Y. MCP-1-Induced ERK/GSK-3β/Snail Signaling Facilitates the Epithelial-Mesenchymal Transition and Promotes the Migration of MCF-7 Human Breast Carcinoma Cells. Cell. Mol. Immunol. 2017, 14, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Toma, L.; Sanda, G.M.; Niculescu, L.S.; Deleanu, M.; Stancu, C.S.; Sima, A.V. Caffeic Acid Attenuates the Inflammatory Stress Induced by Glycated LDL in Human Endothelial Cells by Mechanisms Involving Inhibition of AGE-Receptor, Oxidative, and Endoplasmic Reticulum Stress. Biofactors 2017, 43, 685–697. [Google Scholar] [CrossRef] [PubMed]
- Box, G.P.E.; Wilson, K.B. On the Experimental Attainment of Optimum Conditions. J. R. Stat. Soc. Ser. B 1951, 1, 1–45. [Google Scholar] [CrossRef]
- Lewis, G.A.; Mathieu, D.; Phan, T.L.R. Pharmaceutical Experimental Design; CRC Press: Boca Raton, FL, USA, 1999. [Google Scholar]
- Zhang, J.; Fan, Y.; Smith, E. Experimental Design for the Optimization of Lipid Nanoparticles. J. Pharm. Sci. 2009, 98, 1813–1819. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; van den Driesche, S.; Bunge, F.; Yang, B.; Vellekoop, M.J. Optimization of On-Chip Bacterial Culture Conditions Using the Box-Behnken Design Response Surface Methodology for Faster Drug Susceptibility Screening. Talanta 2019, 194, 627–633. [Google Scholar] [CrossRef] [PubMed]
- Roslan, S.A.; Hassan, M.Z.; Rasid, Z.A.; Bani, N.A.; Sarip, S.; Daud, M.Y.M.; Muhammad-Sukki, F. Mode I Fracture Toughness of Optimized Alkalitreated Bambusa Vulgaris Bamboo by BoxBehnken Design. In Advances in Material Sciences and Engineering; Springer: Berlin/Heidelberg, Germany, 2019; pp. 565–575. [Google Scholar]
- Chung, T.-W.; Moon, S.-K.; Chang, Y.-C.; Ko, J.-H.; Lee, Y.-C.; Cho, G.; Kim, S.-H.; Kim, J.-G.; Kim, C.-H. Novel and Therapeutic Effect of Caffeic Acid and Caffeic Acid Phenyl Ester on Hepatocarcinoma Cells: Complete Regression of Hepatoma Growth and Metastasis by Dual Mechanism. FASEB J. 2004, 18, 1625–1790. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, S.; Gholami, M.H.; Zabolian, A.; Saleki, H.; Farahani, M.V.; Hamzehlou, S.; Far, F.B.; Sharifzadeh, S.O.; Samarghandian, S.; Khan, H.; et al. Caffeic Acid and Its Derivatives as Potential Modulators of Oncogenic Molecular Pathways: New Hope in the Fight against Cancer. Pharmacol. Res. 2021, 171, 105759. [Google Scholar] [CrossRef]
- Kabała-Dzik, A.; Rzepecka-Stojko, A.; Kubina, R.; Jastrzębska-Stojko, Z.; Stojko, R.; Wojtyczka, R.D.; Stojko, J. Migration Rate Inhibition of Breast Cancer Cells Treated by Caffeic Acid and Caffeic Acid Phenethyl Ester: An in Vitro Comparison Study. Nutrients 2017, 9, 1144. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.-L.; Han, N.-Z.; Liu, S.-S. Caffeic Acid Phenethyl Ester Inhibits the Progression of Ovarian Cancer by Regulating NF-ΚB Signaling. Biomed. Pharmacother. 2018, 99, 825–831. [Google Scholar] [CrossRef]
- Teng, Y.N.; Wang, C.; Liao, W.C.; Lan, Y.H.; Hung, C.C. Caffeic Acid Attenuates Multidrug Resistance in Cancer Cells by Inhibiting Efflux Function of Human P-Glycoprotein. Molecules 2020, 25, 247. [Google Scholar] [CrossRef]
- Grunberger, D.; Banerjee, R.; Eisinger, K.; Oltz, E.M.; Efros, L.; Caldwell, M.; Estevez, V.; Nakanishi, K. Preferential Cytotoxicity on Tumor Cells by Caffeic Acid Phenethyl Ester Isolated from Propolis. Experientia 1988, 44, 230–232. [Google Scholar] [CrossRef] [PubMed]
- Secme, M.; Mutlu, D.; Elmas, L.; Arslan, S. Assessing Effects of Caffeic Acid on Cytotoxicity, Apoptosis, Invasion, GST Enzyme Activity, Oxidant, Antioxidant Status and Micro-RNA Expressions in HCT116 Colorectal Cancer Cells. S. Afr. J. Bot. 2023, 157, 19–26. [Google Scholar] [CrossRef]
- Widjaja, A.; Yeh, T.-H.; Ju, Y.-H. Enzymatic Synthesis of Caffeic Acid Phenethyl Ester. J. Chin. Inst. Chem. Eng. 2008, 39, 413–418. [Google Scholar] [CrossRef]
- Schulz, A.; Meyer, F.; Dubrovska, A.; Borgmann, K. Cancer Stem Cells and Radioresistance: DNA Repair and Beyond. Cancers 2019, 11, 862. [Google Scholar] [CrossRef] [PubMed]
- Makena, M.R.; Ranjan, A.; Thirumala, V.; Reddy, A.P. Cancer Stem Cells: Road to Therapeutic Resistance and Strategies to Overcome Resistance. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165339. [Google Scholar] [CrossRef] [PubMed]
- Penalva, R.; Esparza, I.; Agüeros, M.; Gonzalez-Navarro, C.J.; Gonzalez-Ferrero, C.; Irache, J.M. Casein Nanoparticles as Carriers for the Oral Delivery of Folic Acid. Food Hydrocoll. 2015, 44, 399–406. [Google Scholar] [CrossRef]
- Metwally, A.A.; El-Ahmady, S.H.; Hathout, R.M. Selecting Optimum Protein Nano-Carriers for Natural Polyphenols Using Chemoinformatics Tools. Phytomedicine 2016, 23, 1764–1770. [Google Scholar] [CrossRef]
- Hathout, R.M.; Metwally, A.A.; Woodman, T.J.; Hardy, J.G. Prediction of Drug Loading in the Gelatin Matrix Using Computational Methods. ACS Omega 2020, 5, 1549–1556. [Google Scholar] [CrossRef]
- Hathout, R.M.; Metwally, A.A. Towards Better Modelling of Drug-Loading in Solid Lipid Nanoparticles: Molecular Dynamics, Docking Experiments and Gaussian Processes Machine Learning. Eur. J. Pharm. Biopharm. 2016, 108, 262–268. [Google Scholar] [CrossRef]
- Dudkiewicz, A.; Tiede, K.; Loeschner, K.; Helene, L.; Jensen, S.; Jensen, E.; Wierzbicki, R.; Boxall, A.B.A.; Molhave, K. Characterization of Nanomaterials in Food by Electron Microscopy. Trends Anal. Chem. 2011, 30, 28–43. [Google Scholar] [CrossRef]
- Bani, M.S.; Hatamie, S.; Haghpanahi, M.; Bahreinizad, H.; Alavijeh, M.H.S.; Eivazzadeh-Keihan, R.; Wei, Z.H. Casein-Coated Iron Oxide Nanoparticles for in Vitro Hyperthermia for Cancer Therapy. Spin 2019, 9, 1940003. [Google Scholar] [CrossRef]
- Chen, L.; Wei, J.; An, M.; Zhang, L.; Lin, S.; Shu, G.; Yuan, Z.; Lin, J.; Peng, G.; Liang, X.; et al. Casein Nanoparticles as Oral Delivery Carriers of Mequindox for the Improved Bioavailability. Colloids Surf. B Biointerfaces 2020, 195, 111221. [Google Scholar] [CrossRef]
- Masaki, S.; Sugimori, G.; Okamoto, A.; Imose, J.; Hayashi, Y. Effect of Tween 80 on the Growth of Mycobacterium Avium Complex. Microbiol. Immunol. 1990, 34, 653–663. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.J.; Lee, D.W.; Marasini, N.; Poudel, B.K.; Kim, J.H.; Ramasamy, T.; Yoo, B.K.; Choi, H.-G.; Yong, C.S.; Kim, J.O. Optimization of Self-Microemulsifying Drug Delivery System for Telmisartan Using Box—Behnken Design and Desirability Function. J. Pharm. Pharmacol. 2013, 65, 1440–1450. [Google Scholar] [CrossRef]
- Derringer, G.; Suich, R. Simultaneous Optimization of Several Response Variables. J. Qual. Technol. 1980, 12, 214–219. [Google Scholar] [CrossRef]
- Gulzar, M.; Ali, S.; Khan, F.I.; Khan, P.; Taneja, P.; Hassan, M.I. Binding Mechanism of Caffeic Acid and Simvastatin to the Integrin Linked Kinase for Therapeutic Implications: A Comparative Docking and MD Simulation Studies. J. Biomol. Struct. Dyn. 2019, 37, 4327–4337. [Google Scholar] [CrossRef]
- Hathout, R.M.; Saharan, V.A. Computer-Aided Formulation Development. In Computer Aided Pharmaceutics and Drug Delivery; Springer: Singapore, 2022; pp. 73–98. [Google Scholar]
- Ferreira, S.L.C.; Bruns, R.E.; Ferreira, H.S.; Matos, G.D.; David, J.M.; Brandão, G.C.; da Silva, E.G.P.; Portugal, L.A.; dos Reis, P.S.; Souza, A.S.; et al. Box-Behnken Design: An Alternative for the Optimization of Analytical Methods. Anal. Chim. Acta 2007, 597, 179–186. [Google Scholar] [CrossRef]
- Khajvand, T.; Chaichi, M.J.; Nazari, O.; Golchoubian, H. Application of Box-Behnken Design in the Optimization of Catalytic Behavior of a New Mixed Chelate of Copper(II) Complex in Chemiluminescence Reaction of Luminol. J. Lumin. 2011, 131, 838–842. [Google Scholar] [CrossRef]
- Prabhakaran, D.; Basha, C.A.; Kannadasan, T.; Aravinthan, P. Removal of Hydroquinone from Water by Electrocoagulation Using Flow Cell and Optimization by Response Surface Methodology. J. Environ. Sci. Health Part A 2010, 45, 400–412. [Google Scholar] [CrossRef]
- Yildirim-Elikoglu, S.; Erdem, Y.K. Interactions between Milk Proteins and Polyphenols: Binding Mechanisms, Related Changes, and the Future Trends in the Dairy Industry. Food Rev. Int. 2018, 34, 665–697. [Google Scholar] [CrossRef]
- Madhan, B.; Thanikaivelan, P.; Subramanian, V.; Rao, J.R.; Nair, B.U.; Ramasami, T. Molecular Mechanics and Dynamics Studies on the Interaction of Gallic Acid with Collagen-like Peptides. Chem. Phys. Lett. 2001, 346, 334–340. [Google Scholar] [CrossRef]
- Liu, Y.; Xie, M.-X.; Jiang, M.; Wang, Y.-D. Spectroscopic Investigation of the Interaction between Human Serum Albumin and Three Organic Acids. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2005, 61, 2245–2251. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Li, X.; Dai, T.; Hu, P.; Niu, X.; Liu, C.; Chen, J. Binding Mechanism and Antioxidant Capacity of Selected Phenolic Acid-β-Casein Complexes. Food Res. Int. 2020, 129, 108802. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, H.; Jana, S. Evaluation of Physicochemical Properties and Intestinal Permeability of Six Dietary Polyphenols in Human Intestinal Colon Adenocarcinoma Caco-2 Cells. Eur. J. Drug Metab. Pharmacokinet. 2016, 41, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Hafez, S.M.; Hathout, R.M.; Sammour, O.A. Towards Better Modeling of Chitosan Nanoparticles Production: Screening Different Factors and Comparing Two Experimental Designs. Int. J. Biol. Macromol. 2014, 64, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Garg, K.K.; Prasad, B. Development of Box Behnken Design for Treatment of Terephthalic Acid Wastewater by Electrocoagulation Process: Optimization of Process and Analysis of Sludge. J. Environ. Chem. Eng. 2016, 4, 178–190. [Google Scholar] [CrossRef]
- Dudhipala, N.; Janga, K.Y. Lipid Nanoparticles of Zaleplon for Improved Oral Delivery by Box-Behnken Design: Optimization, in Vitro and in Vivoevaluation. Drug Dev. Ind. Pharm. 2017, 43, 1205–1214. [Google Scholar] [CrossRef] [PubMed]
- Safwat, S.; Hathout, R.M.; Ishak, R.A.; Mortada, N.D. Augmented Simvastatin Cytotoxicity Using Optimized Lipid Nanocapsules: A Potential for Breast Cancer Treatment. J. Liposome Res. 2017, 27, 1–10. [Google Scholar] [CrossRef]
- Shadman, M.; Hosseini, M.R.; Zinjenab, Z.T.; Azimi, E. Significant Reduction in Collector Consumption by Implementing Ultrafine Bubbles in Lead and Zinc Rougher Flotation. Powder Technol. 2023, 414, 118096. [Google Scholar] [CrossRef]
- Murphy, S.A. Does Sulfide Affect Short Term Iron Oxidation? Master’s Thesis, University of South Carolina, Columbia, SC, USA, 2012. [Google Scholar]
- Madadlou, A.; Sheehan, D.; Emam-Djomeh, Z.; Mousavi, M.E. Dissociation of Self-Assembled Casein Nanoparticles by Ascending the PH and Sonication. Milchwissenschaft 2012, 67, 155–159. [Google Scholar]
- Dauphas, S.; Mouhous-Riou, N.; Metro, B.; Mackie, A.R.; Wilde, P.J.; Anton, M.; Riaublanc, A. The Supramolecular Organisation of β-Casein: Effect on Interfacial Properties. Food Hydrocoll. 2005, 19, 387–393. [Google Scholar] [CrossRef]
- McCarthy, N.A.; Kelly, A.L.; O’Mahony, J.A.; Fenelon, M.A. Sensitivity of Emulsions Stabilised by Bovine β-Casein and Lactoferrin to Heat and CaCl2. Food Hydrocoll. 2014, 35, 420–428. [Google Scholar] [CrossRef]
- Görner, T.; Gref, R.; Michenot, D.; Sommer, F.; Tran, M.; Dellacherie, E. Lidocaine-Loaded Biodegradable Nanospheres. I. Optimization of the Drug Incorporation into the Polymer Matrix. J. Control. Release 1999, 57, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Parenti, F.; Tassinari, F.; Libertini, E.; Lanzi, M.; Mucci, A. π-Stacking Signature in NMR Solution Spectra of Thiophene-Based Conjugated Polymers. ACS Omega 2017, 2, 5775–5784. [Google Scholar] [CrossRef] [PubMed]
- Soleimanifar, M.; Jafari, S.M.; Assadpour, E. Encapsulation of Olive Leaf Phenolics within Electrosprayed Whey Protein Nanoparticles; Production and Characterization. Food Hydrocoll. 2020, 101, 105572. [Google Scholar] [CrossRef]
- Amighi, F.; Emam-Djomeh, Z.; Labbafi-Mazraeh-Shahi, M. Effect of Different Cross-Linking Agents on the Preparation of Bovine Serum Albumin Nanoparticles. J. Iran. Chem. Soc. 2020, 17, 1223–1235. [Google Scholar] [CrossRef]
- Ahsan, S.M.; Rao, C.M. The Role of Surface Charge in the Desolvation Process of Gelatin: Implications in Nanoparticle Synthesis and Modulation of Drug Release. Int. J. Nanomed. 2017, 12, 795–808. [Google Scholar] [CrossRef] [PubMed]
- Amidi, M.; Romeijn, S.G.; Borchard, G.; Junginger, H.E.; Hennink, W.E.; Jiskoot, W. Preparation and Characterization of Protein-Loaded N-Trimethyl Chitosan Nanoparticles as Nasal Delivery System. J. Control. Release 2006, 111, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Horne, D.S. Casein Interactions: Casting Light on the Black Boxes, the Structure in Dairy Products. Int. Dairy J. 1998, 8, 171–177. [Google Scholar] [CrossRef]
- Pitkowski, A.; Nicolai, T.; Durand, D. Stability of Caseinate Solutions in the Presence of Calcium. Food Hydrocoll. 2009, 23, 1164–1168. [Google Scholar] [CrossRef]
- Phadungath, C. Casein Micelle Structure: A Concise Review. Songklanakarin J. Sci. Technol. 2005, 27, 201–212. [Google Scholar]
- Ashfaq, M.; Talreja, N.; Chauhan, D.; Rodríguez, C.A.; Mera, A.C.; Viswanathan, M.R. A Facile Synthesis of CuBi2O4 Hierarchical Dumbbell-Shaped Nanorod Cluster: A Promising Photocatalyst for the Degradation of Caffeic Acid. Environ. Sci. Pollut. Res. 2022, 29, 53873–53883. [Google Scholar] [CrossRef] [PubMed]
- Gidwani, B.; Vyas, A. Preparation, Characterization, and Optimization of Altretamine-Loaded Solid Lipid Nanoparticles Using Box-Behnken Design and Response Surface Methodology. Artif. Cells Nanomed. Biotechnol. 2016, 44, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Jangde, R.; Elhassan, G.O.; Khute, S.; Singh, D.; Singh, M.; Sahu, R.K.; Jiyauddin, K. Hesperidin-Loaded Lipid Polymer Hybrid Nanoparticles for Topical Delivery of Bioactive Drugs. Pharmaceuticals 2022, 15, 211. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Bajpai, J.; Bajpai, A.K.; Mongre, R.K.; Lee, M.-S. Encapsulation of Cytarabine into Casein Coated Iron Oxide Nanoparticles (CCIONPs) and Study of in Vitro Drug Release and Anticancer Activities. J. Drug Deliv. Sci. Technol. 2020, 55, 101396. [Google Scholar] [CrossRef]
- Pinho, E.; Henriques, M.; Soares, G. Caffeic Acid Loading Wound Dressing: Physicochemical and Biological Characterization. Ther. Deliv. 2014, 5, 1063–1075. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Jiang, T.T.; Yuan, Z.X.; Lu, Y. Self-Assembled Casein Nanoparticles Loading Triptolide for the Enhancement of Oral Bioavailability. Nat. Prod. Commun. 2020, 15, 1–9. [Google Scholar] [CrossRef]
- Chen, J.; Li, S.; Shen, Q. Folic Acid and Cell-Penetrating Peptide Conjugated PLGA-PEG Bifunctional Nanoparticles for Vincristine Sulfate Delivery. Eur. J. Pharm. Sci. 2012, 47, 430–443. [Google Scholar] [CrossRef]
- Priya, P.; Raj, R.M.; Vasanthakumar, V.; Raj, V. Curcumin-Loaded Layer-by-Layer Folic Acid and Casein Coated Carboxymethyl Cellulose/Casein Nanogels for Treatment of Skin Cancer. Arab. J. Chem. 2017, 17, 30140–30145. [Google Scholar] [CrossRef]
- Safwat, S.; Hathout, R.M.; Ishak, R.A.; Mortada, N.D. Elaborated Survey in the Scope of Nanocarriers Engineering for Boosting Chemotherapy Cytotoxicity: A Meta-Analysis Study. Int. J. Pharm. 2021, 610, 121268. [Google Scholar] [CrossRef]
- Borm, P.J.; Robbins, D.; Haubold, S.; Kuhlbusch, T.; Fissan, H.; Donaldson, K.; Schins, R.; Stone, V.; Kreyling, W.; Lademann, J.; et al. The Potential Risks of Nanomaterials: A Review Carried out for ECETOC. Part. Fibre Toxicol. 2006, 3, 11. [Google Scholar] [CrossRef] [PubMed]
- Dreis, S.; Rothweiler, F.; Michaelis, M.; Cinatl, J., Jr.; Kreuter, J.; Langer, K. Preparation, Characterisation and Maintenance of Drug Efficacy of Doxorubicin-Loaded Human Serum Albumin (HSA) Nanoparticles. Int. J. Pharm. 2007, 341, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Hathout, R.M.; Metwally, A.A.; El-Ahmady, S.H.; Metwally, E.S.; Ghonim, N.A.; Bayoumy, S.A.; Erfan, T.; Ashraf, R.; Fadel, M.; El-Kholy, A.I.; et al. Dual stimuli-responsive polypyrrole nanoparticles for anticancer therapy. J. Drug. Deliv. Sci. Technol. 2018, 47, 176–180. [Google Scholar] [CrossRef]
- Huang, C.; Huang, S.; Li, H.; Li, X.; Li, B.; Zhong, L.; Wang, J.; Zou, M.; He, X.; Zheng, H.; et al. The Effects of Ultrasound Exposure on P-Glycoprotein-Mediated Multidrug Resistance In Vitro and In Vivo. J. Exp. Clin. Cancer Res. 2018, 37, 232. [Google Scholar] [CrossRef] [PubMed]
- Rezaei-seresht, H.; Cheshomi, H.; Falanji, F.; Movahedi-Motlagh, F.; Hashemian, M.; Mireskandari, E. Cytotoxic Activity of Caffeic Acid and Gallic Acid against MCF-7 Human Breast Cancer Cells: An in Silico and In Vitro Study. Avicenna J. Phytomed. 2019, 9, 574–586. [Google Scholar] [CrossRef] [PubMed]
- Kerr, J.F.R.; Wyllie, A.H.; Currie, A.R. Apoptosis: A Basic Biological Phenomenon with Wideranging Implications in Tissue Kinetics. Br. J. Cancer 1972, 26, 239–257. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.C.; Strasser, A. BH3-Only Proteins—Essential Initiators of Apoptotic Cell Death. Cell 2000, 103, 839–842. [Google Scholar] [CrossRef] [PubMed]
- Moll, U.M.; Zaika, A. Nuclear and Mitochondrial Apoptotic Pathways of P53. FEBS Lett. 2001, 493, 65–69. [Google Scholar] [CrossRef]
- Gartel, A.L.; Tyner, A.L. The Role of the Cyclin-Dependent Kinase Inhibitor P21 in Apoptosis 1 Supported in Part by NIH Grant R01 DK56283 (to A. L. T.) for the P21 Research and Campus Research Board and Illinois Department of Public Health Penny Severns Breast and Cervical Cancer. Mol. Cancer Ther. 2002, 1, 639–649. [Google Scholar]
- Chen, J.; Jackson, P.K.; Kirschner, M.W.; Dutta, A. Separate Domains of P21 Involved in the Inhibition of Cdk Kinase and PCNA. Nature 1995, 374, 386–388. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Published by MDPI on behalf of the Österreichische Pharmazeutische Gesellschaft. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).