A Network Pharmacology and Molecular Docking Technology to Identify and Explore Mechanism of Bioactive Components of Fucus vesiculosus against Gut Disorders
Abstract
1. Introduction
2. Materials and Methods
2.1. Active Components and Corresponding Target Selection
2.2. Procurement of Candidate Gene and Disease-Associated Targets Genes
2.3. Protein-Protein Interaction (PPI) Construction Network
2.4. GO Enrichment and KEGG Pathway Analysis
2.5. Drug-Target Pathways Network Construction
2.6. Molecular Docking Studies
3. Results and Discussion
3.1. F. vesiculosus Bioactive Components and Target Gene Related to Disorder Identification
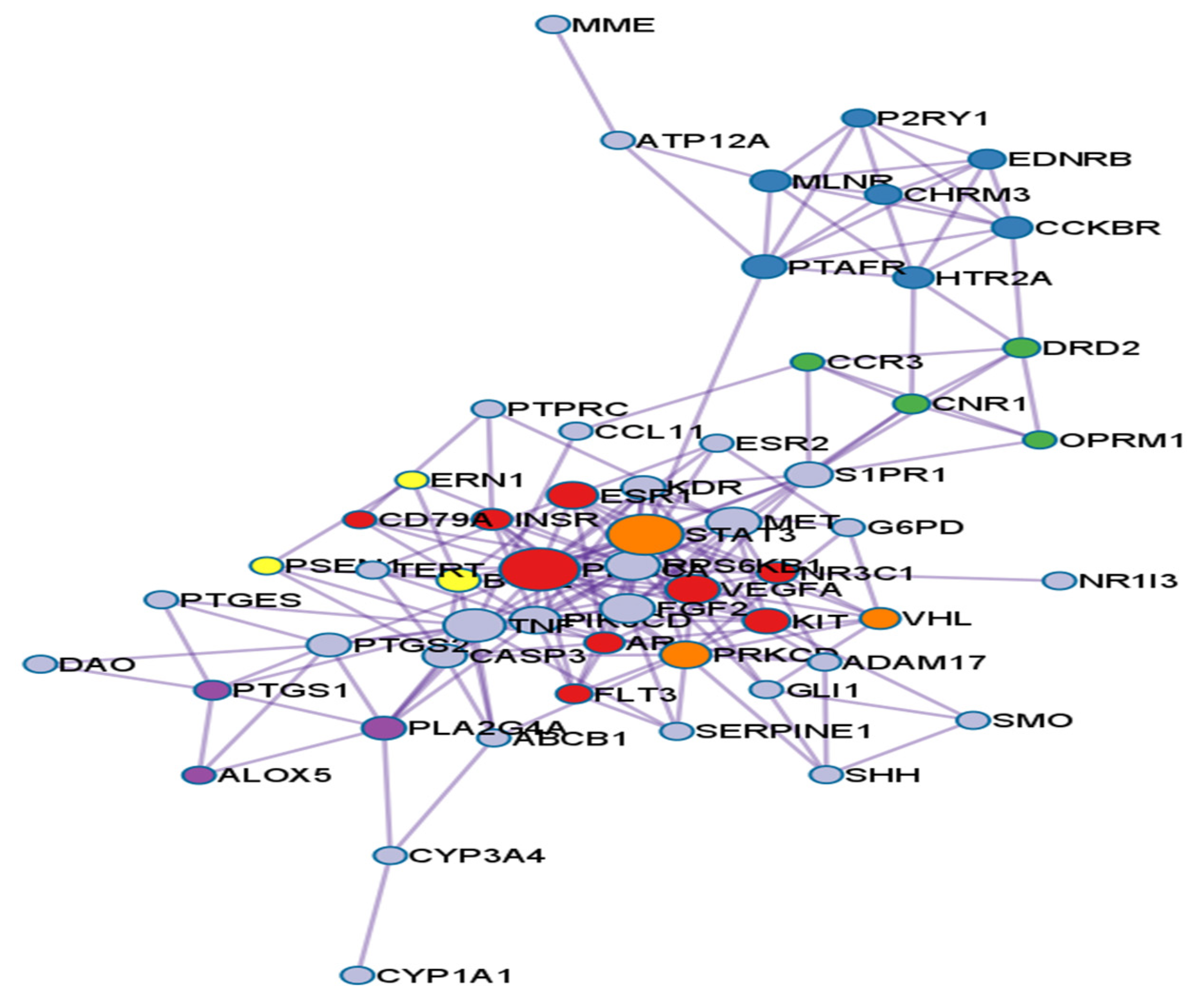
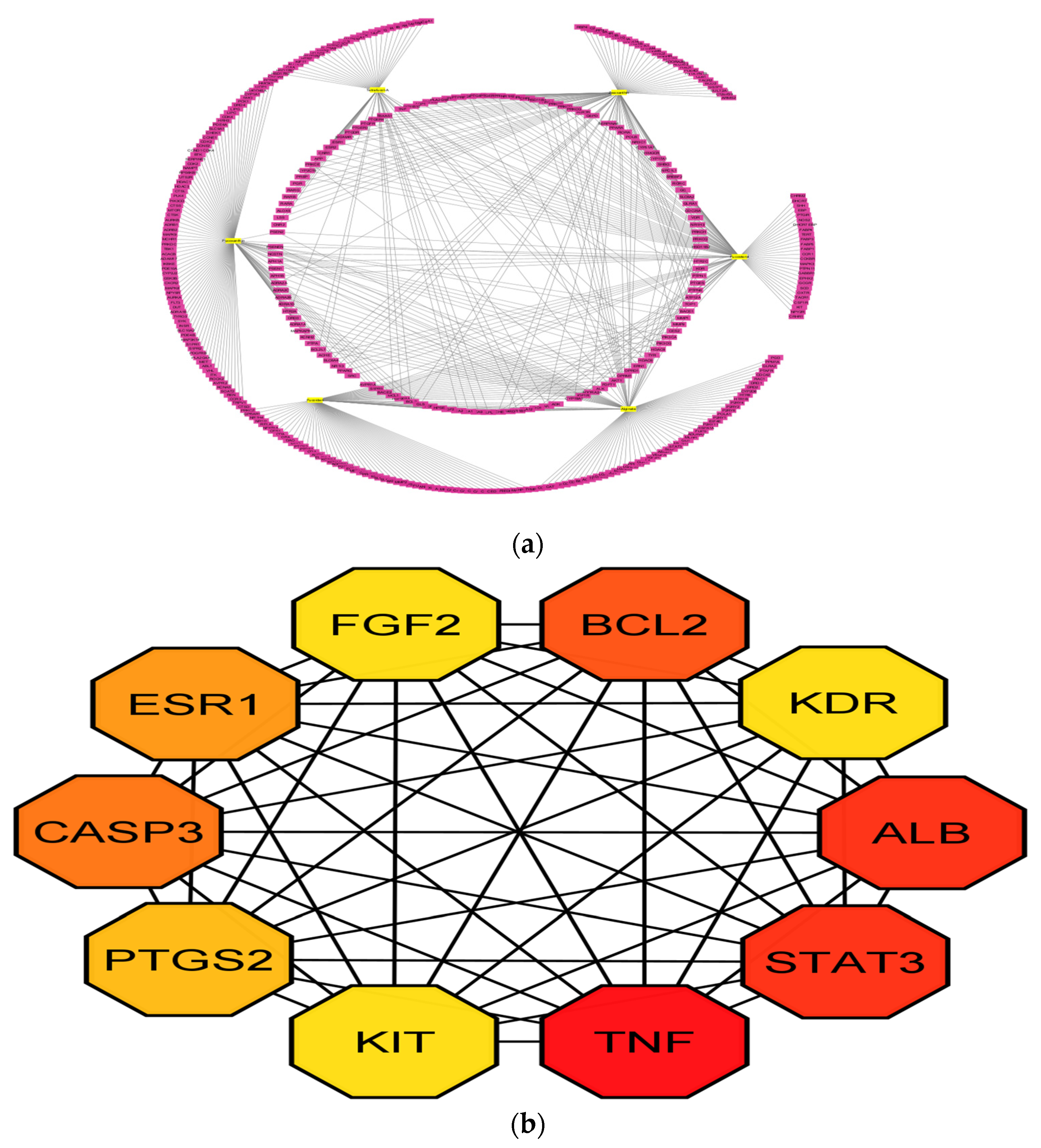
3.2. PPI Network Analysis
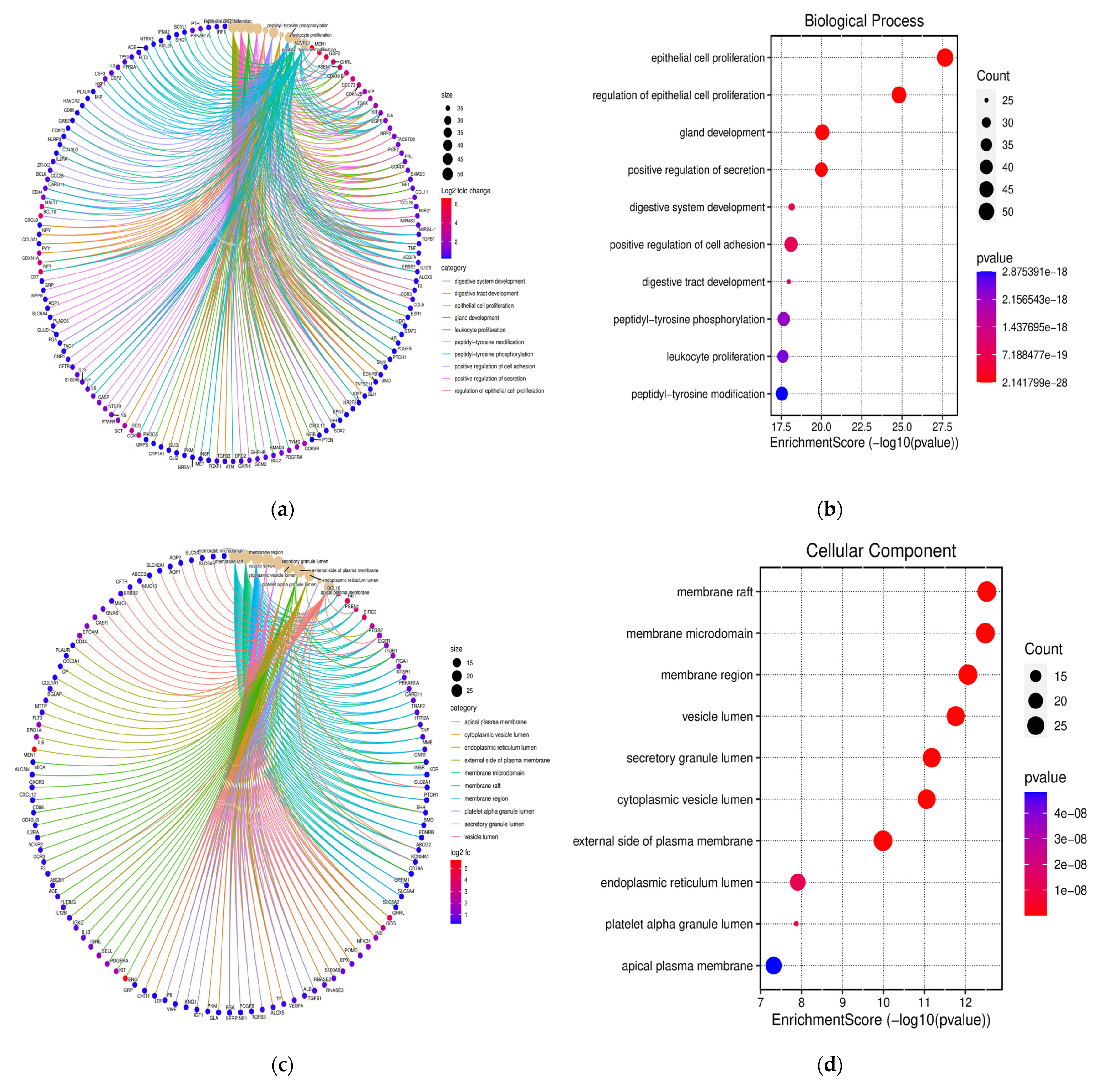
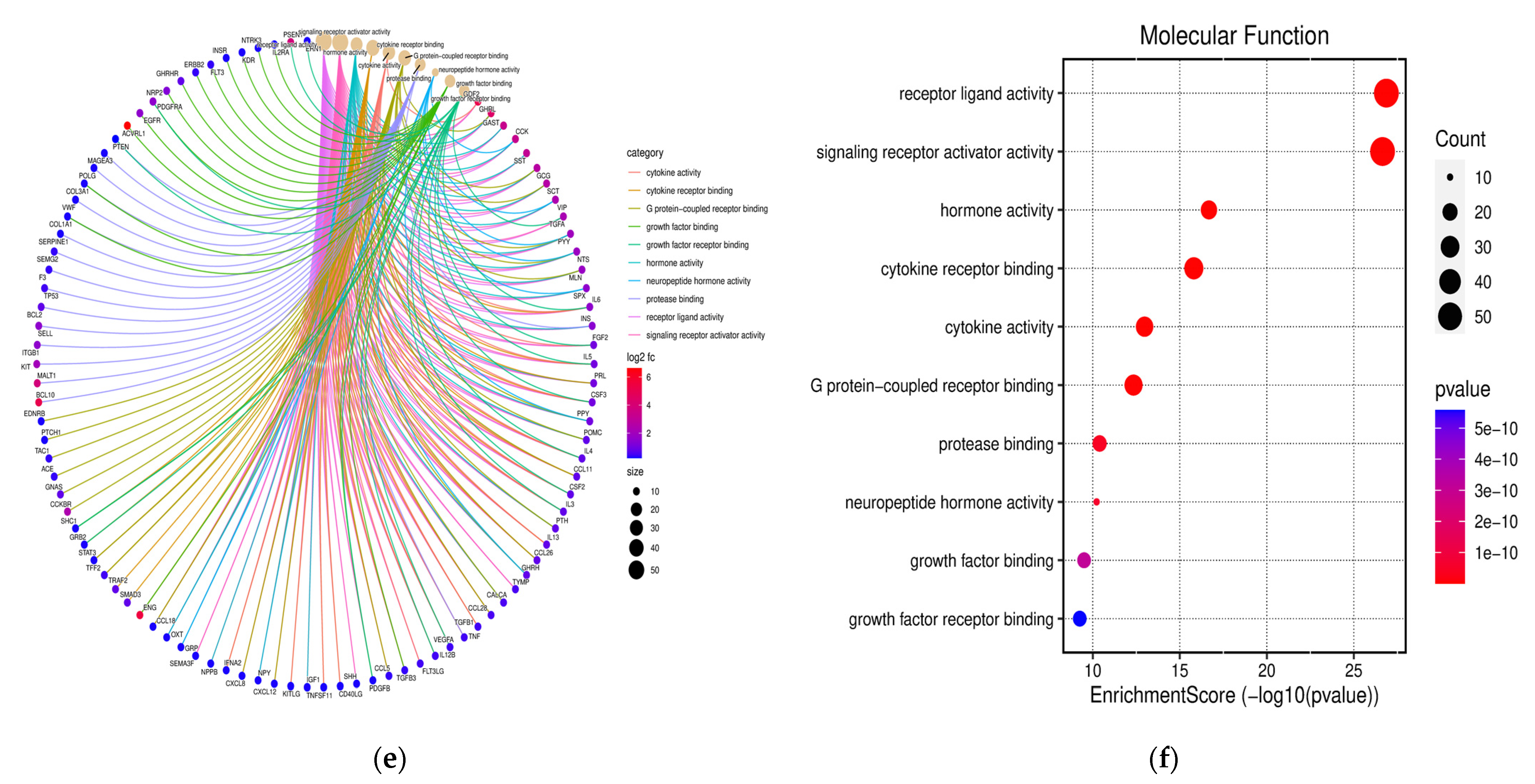
3.3. Functional Annotation and GO Analysis
3.4. KEGG Pathway Examination
3.5. Compound-Protein/Target Interaction (CPI) Network
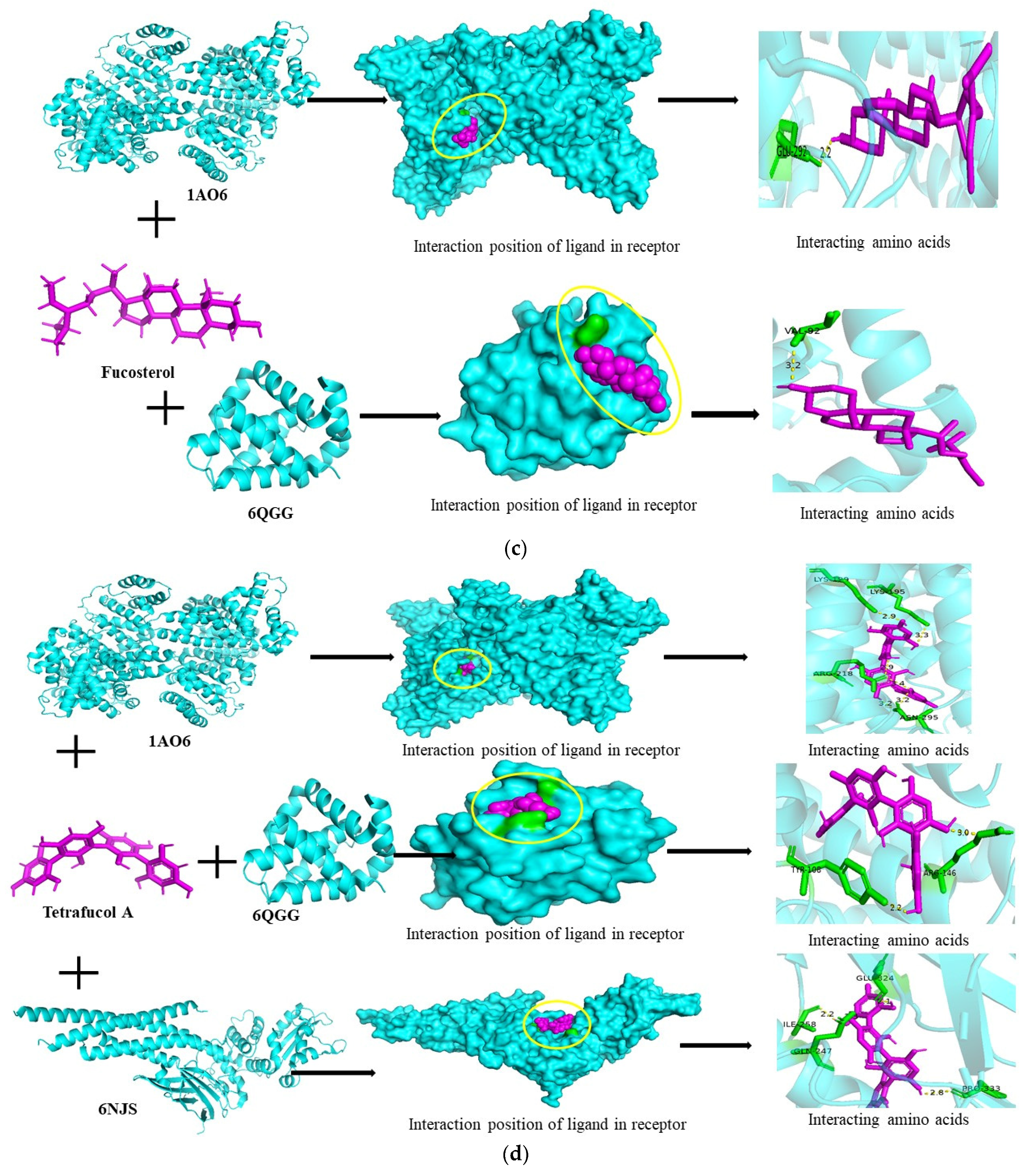
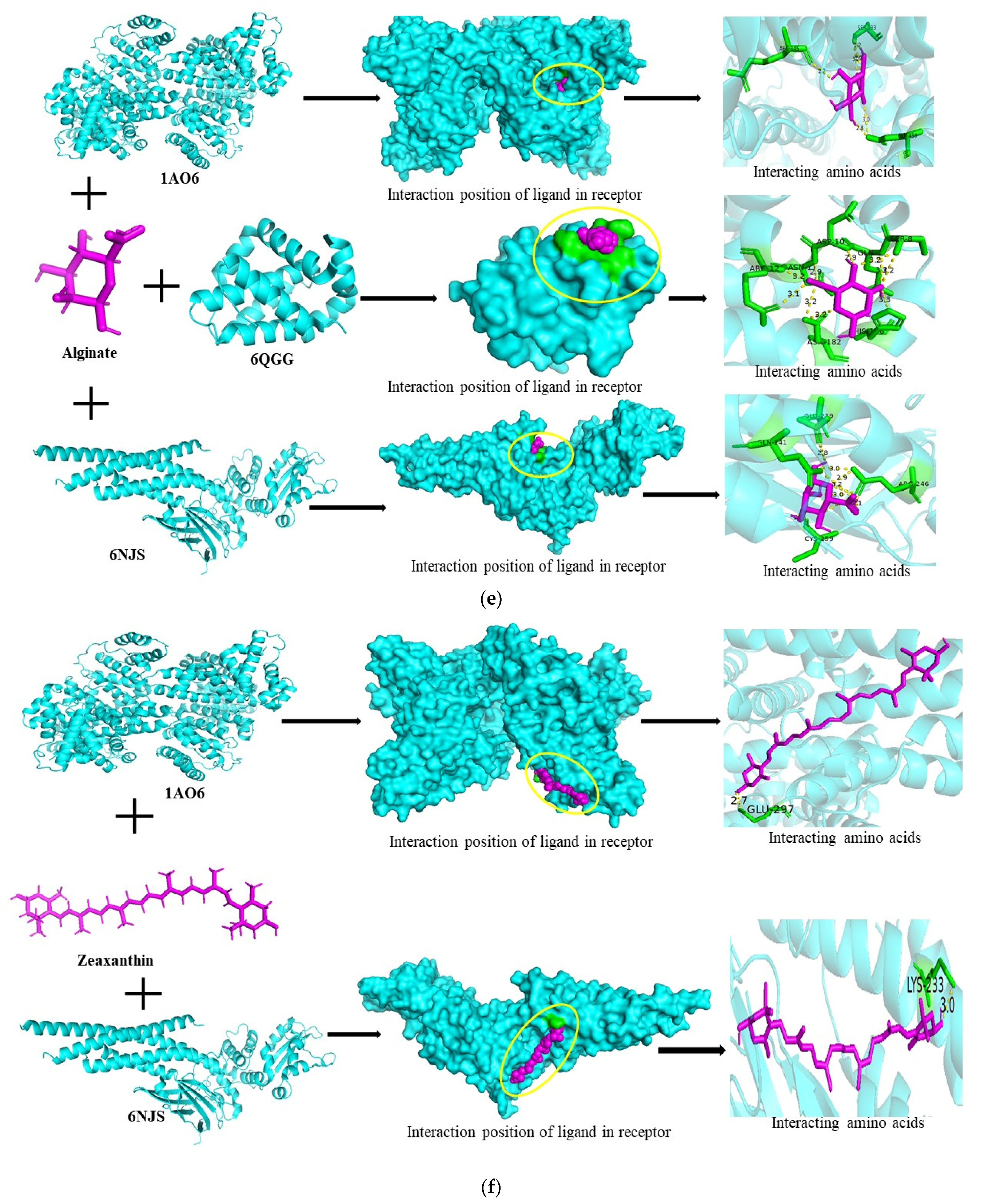
3.6. Molecular Docking Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Beitz, D.C. Physiological and metabolic systems important to animal growth: An overview. J. Anim. Sci. 1985, 61, 1–20. [Google Scholar] [CrossRef]
- Owens, F.N.; Dubeski, P.; Hanson, C.F. Factors that alter the growth and development of ruminants. J. Anim. Sci. 1993, 71, 3138–3150. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.B.; Rodger, J.; Blache, D. Nutritional and environmental effects on reproduction in small ruminants. Reprod. Fertil. Dev. 2004, 16, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Holdt, S.L.; Kraan, S. Bioactive compounds in seaweed: Functional food applications and legislation. J. Appl. Phycol. 2011, 23, 543–597. [Google Scholar] [CrossRef]
- Simpi, C.C.; Nagathan, C.V.; Karajgi, S.R.; Kalyane, N.V. Evaluation of marine brown algae Sargassum ilicifolium extract for analgesic and anti-inflammatory activity. Pharmacogn. Res. 2013, 5, 146–149. [Google Scholar] [CrossRef]
- Subramaniam, S.; Selvaduray, K.R.; Radhakrishnan, A.K. Bioactive compounds: Natural defense against cancer? Biomolecules 2019, 9, 758. [Google Scholar] [CrossRef]
- Lin, S.; Wang, Z.; Lam, K.L.; Zeng, S.; Tan, B.K.; Hu, J. Role of intestinal microecology in the regulation of energy metabolism by dietary polyphenols and their metabolites. Food Nutr. Res. 2019, 63, 1–12. [Google Scholar] [CrossRef]
- Fitton, J.H. Therapies from fucoidan; Multifunctional marine polymers. Mar. Drugs 2011, 9, 1731–1760. [Google Scholar] [CrossRef]
- Vo, T.S.; Kim, S.K. Fucoidans as a natural bioactive ingredient for functional foods. J. Funct. Foods 2013, 5, 16–27. [Google Scholar] [CrossRef]
- Li, B.; Lu, F.; Wei, X.J.; Zhao, R.X. Fucoidan: Structure and bioactivity. Molecules 2008, 13, 1671–1695. [Google Scholar] [CrossRef]
- Ikeguchi, M.; Saito, H.; Miki, Y.; Kimura, T. Effect of fucoidan dietary supplement on the chemotherapy treatment of patients with unresectable advanced gastric cancer. J. Cancer Ther. 2015, 6, 1020–1026. [Google Scholar] [CrossRef]
- Fitton, J.H.; Dell’Acqua, G.; Gardiner, V.A.; Karpiniec, S.S.; Stringer, D.N.; Davis, E. Topical benefits of two fucoidan-rich extracts from marine macroalgae. Cosmetics 2015, 2, 66–81. [Google Scholar] [CrossRef]
- Palanisamy, S.; Vinosha, M.; Marudhupandi, T.; Rajasekar, P.; Prabhu, N.M. Isolation of fucoidan from Sargassum polycystum brown algae: Structural characterization, in vitro antioxidant and anticancer activity. Int. J. Biol. Macromol. 2017, 102, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Flannigan, K.L.; Geem, D.; Harusato, A.; Denning, T.L. Intestinal antigen-presenting cells Key regulators of immune homeostasis and inflammation. Am. J. Pathol. 2015, 185, 1809–1819. [Google Scholar] [CrossRef]
- Lebedev, K.A.; Poniakina, I.D. Immunophysiology of epithelial cells and pattern-recognition receptors. Fiziol. Cheloveka 2006, 32, 114–126. [Google Scholar] [CrossRef]
- Fukata, M.; Arditi, M. The role of pattern recognition receptors in intestinal inflammation. Mucosal Immunol. 2013, 6, 451–463. [Google Scholar] [CrossRef]
- Ikeda-Ohtsubo, W.; Lopez Nadal, A.; Zaccaria, E.; Iha, M.; Kitazawa, H.; Kleerebezem, M.; Brugman, S. Intestinal microbiota and immune modulation in zebrafish by fucoidan from Okinawa mozuku (Cladosiphon okamuranus). Front. Nutr. 2020, 7, 67. [Google Scholar] [CrossRef]
- Liu, S.; Wu, J.; Zhu, Y.; Zhou, W.; Liu, X.; Fu, C.; Ding, Z.; Xu, L.; Zhang, Y.; Meng, Z.; et al. Network pharmacology-based approach to investigate the mechanisms of Shenqi Fuzheng injection in the treatment of breast cancer. Eur. J. Integr. Med. 2020, 34, 101064. [Google Scholar] [CrossRef]
- Song, X.; Zhang, Y.; Dai, E.; Wang, L.; Du, H. Prediction of triptolide targets in rheumatoid arthritis using network pharmacology and molecular docking. Int. Immunopharmacol. 2020, 80, 106179. [Google Scholar] [CrossRef]
- Kim, S.K.; Lee, S.; Lee, M.K.; Lee, S. A systems pharmacology approach to investigate the mechanism of Oryeong-sanformula for the treatment of hypertension. J. Ethnopharmacol. 2019, 244, 112–129. [Google Scholar] [CrossRef]
- Cui, Q.; Zhang, Y.L.; Ma, Y.H.; Yu, H.Y.; Zhao, X.Z.; Zhang, L.H.; Ge, S.Q.; Zhang, G.W.; Qin, X.D. A network pharmacology approach to investigate the mechanism of Shuxuening injection in the treatment of ischemic stroke. J. Ethnopharmacol. 2020, 257, 112891. [Google Scholar] [CrossRef] [PubMed]
- Gfeller, D.; Grosdidier, A.; Wirth, M.; Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: A web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014, 42, W32–W38. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Kohl, M.; Wiese, S.; Warscheid, B. Cytoscape: Software for visualization and analysis of biological networks (M)//data mining in proteomics. Methods Mol. Biol. 2011, 96, 291–303. [Google Scholar]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock 4 and AutoDock Tools 4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Shivanika, C.; Kumar, S.D.; Ragunathan, V.; Tiwari, P.; Sumitha, A.; Devi, P.B. Molecular docking, validation, dynamics simulations, and pharmacokinetic prediction of natural compounds against the SARS-CoV-2 main-protease. J. Biomol. Struct. Dyn. 2022, 40, 585–611. [Google Scholar] [CrossRef]
- Chen, L.; Deng, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Inflammatory Responses and Inflammation-Associated Diseases in Organs. Oncotarget 2018, 9, 7204–7218. [Google Scholar] [CrossRef]
- Jounai, N.; Kobiyama, K.; Takeshita, F.; Ishii, K.J. Recognition of Damage-Associated Molecular Patterns Related to Nucleic Acids during Inflammation and Vaccination. Front. Cell. Infect. Microbiol. 2012, 2, 168. [Google Scholar] [CrossRef]
- Ross, J. The Biology of the Macrophage; ScienceOpen, Inc.: Burlington, MA, USA, 2002. [Google Scholar]
- Yu, R.X.; Hu, X.M.; Xu, S.Q.; Jiang, Z.J.; Yang, W. Effects of Fucoxanthin on Proliferation and Apoptosis in Human Gastric Adenocarcinoma MGC-803 Cells via JAK/STAT Signal Pathway. Eur. J. Pharmacol. 2011, 657, 10–19. [Google Scholar] [CrossRef]
- Yu, R.X.; Yu, R.T.; Liu, Z. Inhibition of Two Gastric Cancer Cell Lines Induced by Fucoxanthin Involves Downregulation of Mcl-1 and STAT3. Hum. Cell 2018, 31, 50–63. [Google Scholar] [CrossRef]
- Zhu, Y.; Cheng, J.; Min, Z.; Yin, T.; Zhang, R.; Zhang, W.; Hu, L.; Cui, Z.; Gao, C.; Xu, S.; et al. Effects of Fucoxanthin on Autophagy and Apoptosis in SGC-7901cells and the Mechanism. J. Cell. Biochem. 2018, 119, 7274–7284. [Google Scholar] [CrossRef] [PubMed]
- Das, S.K.; Hashimoto, T.; Shimizu, K.; Yoshida, T.; Sakai, T.; Sowa, Y.; Komoto, A.; Kanazawa, K. Fucoxanthin Induces Cell Cycle Arrest at G0/G1 Phase in Human Colon Carcinoma Cells through Up-Regulation of p21WAF1/Cip1. Biochim. Biophys. Acta 2005, 1726, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Kotake-Nara, E.; Asai, A.; Nagao, A. Neoxanthin and Fucoxanthin Induce Apoptosis in PC-3 Human Prostate Cancer Cells. Cancer Lett. 2005, 220, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Bteich, M. An overview of albumin and alpha-1-acid glycoprotein main characteristics: Highlighting the roles of amino acids in binding kinetics and molecular interactions. Heliyon 2019, 5, e02879. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.B.; Davidson, J.; Chen, I.; Davis, B.; Dokurno, P.; Graham, C.J.; Harris, R.; Jordan, A.; Matassova, N.; Pedder, C.; et al. Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1. ACS Omega 2019, 4, 8892–8906. [Google Scholar] [CrossRef]
- Lee, M.C.; Huang, Y.C. Soluble eggshell membrane protein-loaded chitosan/fucoidan nanoparticles for treatment of defective intestinal epithelial cells. Int. J. Biol. Macromol. 2019, 131, 949–958. [Google Scholar] [CrossRef]
- Wang, L.; Ai, C.; Wen, C.; Qin, Y.; Liu, Z.; Wang, L.; Gong, Y.; Su, C.; Wang, Z.; Song, S. Fucoidan isolated from Ascophyllum nodosum alleviates gut microbiota dysbiosis and colonic inflammation in antibiotic-treated mice. Food Funct. 2020, 11, 5595–5606. [Google Scholar] [CrossRef]
- Apostolova, E.; Lukova, P.; Baldzhieva, A.; Katsarov, P.; Nikolova, M.; Iliev, I.; Peychev, L.; Trica, B.; Oancea, F.; Delattre, C.; et al. Immunomodulatory and Anti-Inflammatory Effects of Fucoidan: A Review. Polymers 2020, 12, 2338. [Google Scholar] [CrossRef]
- Kim, I.H.; Nam, T.J. Fucoidan downregulates insulin-like growth factor-I receptor levels in HT-29 human colon cancer cells. Oncol. Rep. 2018, 39, 1516–1522. [Google Scholar] [CrossRef]
- Bandaranayake, W.M. The nature and role of pigments of marine invertebrates. Nat. Prod Rep. 2006, 23, 223–255. [Google Scholar] [CrossRef]
- Mondal, A.; Bose, S.; Banerjee, S.; Patra, J.K.; Malik, J.; Mandal, S.K.; Kilpatrick, K.L.; Das, G.; Kerry, R.G.; Fimognari, C.; et al. Marine cyanobacteria and microalgae metabolites—A rich source of potential anticancer drugs. Mar. Drugs 2020, 18, 476. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.X.; Han, Y.L.; Zhu, G.F.; Huang, L.Q.; Deng, Y.Y.; Wang, Q.S.; Jiang, W.Q.; Wen, M.Y.; Han, Q.P.; Xie, D.; et al. Hypertonic saline attenuates expression of Notch signaling and proinflammatory mediators in activated microglia in experimentally induced cerebral ischemia and hypoxic BV-2 microglia. BMC Neurosci. 2017, 18, 32. [Google Scholar] [CrossRef] [PubMed]
- Shabab, T.; Khanabdali, R.; Moghadamtousi, S.Z.; Kadir, H.A.; Mohan, G. Neuroinflammation pathways: A general review. Int. J. Neurosci. 2017, 127, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Teismann, P.; Sathe, K.; Bierhaus, A.; Leng, L.; Martin, H.L.; Bucala, R.; Weigle, B.; Nawroth, P.P.; Schulz, J.B. Receptor for advanced glycation endproducts (RAGE) deficiency protects against MPTP toxicity. Neurobiol. Aging 2012, 33, 2478–2490. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Zhou, H.; Xu, R.; Zhao, Y.; Chinnaswamy, K.; McEachern, D.; Chen, J.; Yang, C.Y.; Liu, Z.; Wang, M.; et al. A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo. Cancer Cell 2019, 36, 498–511.e17. [Google Scholar] [CrossRef]
- Diago, K.; Yamada, C.; Yamaji, M.; Nakagiri, N.; Okada, M.; Komiya, H.; Horiuchi, Y.; Miyazato, T. Pharmacological studies of sodium alginate. V. Effect of sodium alginate on platelet aggregation. Yakugaku Zasshi 1985, 105, 171–182. [Google Scholar] [CrossRef]
- Yamamoto, A.; Itoh, T.; Nasu, R.; Kajiwara, E.; Nishida, R. Sodium alginate inhibits methotrexate-induced gastrointestinal mucositis in rats. Biol. Pharm. Bull. 2013, 36, 1528–1534. [Google Scholar] [CrossRef]
- Yamamoto, A.; Itoh, T.; Nasu, R.; Nishida, R. Sodium alginate ameliorates indomethacin-induced gastrointestinal mucosal injury via inhibiting translocation in rats. World J. Gastroenterol. 2014, 20, 2641–2652. [Google Scholar] [CrossRef]
- Guan, Q.; Zhang, J. Recent advances: The imbalance of cytokines in the pathogenesis of inflammatory bowel disease. Mediat. Inflamm. 2017, 2017, 4810258. [Google Scholar] [CrossRef]
- Ying, C.; Chen, L.; Wang, S.; Mao, Y.; Ling, H.; Li, W.; Zhou, X. Zeaxanthin ameliorates high glucose-induced mesangial cell apoptosis through inhibiting oxidative stress via activating AKT signalling-pathway. Biomed. Pharmacother. 2017, 90, 796–805. [Google Scholar] [CrossRef]
- Reif, S.; Klein, I.; Lubin, F.; Farbstein, M.; Hallak, A.; Gilat, T. Pre-illness dietary factors in inflammatory bowel disease. Gut 1997, 40, 754–760. [Google Scholar] [CrossRef] [PubMed]
- Lubin, F.; Rozen, P.; Arieli, B.; Farbstein, M.; Knaani, Y.; Bat, L. Nutritional and lifestyle habits and water-fiber interaction in colorectal adenoma etiology. Cancer Epidemiol. Biomark. Prev. 1997, 6, 79–85. [Google Scholar]
- Głąbska, D.; Guzek, D.; Zakrzewska, P.; Lech, G. Intake of Lutein and Zeaxanthin as a Possible Factor Influencing Gastrointestinal Symptoms in Caucasian Individuals with Ulcerative Colitis in Remission Phase. J. Clin. Med. 2019, 8, 77. [Google Scholar] [CrossRef] [PubMed]



| Compound_ID | Active Compound | DL | OB% | Structure |
|---|---|---|---|---|
| 129532628 | Fucoidan | −1.63 | 0.56 |  |
| 71308278 | Tetrafucol A | −1.82 | 0.17 | 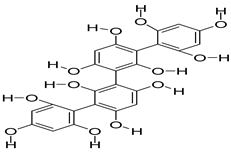 |
| 5281239 | Fucoxanthin | −0.35 | 0.17 | 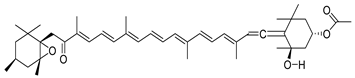 |
| 5281328 | Fucosterol | 0.85 | 0.55 | 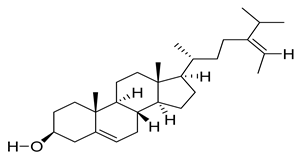 |
| 5280899 | Zeaxanthin | −0.18 | 0.17 | 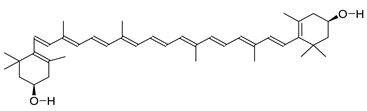 |
| 5102882 | Alginate | −0.08 | 0.11 | 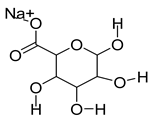 |
| Standard Parameters | Fucoxanthin | Fucoidan | Fucosterol | Tetrafucol A | Sodium Alginate | Zeaxanthin |
|---|---|---|---|---|---|---|
| GI absorption | Low | High | Low | Low | Low | Low |
| BBB | No | No | Yes | Yes | No | No |
| P-gp substrate | Yes | No | No | No | Yes | Yes |
| CYP1A2 inhibitor | No | No | No | No | No | No |
| CYP2C19 inhibitor | No | No | No | No | No | No |
| CYP2C9 inhibitor | No | No | No | Yes | No | No |
| CYP2D6 inhibitor | No | No | No | No | No | No |
| CYP3A4 inhibitor | No | No | No | No | No | No |
| Log Kp (Skin permeation; cm/s) | −4.60 | −8.56 | −2.53 | −7.54 | −11.71 | −2.02 |
| Toxicity | ||||||
| Hepatoxicity | No | No | No | No | No | No |
| Carcinogenicity | Yes | Yes | No | No | No | No |
| Immunotoxicity | Yes | No | Yes | No | No | No |
| Mutangenicity | No | No | No | No | No | No |
| Cytotoxicity | No | No | No | No | No | No |
| Uniprot ID | Gene Symbol | Target Name | Degree | Betweenness |
|---|---|---|---|---|
| 2099 | ESR1 | Estrogen Receptor 1 | 168 | 166,699.5 |
| 5290 | PIK3CA | Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Alpha | 131 | 117,015 |
| 6774 | STAT3 | Signal transducer and activator of transcription 3 | 128 | 93,667.13 |
| 367 | AR | Androgen receptor | 107 | 68,610.89 |
| 2908 | NR3C1 | Nuclear receptor subfamily 3 group C member 1 | 80 | 41,933.35 |
| 2735 | GLI1 | Glioma-associated oncogene homologue 1 | 77 | 64,882.65 |
| 836 | CASP3 | cysteine-aspartic acid protease | 76 | 52,311.44 |
| 4233 | MET | Mesenchymal Epithelial Transition | 76 | 42,045.77 |
| 3791 | KDR | Kinase inserts domain receptor | 70 | 41,273.74 |
| 6198 | RPS6KB1 | Ribosomal protein S6 kinase B1 | 51 | 38,342.19 |
| 5580 | PRKCD | Protein kinase C delta | 50 | 26,321.13 |
| 1576 | CYP3A4 | Cytochrome P450 3A4 | 49 | 26,557.22 |
| 7124 | TNF | Tumor Necrosis Factor | 47 | 33,392.7 |
| 3643 | INSR | Insulin Receptor | 43 | 19,377.29 |
| 3815 | KIT | KIT Proto-Oncogene, Receptor Tyrosine Kinase | 40 | 14,474.71 |
| 5663 | PSEN1 | Presenilin-1 | 38 | 27,637.65 |
| 1543 | CYP1A1 | Cytochrome P450 Family 1 Subfamily A Member 1 | 36 | 30,323.08 |
| 2100 | ESR2 | Estrogen receptor 2 | 35 | 5959.107 |
| 7015 | TERT | Telomerase reverse transcriptase | 34 | 25,487.66 |
| 7422 | VEGFA | Vascular endothelial growth factor A | 31 | 15,861.87 |
| 5788 | PTPRC | Protein tyrosine phosphatase receptor type C | 29 | 20,399.73 |
| 596 | BCL2 | B-cell lymphoma 2 | 28 | 19,894.33 |
| 2247 | FGF2 | Fibroblast growth factor 2 | 25 | 12,124.24 |
| 5293 | PIK3CD | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta | 25 | 1706.552 |
| 6469 | SHH | Sonic Hedgehog Signaling Molecule | 23 | 15,708.06 |
| 1232 | CCR3 | C-C motif chemokine receptor 3 | 23 | 27,637.78 |
| 6608 | SMO | Smoothened, frizzled class receptor | 19 | 10,874.2 |
| 7428 | VHL | Von Hippel-Lindau tumor suppressor | 19 | 12,073.5 |
| 2322 | FLT3 | FMS-like tyrosine kinase 3 | 18 | 3634.476 |
| 213 | ALB | Albumin | 17 | 16,632 |
| 1910 | EDNRB | Endothelin Receptor Type B | 15 | 5792.32 |
| 5054 | SERPINE1 | Endothelial plasminogen activator inhibitor 1 | 14 | 17,182.99 |
| 5295 | PIK3R1 | Phosphoinositide-3-kinase regulatory subunit 1 | 13 | 6946.97 |
| 1901 | S1PR1 | Sphingosine-1-phosphate receptor 1 | 13 | 2106.34 |
| 1813 | DRD2 | Dopamine receptor D2 | 12 | 6695.78 |
| 973 | CD79A | Cluster of differentiation 79 associated protein alpha | 12 | 3759.8 |
| 1268 | CNR1 | Cannabinoid Receptor 1 | 12 | 3593.6 |
| 207 | AKT1 | AKT serine/threonine kinase 1 | 11 | 14,063.64 |
| 3156 | HMGCR | 3-hydroxy-3-methylglutaryl-CoA reductase | 10 | 7971.86 |
| 5595 | MAPK3 | Mitogen-activated protein kinase 3 | 10 | 7717.44 |
| 3320 | HSP90AA1 | Heat shock protein 90 alpha family class A member 1 | 10 | 6916.55 |
| 6868 | ADAM17 | A disintegrin and metalloprotease 17 | 10 | 5706.29 |
| 6714 | SRC | Proto-oncogene tyrosine-protein kinase | 10 | 3812.01 |
| 2885 | GRB2 | Growth factor receptor bound protein 2 | 10 | 3631.08 |
| Gene Name | Compound | Score | Pathways |
|---|---|---|---|
| TNF | Fucosterol | 46 | Regulation of epithelial cell proliferation, gland development, positive regulation of secretion, digestive system development, digestive tract development, peptidyl-tyrosine phosphorylation, activation of protein kinase activity, positive regulation of secretion by cell, leukocyte cell-cell adhesion, positive regulation of peptidyl-tyrosine phosphorylation, regulation of peptide secretion, protein kinase B signaling, positive regulation of MAP kinase activity, positive regulation of peptide secretion, hormone transport, regulation of response to wounding, membrane microdomain, cytokine activity, TNF signaling pathway, NF-kappa B signaling pathway, inflammatory bowel disease |
| STAT3 | Sodium Alginate/Fucoxanthin/Tetrafucol A/Zeaxanthin | 39 | Positive regulation of epithelial cell proliferation, peptidyl-tyrosine phosphorylation, positive regulation of peptidyl-tyrosine phosphorylation, T cell activation, regulation of inflammatory response, G protein-coupled receptor binding, inflammatory bowel disease |
| ALB | Fucoxanthin/Tetrafucol A/Fucosterol/Zeaxanthin/Fucoidan | 39 | Blood microparticle, vesicle lumen, secretory granule lumen, cytoplasmic vesicle lumen, endoplasmic reticulum lumen, drug binding |
| BCL2 | Fucoxanthin/Tetrafucol A/Fucosterol/Fucoidan | 36 | Gland development, digestive system development, digestive tract development, response to nutrient levels, ossification, positive regulation of lymphocyte and leukocyte activation, multi-multicellular organism process, positive regulation of cell activation, T cell activation, membrane raft, focal adhesion |
| CASP3 | Fucoidan | 34 | Signaling receptor activator activity |
| ESR1 | Tetrafucol A | 33 | Multi-multicellular organism process, epithelial tube morphogenesis, regulation of inflammatory response, DNA-binding transcription factor binding |
| PTGS2 | Fucoidan/Fucoxanthin/Sodium alginate | 29 | Response to nutrient levels, regulation of blood pressure, ossification, multi-multicellular organism process, response to lipopolysaccharide, regulation of inflammatory response, muscle cell proliferation, response to hypoxia, response to corticosteroid, plasma membrane raft, arachidonic acid metabolism, gastric acid secretion |
| KDR | Fucosterol | 25 | Positive regulation of epithelial cell proliferation, peptidyl-tyrosine phosphorylation, protein kinase B signaling, epithelial tube morphogenesis, positive regulation of ERK1 and ERK2 cascade, membrane microdomain, membrane region, protein tyrosine kinase activity, focal adhesion |
| FGF2 | Fucoidan/Fucoxanthin | 25 | Regulation of epithelial cell proliferation, activation of protein kinase activity, protein kinase B signaling, positive regulation of MAP kinase activity, positive regulation of ERK1 and ERK2 cascade, muscle cell proliferation, transcription coregulator activity |
| KIT | Fucosterol/Fucoxanthin/Tetrafucol A | 25 | Epithelial cell proliferation, digestive system development, digestive tract development, peptidyl-tyrosine phosphorylation, activation of protein kinase activity, positive regulation of peptidyl-tyrosine phosphorylation, protein kinase B signaling, positive regulation of MAP kinase activity, protein tyrosine kinase activity |
| Ligand Name | Protein Name | Docking Speed | Binding Energy (Kcal/mol) | Interacting Residues |
|---|---|---|---|---|
| Fucoidan | 1AO6 | −1,240,832,140 | −5.8 | SER-193, GLN-459, ASP-108 |
| 1NME | 521,076,508 | −0.0 | No binding | |
| 6NJS | 1,503,384,624 | −4.9 | LYS-370, ASP-371 | |
| 6QGG | −1,184,102,352 | −5.5 | ARG-12, ASN-11, ASP-10, ASN-182, GLN-9, SER-8, HIS-186 | |
| 6RMJ | 1,792,153,888 | −0.0 | No binding | |
| Alginate | 1AO6 | −1,949,256,596 | −6.0 | ARG-145, SER-193, GLN-459 |
| 1NME | −1,074,133,996 | +0.0 | No binding | |
| 6NJS | 1,628,093,584 | −5.5 | GLN-239, GLN-141, ARG-246, CYS-259 | |
| 6QGG | 309,544,064 | −4.9 | ARG-12, ASN-11, ASP-10, SER-8, HIS-186, ASN-182, GLN-9 | |
| 6RMJ | 721,266,312 | +0.0 | No binding | |
| Fucosterol | 1AO6 | −339,712,888 | −5.7 | GLN-292 |
| 1NME | 1,358,786,528 | +0.0 | No binding | |
| 6NJS | 265,472,888 | −6.2 | Hydrophobic interaction | |
| 6QGG | 1,571,179,760 | −6.8 | VAL-92 | |
| 6RMJ | 709,042,400 | −0.0 | No binding | |
| Fucoxanthin | 1AO6 | 26,901,568 | −8.3 | ARG-186, ARG-114 |
| 1NME | −1,890,328,104 | −0.0 | No binding | |
| 6NJS | 1,489,833,176 | −6.8 | GLU-506 | |
| 6QGG | 1,833,423,984 | −8.1 | SER-129 | |
| 6RMJ | 771,959,900 | −0.0 | No binding | |
| Tetrafucol A | 1AO6 | 2,005,618,124 | −8.3 | LYS-199, LYS-195, ARG-218, ASN-295 |
| 1NME | −1,003,280,536 | −0.0 | No binding | |
| 6NJS | 1,664,256,024 | −7.7 | GLU-324, ILE-258, GLN-247, PRO-333 | |
| 6QGG | −429,812,260 | −7.5 | TYR-108, ARG-146 | |
| 6RMJ | 1,619,515,552 | −0.0 | No binding | |
| Zeaxanthin | 1AO6 | 612,375,932 | −6.2 | GLU-297 |
| 1NME | 910,425,852 | −0.0 | No binding | |
| 6NJS | −675,009,912 | −7.3 | LYS-233 | |
| 6QGG | −734,827,780 | −6.6 | Hydrophobic interaction | |
| 6RMJ | −178,329,644 | +0.0 | No binding |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Published by MDPI on behalf of the Österreichische Pharmazeutische Gesellschaft. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anjum, V.; Bagale, U.; Kadi, A.; Potoroko, I. A Network Pharmacology and Molecular Docking Technology to Identify and Explore Mechanism of Bioactive Components of Fucus vesiculosus against Gut Disorders. Sci. Pharm. 2024, 92, 49. https://doi.org/10.3390/scipharm92030049
Anjum V, Bagale U, Kadi A, Potoroko I. A Network Pharmacology and Molecular Docking Technology to Identify and Explore Mechanism of Bioactive Components of Fucus vesiculosus against Gut Disorders. Scientia Pharmaceutica. 2024; 92(3):49. https://doi.org/10.3390/scipharm92030049
Chicago/Turabian StyleAnjum, Varisha, Uday Bagale, Ammar Kadi, and Irina Potoroko. 2024. "A Network Pharmacology and Molecular Docking Technology to Identify and Explore Mechanism of Bioactive Components of Fucus vesiculosus against Gut Disorders" Scientia Pharmaceutica 92, no. 3: 49. https://doi.org/10.3390/scipharm92030049
APA StyleAnjum, V., Bagale, U., Kadi, A., & Potoroko, I. (2024). A Network Pharmacology and Molecular Docking Technology to Identify and Explore Mechanism of Bioactive Components of Fucus vesiculosus against Gut Disorders. Scientia Pharmaceutica, 92(3), 49. https://doi.org/10.3390/scipharm92030049






