Abstract
The algae Kappaphycus alvarezii is considered an important raw material for industrial practices, producing high economic value of various derived products. However, the quality of this commodity, which can be indicated by the level of phenolic compounds, may vary due to growth factors, including cultivation sites. An analytical UV-Vis spectrophotometry method coupled with chemometrics was proposed to standardize the red alga based on the content of phenolic compounds. The correlation between the UV-Vis spectra and UPLC–PDA results, combined with a multivariate calibration of the K. alvarezii extracts, was analyzed. The extracts were prepared using an ultrasound-based technique and subsequently subjected to UV-Vis spectral measurements at 200–800 nm and UPLC–PDA at 260 and 330 nm. Chemometric techniques and partial least squares (PLS) were applied to the acquired data to build a reliable analysis of the phenolics in the K. alvarezii extracts. The result showed that the wavelength combination of 200–450 and 600–690 nm provided a valid method for quantitative analysis of the studied phenolics that belong to hydroxybenzoic acid, hydroxycinnamic acid, and flavonoid with a coefficient of regression (R2) > 0.96 in the calibration and validation models, along with an RMSEC and RMSEP value < 8%. The method was then employed to characterize the K. alvarezii samples from 13 different cultivation areas. Principal component analysis (PCA) generated principal components that produced a clear distribution among the samples of K. alvarezii based on phenolic compounds corresponding to the geographical origin.
1. Introduction
Indonesia is the second-largest producer country of raw macroalgae after China, which supplies 25% of the world’s Kappaphycus alvarezii [1]. Several Southeast Asian countries, including Indonesia, Malaysia, and the Philippines, provide sheltered areas favorable for the cultivation of K. alvarezii [2]. In Indonesia, the cultivation regions cover the main islands such as Sumatra, Java, Sulawesi, Bali, and Lombok. In addition to its common use as a raw material for the production of carrageenan and agar, K. alvarezii has the potential to become one of the ingredients of functional foods due to the presence of bioactive phenolic compounds [3]. Several derivatives of cinnamic acid, hydroxybenzoic acid, and flavonoids are known to be naturally presented in K. alvarezii [4]. However, different cultivation sites influence the composition and levels of the phenolic compounds in K. alvarezii [5]. The fact that K. alvarezii is an essential raw material for industrial practices endorses the necessity to develop reliable analytical methods to standardize this red alga based on the level of phenolic compounds. To standardize the quality of K. alvarezii, information about the levels of total and individual phenolics could be used to establish fingerprinting data [6].
Chromatographic techniques such as high-performance liquid chromatography (HPLC) and ultra-performance liquid chromatography (UPLC) have been developed for phenolic compounds determination in plant sources [7,8]. The UPLC–PDA technique has successfully characterized the phenolic acids and flavonoids from herbal plants [9]. Additionally, the UPLC–PDA was employed to characterize and quantify the phenolic compounds in K. alvarezii due to a faster separation than the HPLC. Henceforth, the analyses can be performed in reduced time with an increase in resolution. [10].
In recent years, the Fourier transform infrared (FTIR) and ultraviolet-visible (UV-Vis) spectroscopy techniques for data fingerprinting have attracted much attention. First, because they provide information rapidly yet reliably, and second because most organic compounds produce signals in the IR or UV-Vis spectroscopic ranges. Moreover, their combination with multivariate statistical analysis allows for the authentication of plant origins [11], the detection of adulteration [12], and the prediction of food quality parameters [13]. UV-Visible spectroscopy appears to be suitable for analyzing phenolic compounds considering phenolic compounds have a strong capacity to absorb both UV and visible radiation [14]. UV-Vis spectrophotometry is known to have better ability than FTIR as an analytical method to authenticate and predict the content of phenolic compounds in green tea with the help of the partial least squares (PLS) regression model [15].
The UV-Vis spectrum of plant extracts usually contains information from hundreds of different compounds at different levels. Because of the broad absorption ranges from individual compounds, it is not easy to find specified wavelengths showing information from individual compounds. However, a combination of several different individual wavelengths can provide specific information about individual components in the samples [16]. Partial least squares (PLS) regression and principal component analysis (PCA) are the most commonly applied methods to establish the correlation between the spectroscopic data and specific values for individual components, as well as to develop fingerprints allowing for characterization in food products [17,18]. PLS is known to have the higher predictive ability of the models [19]. Therefore, this manuscript discloses the application of multivariate calibration to correlate the spectroscopic (UV-Vis) spectra with the individual phenolic compounds provided by the chromatographic analyses (UPLC–PDA) for the standardization of K. alvarezii at different cultivation sites.
2. Materials and Methods
2.1. Sampling and Sample Preparation
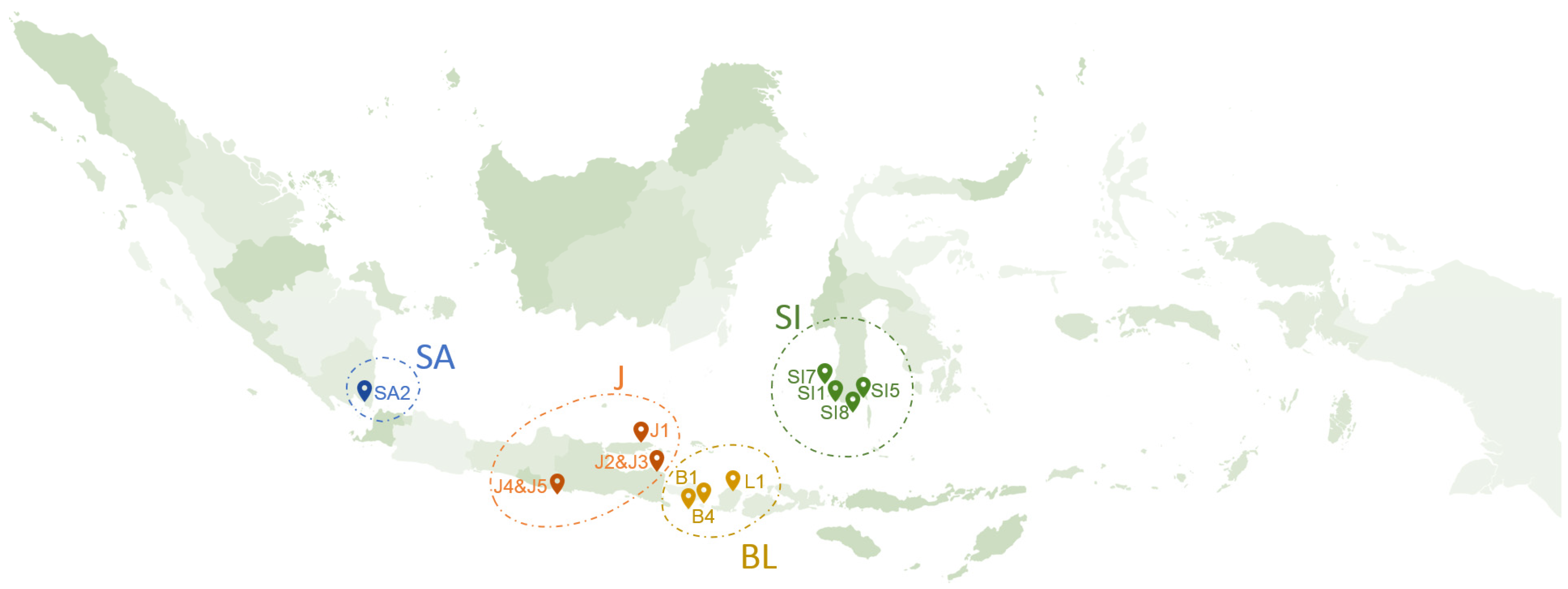
The fresh K. alvarezii from Jepara Brackish Water Aquaculture Center was used to build a predictive model for the studied phenolic compounds. For method application, samples of 13 freshly harvested K. alvarezii were collected from different geographical regions in Indonesia (Figure 1), including Sumatera (SA): Teluk Pandan (SA2); Java (J): Pacitan (J4 and J5), Banyuwangi (J2 and J3), and Sumenep (J1); Bali and Lombok (BL): Nusa Penida (B4), Lembongan (B1), and Lombok (L1); and Celebes (SI): Tanakeke island (Sl7), Bantaeng (Sl5), Puntondo (Sl1), and Djene Ponto (Sl8). The samples were harvested from August 2020 to January 2021. The fresh K. alvarezii samples were freeze-dried for 48 h, ground into powder, and stored at 4 °C in a refrigerator until further treatment.
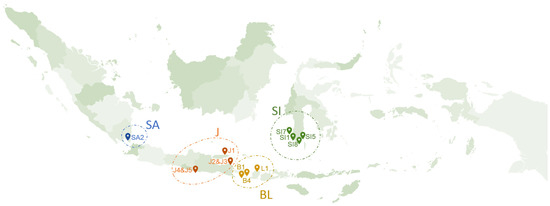
Figure 1.
Sampling illustration at the cultivation sites, Sumatera (SA): Teluk Pandan (SA2); Java (J): Pacitan (J4 and J5), Banyuwangi (J2 and J3), and Sumenep (J1); Bali and Lombok (BL): Nusa Penida (B4), Lembongan (B1), and Lombok (L1); and Celebes (SI): Tanakeke island (Sl7), Bantaeng (Sl5), Puntondo (Sl1), and Djene Ponto (Sl8).
Approximately 1 g of the finely ground K. alvarezii sample was weighed. The sample was then extracted using different conditions of ultrasound-assisted extraction (UP200St, Hielscher Ultrasonics GmbH, Teltow, Germany) to obtain extracts that varied in the content of phenolic compounds. The extraction conditions used are described in Table 1. Subsequently, the real sample application was performed by setting the ultrasonic device at 26 kHz frequency, 100% ultrasound power (200 W), 0.6 s−1 pulse duty-cycle, and 56.4 °C extraction temperature and by employing extraction solvent 50% ethanol in water with a solvent to sample ratio of 1:25. The obtained extract was concentrated using a rotary evaporator under vacuum and adjusted to 5 mL with a fresh solvent. The extract was then passed through a 0.45 μm nylon filter before being sent to the detection system.

Table 1.
Ultrasound-assisted conditions to prepare extracts from K. alvarezii with different concentrations of phenolic compound.
2.2. UPLC-PDA
The chromatographic analyses were performed using ACQUITY UPLC H-Class equipment. The UPLC system was managed by Empower 3 Chromatography Data (Waters Corporation, Milford, MA, USA). The detector was an ACQUITY UPLC photodiode array (PDA). The PDA was set to three-dimensional (3D) scan mode for compound identification, capturing 40 points per second from 200 to 400 nm. In the case of compound quantification, a two-dimensional (2D) scan of PDA at 80 points per second collection data rate at a fixed wavelength provided maximum absorbance of the corresponding compounds (260 and 330 nm).
The separations of phenolic compounds in 3.0 µL injected samples were performed using a reversed-phase column at a temperature of 47 °C. A particle-based (PB) column, ACQUITY UPLC ethylene bridging hybrid (BEH, Waters Corporation, Wexford, Ireland), was used. The PB column was 100 mm long, with a 2.1 mm inner diameter and a particle size of 1.7 µm. The mobile phases consisted of two solvents: phase A (2% acetic acid in water) and phase B (2% acetic acid in acetonitrile). The 4.0 min gradient program was as follows (%B): 0–3 min, 4.1–50.2%; 3–4 min, 50.2–100%. The flow rate was set at 0.64 mL min−1. After the analysis, the columns were washed for 3 min with phase B. The following injection was performed with 3 min to equilibrate [8].
2.3. UV–Vis Spectra Acquisition
A UV-Vis spectrophotometer (Genesys 10S UV-Vis, Thermo Fisher, Tianjin, China) was used to measure the spectra of the K. alvarezii extracts. The spectrum was recorded between 200 nm and 800 nm (at 1 nm intervals). Each sample was measured in duplicate. UV-Vis spectra were exported from the Thermo Fisher UV-Vis spectrophotometer in .csv format. The data was arranged into a matrix of 46 (sample) × 601 (absorbance) for the calibration model and 14 (sample) × 601 (absorbance) for the real sample application.
2.4. Multivariate Calibration Analysis
The data matrix containing 46 (sample) × 601 (absorbance) was imported into Unscrambler × 10.4 (Camo Software AS, Oslo, Norway) for data processing and further regression analysis. The effects of two different processing methods on the PLS regression were compared, including the combination of smoothing with the first derivative using the Savitzky–Golay method with first-order polynomials through 11 smoothing second derivatives using the Savitzky–Golay method with second-order polynomials through 13 points. Prior to PLS analysis, both the raw and modified spectral data were mean-centered. The optimum processing method was used for further analysis.
UPLC–PDA data at 260 nm and 330 nm (Y) and UV–Vis spectra (X) were used to create a PLS regression model to predict the phenolic compound of K. alvarezii, with the optimum range of the selected wavelength. The Kennard–Stone technique was used to partition the data matrices into calibration and validation sets. The calibration set was used to construct and optimize the model, while the validation set was used to assess the model’s prediction performance. The performance of the model was assessed by the parameters of the determination coefficient of calibration (R2c), cross-validation (R2cv) and prediction (R2p), and the root mean square error of calibration (RMSEC), cross-validation (RMSECV), and prediction (RMSEP).
2.5. Real Sample Application
The developed PLS model was then applied to the real samples. PCA was created to visualize the data structure based on the original group with the selected wavelength range. On the other hand, the PLS developed model was used to predict phenolic compounds on 13 real samples. Then, PCA and CA were constructed to analyze the correlation between geographical origin and the phenolic compound.
3. Result and Discussion
3.1. Identification of Phenolic Compounds in the K. alvarezii Extracts
The chromatographic system (UPLC-PDA) used in this study provided sufficient separation of individual compounds in the K. alvarezii extracts. Each resulting peak in the chromatogram was checked to identify the compound based on the full UV spectra recorded by the PDA detection system. General identification was performed by comparing the resulting spectra of the injected sample with the chromophore spectra of the phenolic backbone. Two peaks were identified as derivatives of hydroxycinnamic acids (HCA1 and HCA2), while the others were derivatives of hydroxybenzoic acid (HBA) and flavonoid. Because of the similarities between the UV-Vis spectra by several hydroxybenzoic derivatives and flavonoid, absorbance above 350 nm was measured. The two hydroxycinnamic acid-derived compounds were the major peaks on the extracting chromatogram at 330 nm, and hence were quantified at this wavelength. Meanwhile, the typical channel for quantifying compounds derived from hydroxybenzoic acid (HBA) and flavonoid was 260 nm.
3.2. UV-Vis Spectra of Phenolic Compounds in the K. alvarezii Extracts
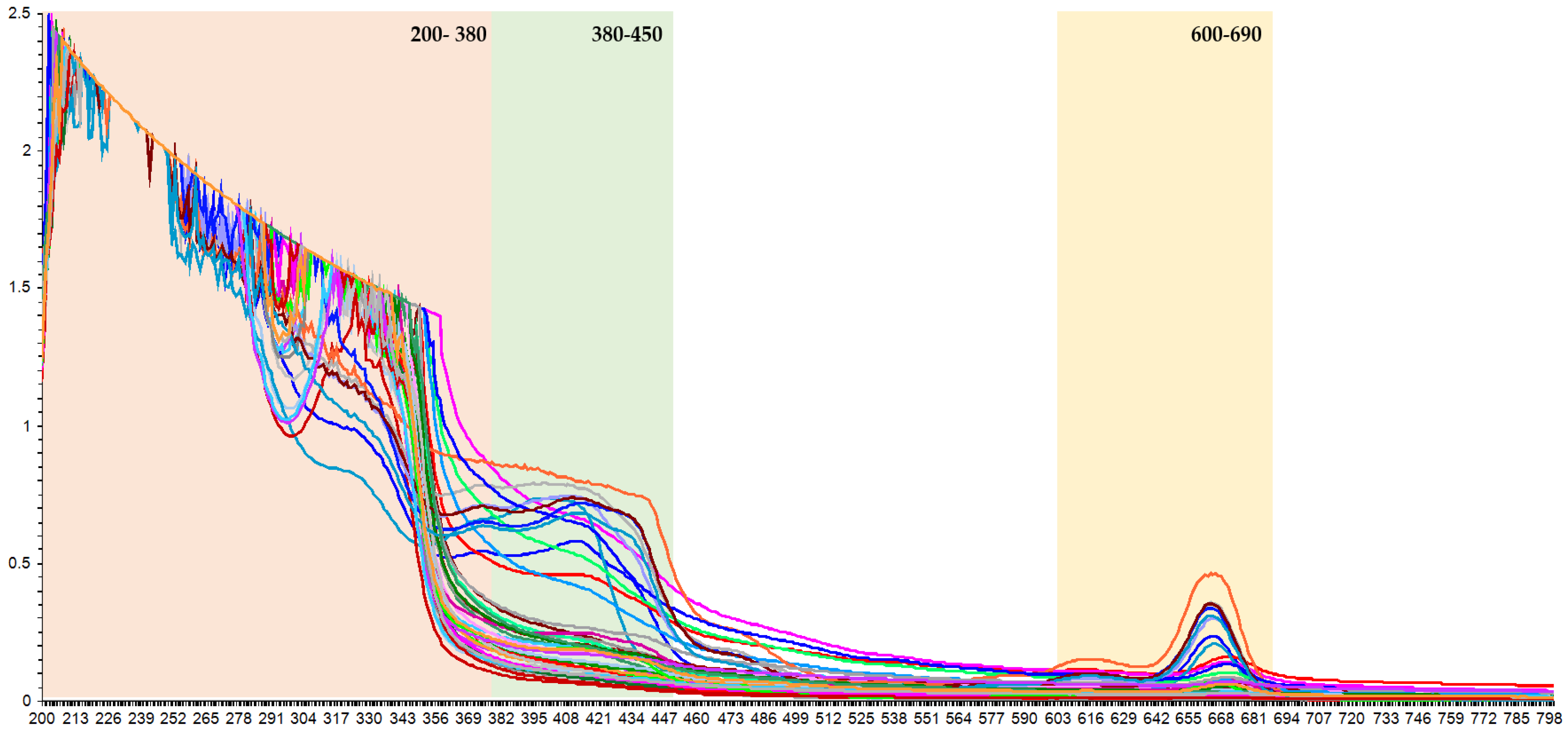
Several extractions conditions, including ultrasound power, extraction temperature, and solvent composition [20], produced extracts with different levels of individual phenolic compounds (Table 2). A set of extracts with different levels of phenolic compounds was needed to develop a correlation model between the UV-Vis spectra and the results from the UPLC-PDA system. Since the chromophores of phenolic compounds have a high capacity to absorb UV-Vis radiation, the UV-Vis spectra provide sufficient information on the specific numbers of individual and family compounds in the extracts. Therefore, the information recorded in the UV-Vis spectra should enable the determination of their levels in the K. alvarezii extract samples. If this is the case, a rapid analytical technique that is simple to apply yet has a low solvent consumption should perhaps be developed. Hence, this study developed a rapid analytical method based on UV-Vis spectrophotometry by comparing the spectroscopic results with those generated by the UPLC-PDA. However, the recorded data for the full spectra from the UV-Vis spectrophotometry (Figure 2) required chemometrics to develop the method and interpret the results.

Table 2.
Predictive performance of the PLS regression models for phenolic compounds in K. alvarezii extracts.
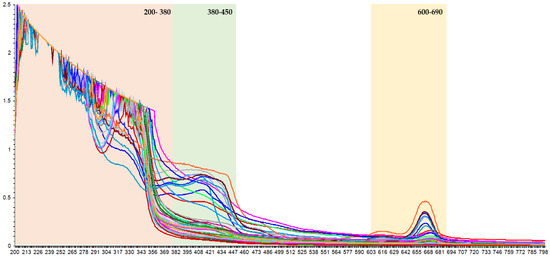
Figure 2.
The basic ATR UV-Vis spectra correction of K. alvarezii extracts.
In order to obtain a sensitive measurement, the spectral data must be selected for a wavelength or range of wavelengths providing capability in detecting different levels of phenolics in the extract. The spectra revealed three distinct peaks in the UV-Vis region, with optimum absorption rangea at 200–450 nm and 600–690 nm (Figure 2), corresponding to phenolic acid and flavonoid derivatives. Hydroxybenzoic acid and flavonol groups demonstrated a strong single absorption band at 280 nm, while hydroxycinnamic acid showed an absorption band around 320 nm [21].
3.3. K. alvarezii Description Based on the Spectroscopic Properties
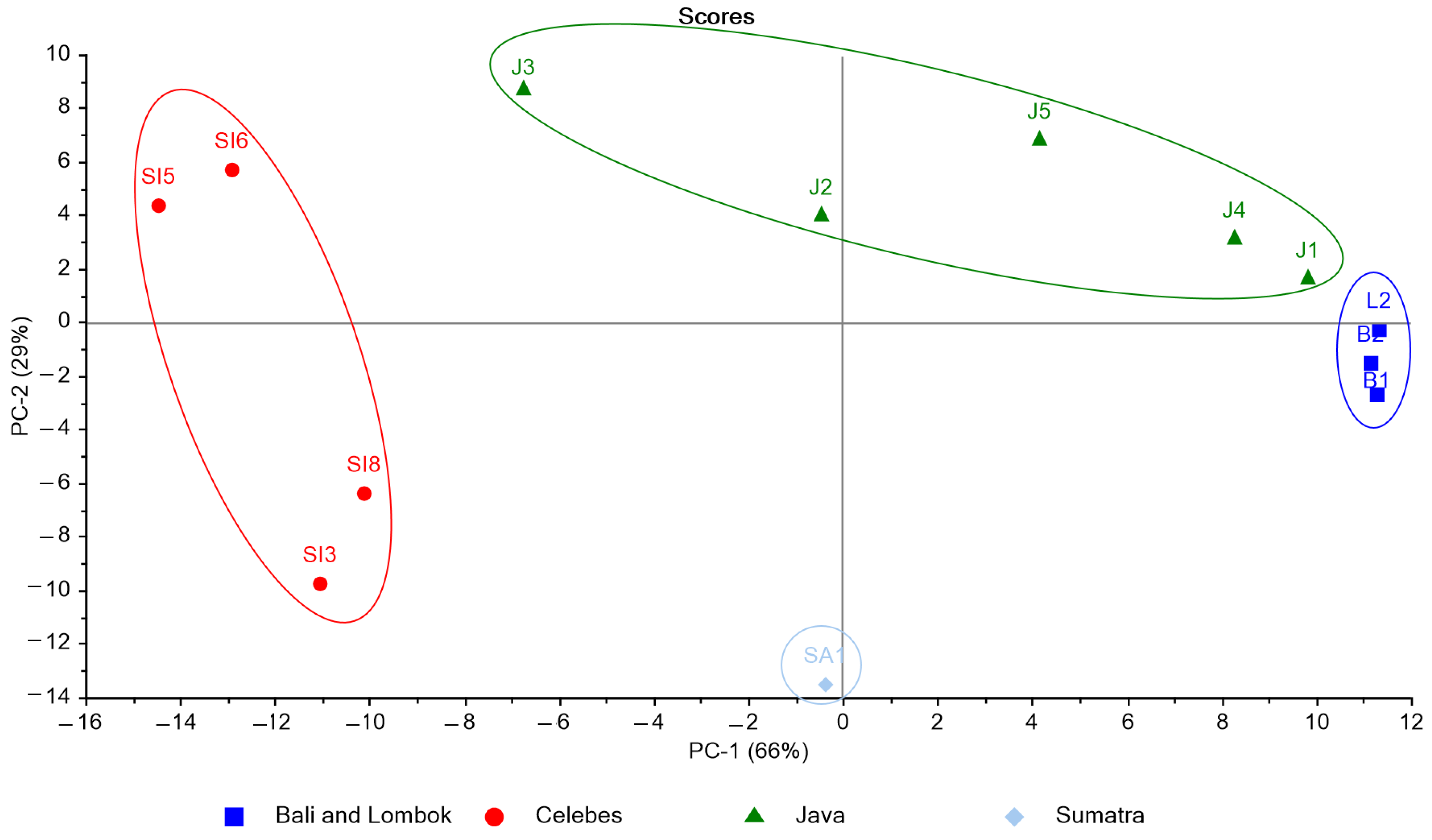
A non-supervised exploratory PCA was performed on the acquired spectra by the UV-Vis spectrophotometry method to assess the possibility of describing the data distribution of K. alvarezii collected from 13 different growing locations. The raw data of the samples was measured at 200–800 nm. Subsequently, the working range was selected for the wavelength prior to the principal component analysis (PCA), employing a cluster analysis on the resulting variables. The combined wavelength regions of 200–450 nm and 600–690 nm were then selected, thus consisting of 342 variables. From the PCA result, two components were extracted that accounted for 95% of the variability in the original data (Figure 3).
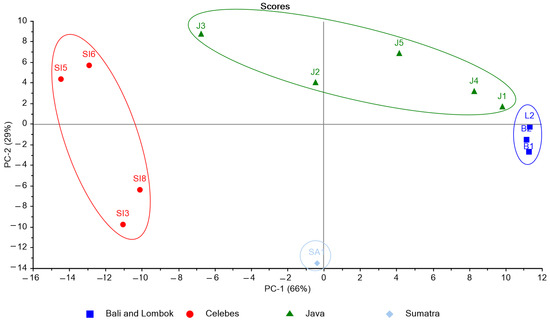
Figure 3.
PCA score plot for spectral data of K. alvarezii collected from different cultivation sites.
On PC1, the sample from the island of Java was distributed both in the positive (J2 and J3) and negative (J1, J4, J5) axis. This discrepancy can arise because J2 and J3 were collected in the Indian ocean, whereas the other samples were collected in the Java Sea. Red algae grow naturally in sands that are frequently mixed with mud, shell fragments, or coral. The Indian Ocean is an area of a high abundance of corals with massive waves. In contrast, the Java Sea shelf is composed mainly of sand and mud. Therefore, the production of several metabolites by the algae can be different due to different growing areas [22]. The score plot revealed that the spectroscopy properties of the K. alvarezii extracts could be used to describe the distribution of the studied samples in the PC. The resulting distribution by PCA explained four classifications representing the five large islands of Indonesia: Celebes, Java, Bali, Lombok, and Sumatra. The 13 samples from different growing sites were 100% correctly classified into their corresponding island. The classification of K. alvarezii based on the origin of the cultivation island might be caused by differences in the composition of the phenolic compounds naturally present in the samples.
3.4. Calibration and Validation of PLS Regression
The correlation between the UPLC–PDA data (Y) and UV-Vis spectra (X) in a specific wavelength range was determined using partial least squares (PLS). The regression model was first generated using the whole data set. Then, to eliminate noise from non-essential spectroscopic ranges, particular spectroscopic ranges were analyzed. The PLS was priorly used in selecting the spectroscopic ranges of UV-Vis spectra corresponding to the levels of phenolic compounds based on the UPLC–PDA data. As the X-loading weights (Figure S1) from the PLS analysis are useful for detecting important variables, this approach extracted the factors that allowed for more influence than using the complete spectra.
If a variable has a significant positive or negative loading weight, the variable is important for the corresponding component. Based on the plot, it is known that the wavelength range of 200–450 and 600–690 included important variables in the correlation with phenolic compounds in the UPLC–PDA data. The wavelength range can therefore be used as a critical variable in establishing a robust regression model.
The most suited model to predict the level of studied phenolic compounds was selected based on the high value of the coefficient of determination (R2) with low error (RMSE). The R2 values greater than 0.9 indicated an excellent model, while values between 0.8 and 0.9 were considered acceptable. Table 2 compiles the model performance of the PLS result for some selected wavelength ranges.
PLS regression models were used to predict the levels of HCA1 and HCA2 using UV-Vis spectra at combined wavelength regions of 200–450 and 600–690 nm, as indicated by the high values of R2C, R2CV, and R2P with low RMSEC, RMSECV, and RMSEP. Alternatively, the levels of HBA and flavonoids were predicted using a combined wavelength region of 380–200 nm. Former research revealed the efficacious employment of the PLS regression models based on UV-Vis spectra of a specific wavelength range. The selected wavelength range was confirmed to be useful for real-world sample classification, and the developed model can be utilized to predict the levels of phenolic acid and flavonoid derivatives [13,23].
3.5. Phenolic Compounds Measurement in K. alvarezii
The developed calibration model was applied to measure the levels of HCA1, HCA2, HBA, and flavonoid in the studied samples from different geographical origins. This experiment aimed to confirm the reliability of the proposed UV-Vis spectrophotometry method combined with the developed PLS regression model. Table 3 shows the level of four studied compounds estimated by the proposed method in 13 samples from different growing locations. The values to distinguish the levels of phenolic compounds indicated the prediction for chromatographic responses (area of corresponding peaks) by the PLS regression model. The table also displays the results of the mean absolute error (MAPE) calculations. MAPE is a relative error indicator that expresses the percentage of inaccuracy in estimating or predicting results compared to the actual results [24]. According to the table, all the investigated compounds had a MAPE of less than 15%, indicating good predictive accuracy. As per the prior study, if the MAPE value is less than 10%, the method’s accuracy is excellent, and if it is between 10% and 20%, the method’s accuracy is good [25].

Table 3.
The level of the studied phenolic compounds in K. alvarezii by the developed PLS models.
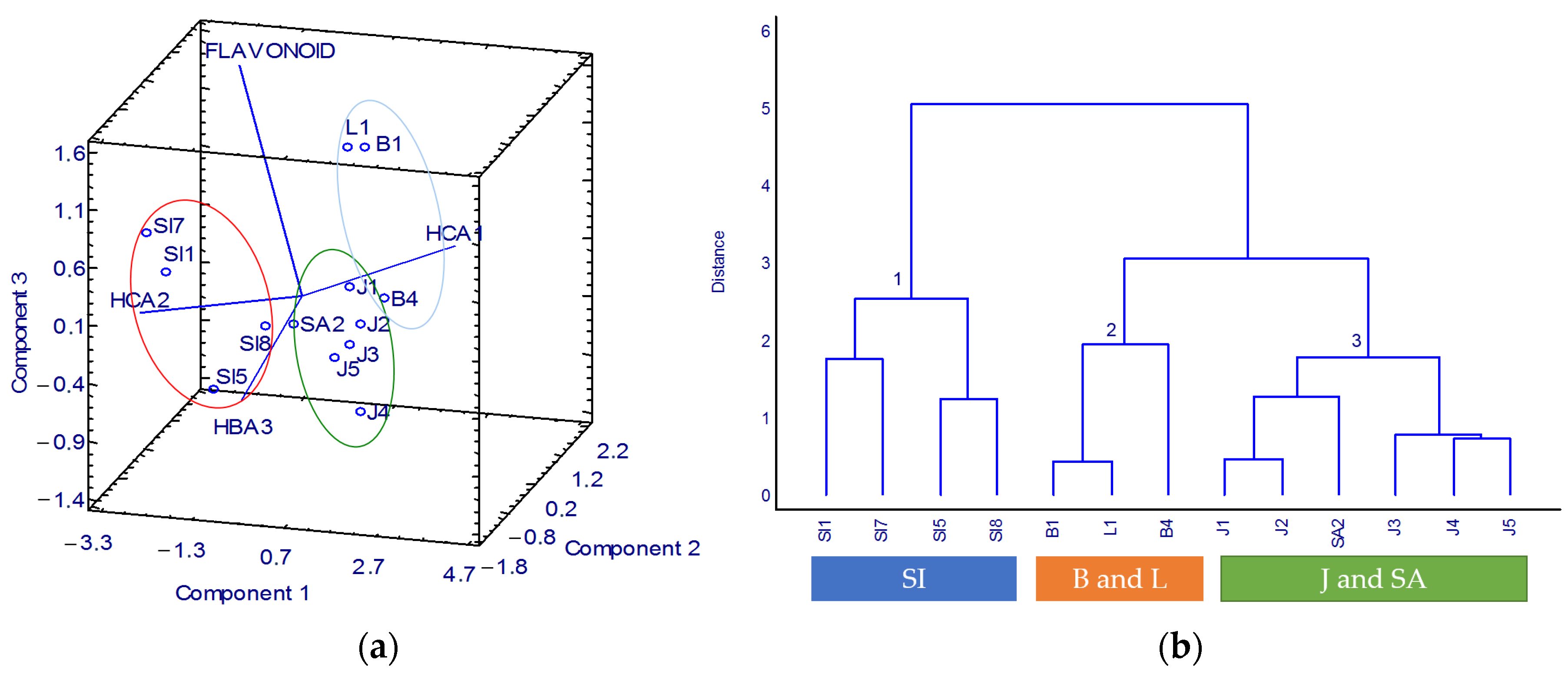
To evaluate the potential of determining important compounds in each group of cultivation sites, PCA was performed on the predicted levels of phenolic compounds in the K. alvarezii samples. The PCA biplot of the correlation load was selected over the conventional loading plot for a more straightforward interpretation of the correlation between the phenolic compounds and K. alvarezii cultivation sites (Figure 4a). According to the PCA 3D biplot, the principal components of PC1, PC2, and PC3 explained 70.35%, 17.46%, and 11.26% of the total variation of the data, respectively.
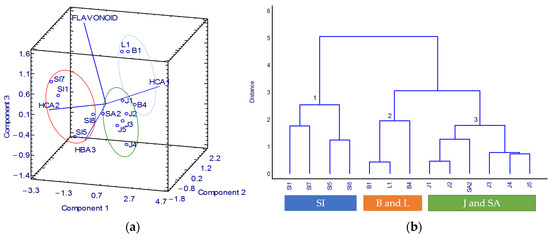
Figure 4.
Correlation of geographical origin with specific phenolic compound: (a) PCA 3D biplot and (b) dendrogram. SI: Celebes, B: Bali, L: Lombok, J: Java, SA: Sumatra.
The positive axis of PC3 is represented by HCAs and flavonoid compounds, whereas HBA compounds represent the negative axis. Two samples from Celebes, Djene Ponto (Sl8) and Bantaeng (Sl5), were on the PC3 negative axis, while the other two samples, Tanakeke Island (Sl7) and Puntondo (Sl1), were in the opposite quadrant. Even though the aforementioned four samples were originally from the same island, Sl7 and Sl1 were closely related to the HCA1, while Sl8 and Sl5 were described by the HBA derivative. The different key compounds were most likely due to their different cultivation sites. S18 and S15 were cultivated in a bay on the south side of Celebes Island. In comparison, SI7 and SI1 were collected from a strait where the sea was mainly composed of karst rock. In the synthesis of phenolic compounds, the amount of nutrients in the water is a crucial aspect. The inadequate nutritional content of karst soil has been reported [26]. As a result, the phenolic levels, including the HBA and HCA, were also varied.
Flavonoid is located on the positive axis of PC3. The samples from Bali and Lombok islands were included in this specific axis. Therefore, the samples from Bali and Lombok can be classified as flavonoid-rich samples. The islands of Bali and Lombok have excellent K. alvarezii productivity as the cultivation sites provide a living environment that suits the growth of K. alvarezii. Calm seas with high salinity and plenty of light are some of the characteristics [27].
Based on PC1, the compounds of HCA derivatives were separated by positive (HCA1) and negative (HCA2) axes altogether with samples from Java and Sumatra. This PCA result implies that samples from Java and Sumatra can be distinguished by these HCA compounds.
The cluster analysis (CA) outcomes confirmed the PCA results. As shown in the dendrogram (Figure 4b), the hierarchical clusters clearly identified three groups. All the samples from Celebes Island with high HBA compounds were in the first group. The second group consisted of the flavonoid-rich samples from Bali and Lombok. The samples from Java and Sumatra islands highly linked with HCA derivatives were in the third group. Thereby, the developed UV-Vis spectrophotometry method combined with chemometrics was confirmed as successfully applied to determine the quality of K. alvarezii based on phenolic compounds corresponding to the geographical origin.
4. Conclusions
Phenolic compounds in K. alvarezii extracts analyzed by UPLC–PDA consisted of phenolic acids (hydroxycinnamic and hydroxybenzoic acids) and flavonoid. The chemometrics were useful for developing the right relationship between the results of UV-Vis spectrometry and the UPLC-PDA. As combined with PLS regression based on the spectral data of the absorption range at 200–450 nm and 600–690 nm, the UV–Vis spectrometry method has been developed to predict the level of phenolic compounds. The basic standardization of Indonesian K. alvarezii was then defined by the developed method that confirmed the level and composition of phenolic acids and flavonoid that should be contained in the matrices. Additionally, PCA generated principal components that produced a clear distribution among the K. alvarezii samples based on the phenolic compounds corresponding to the geographical origin. Thus, the new UV-Vis spectrometry method was demonstrated as a reliable analytical approach to standardize the K. alvarezii samples.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/scipharm89040047/s1, Figure S1: PLS analysis X-loading weights of UV-Vis Spectra.
Author Contributions
Conceptualization, S.M. and W.S.; methodology, M.P. and W.S.; software, S.M.; validation, S.A., M.P. and W.S.; formal analysis, S.M., V.G.P.P., M.P. and W.S.; investigation, S.M., V.G.P.P., W.C. and C.C.; resources, M.P., V.G.P.P. and W.S.; data curation, S.M. and W.S.; writing—original draft preparation, S.M. and W.C.; writing—review and editing, W.S., M.P. and S.A.; visualization, S.M.; supervision, S.A. and W.S.; project administration, W.S.; funding acquisition, W.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research forms part of the National Research Priority (PRN) project on the standardization of Indonesian algae funded by the BOPTN Research Grant 006/E4.1/AK.04.PRN/2021 from the Directorate General of Higher Education, Ministry of Education and Culture, Republic of Indonesia. The APC was supported by Universitas Gadjah Mada through the Final Project Recognition Program (Rekognisi Tugas Akhir, RTA) 2020 with assignment number 3143/UN1.P.III/DIT-LIT/PT/2021.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- World Bank Group. Seaweed Aquaculture for Food Security, Income Generation and Environmental Health in Tropical Developing Countries. World Bank USA 2016. [Google Scholar] [CrossRef]
- Hayashi, L.; Hurtado, A.Q.; Msuya, F.E.; Bleicher-Lhonneur, G.; Critchley, A.T. Kappaphycus Farming: Prospects and Constraints; Springer Nature Switzerland: Cham, Switzerland, 2007; Volume 15, ISBN 9789048185696. [Google Scholar]
- Rengasamy, K.R.; Mahomoodally, M.F.; Aumeeruddy, M.Z.; Zengin, G.; Xiao, J.; Kim, D.H. Bioactive compounds in seaweeds: An overview of their biological properties and safety. Food Chem. Toxicol. 2020, 135, 111013. [Google Scholar] [CrossRef]
- Sumayya, S.; Murugan, K. Phytochemical screening, RP-HPLC and FTIR analysis of Kappaphycus alvarezii (Doty) Doty EX P.C Silva: Macro red algae. J. Pharmacogn. Phytochem. 2017, 6, 325–330. [Google Scholar]
- Stengel, D.B.; Connan, S.; Popper, Z.A. Algal chemodiversity and bioactivity: Sources of natural variability and implications for commercial application. Biotechnol. Adv. 2011, 29, 483–501. [Google Scholar] [CrossRef]
- Berrueta, L.A.; Alonso-Salces, R.M.; Héberger, K. Supervised pattern recognition in food analysis. J. Chromatogr. A 2007, 1158, 196–214. [Google Scholar] [CrossRef]
- Bae, I.K.; Ham, H.M.; Jeong, M.H.; Kim, D.H.; Kim, H.J. Simultaneous determination of 15 phenolic compounds and caffeine in teas and mate using RP-HPLC/UV detection: Method development and optimization of extraction process. Food Chem. 2015, 172, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Setyaningsih, W.; Saputro, I.E.; Carrera, C.A.; Palma, M.; García-Barroso, C. Fast Determination of Phenolic Compounds in Rice Grains by Ultraperformance Liquid Chromatography Coupled to Photodiode Array Detection: Method Development and Validation. J. Agric. Food Chem. 2019, 67, 3018–3027. [Google Scholar] [CrossRef] [PubMed]
- Mandhania, S.; Pal, A.; Saharan, V. Simultaneous estimation of twenty eight phenolic compounds by a novel and expeditious method developed on quaternary ultra-performance liquid chromatography system with a photodiode array detector. Biomolecules 2020, 10, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gumustas, M.; Kurbanoglu, S.; Uslu, B.; Ozkan, S.A. UPLC versus HPLC on Drug Analysis: Advantageous, Applications and Their Validation Parameters; Springer Nature Switzerland: Cham, Switzerland, 2013; Volume 76, ISBN 1033701324778. [Google Scholar]
- Palacios-Morillo, A.; Alcázar, Á.; De Pablos, F.; Jurado, J.M. Differentiation of tea varieties using UV-Vis spectra and pattern recognition techniques. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 103, 79–83. [Google Scholar] [CrossRef]
- Riswahyuli, Y.; Rohman, A.; Setyabudi, F.M.C.S.; Raharjo, S. Indonesian wild honey authenticity analysis using attenuated total reflectance-fourier transform infrared (ATR-FTIR) spectroscopy combined with multivariate statistical techniques. Heliyon 2020, 6, e03662. [Google Scholar] [CrossRef]
- Song, X.C.; Canellas, E.; Asensio, E.; Nerín, C. Predicting the antioxidant capacity and total phenolic content of bearberry leaves by data fusion of UV–Vis spectroscopy and UHPLC/Q-TOF-MS. Talanta 2020, 213, 120831. [Google Scholar] [CrossRef]
- Boulet, J.C.; Ducasse, M.A.; Cheynier, V. Ultraviolet spectroscopy study of phenolic substances and other major compounds in red wines: Relationship between astringency and the concentration of phenolic substances. Aust. J. Grape Wine Res. 2017, 23, 193–199. [Google Scholar] [CrossRef]
- Aboulwafa, M.M.; Youssef, F.S.; Gad, H.A.; Sarker, S.D.; Nahar, L.; Al-Azizi, M.M.; Ashour, M.L. Authentication and discrimination of green tea samples using UV–vis, FTIR and HPLC techniques coupled with chemometrics analysis. J. Pharm. Biomed. Anal. 2019, 164, 653–658. [Google Scholar] [CrossRef]
- Shi, H.; Yu, P. Comparison of grating-based near-infrared (NIR) and Fourier transform mid-infrared (ATR-FT/MIR) spectroscopy based on spectral preprocessing and wavelength selection for the determination of crude protein and moisture content in wheat. Food Control 2017, 82, 57–65. [Google Scholar] [CrossRef]
- Rohaeti, E.; Muzayanah, K.; Septaningsih, D.A.; Rafi, M. Fast analytical method for authentication of chili powder from synthetic dyes using uv-vis spectroscopy in combination with chemometrics. Indones. J. Chem. 2019, 19, 668–674. [Google Scholar] [CrossRef]
- Liu, L.; Zuo, Z.-t.; Wang, Y.-z.; Xu, F.-r. A fast multi-source information fusion strategy based on FTIR spectroscopy for geographical authentication of wild Gentiana rigescens. Microchem. J. 2020, 159, 105360. [Google Scholar] [CrossRef]
- Sampaio, P.S.; Soares, A.; Castanho, A.; Almeida, A.S.; Oliveira, J.; Brites, C. Optimization of rice amylose determination by NIR-spectroscopy using PLS chemometrics algorithms. Food Chem. 2018, 242, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Gam, D.H.; Kim, S.Y.; Kim, J.W. Optimization of ultrasound-assisted extraction condition for phenolic compounds, antioxidant activity, and epigallocatechin gallate in lipid-extracted microalgae. Molecules 2020, 25, 454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez, E.; Arqués, J.L.; Rodríguez, R.; Nuñez, M.; Medina, M.; Talarico, T.L.; Casas, I.A.; Chung, T.C.; Dobrogosz, W.J.; Axelsson, L.; et al. The Role of UV-Visible Spectroscopy for Phenolic Compounds Quantification in Winemaking. Intech 2011, 32, 137–144. [Google Scholar]
- Widyartini, D.S.; Widodo, P.; Susanto, A.B. Thallus variation of Sargassum polycystum from Central Java, Indonesia. Biodiversitas 2017, 18, 1004–1011. [Google Scholar] [CrossRef]
- Tian, W.; Chen, G.; Gui, Y.; Zhang, G.; Li, Y. Rapid quantification of total phenolics and ferulic acid in whole wheat using UV–Vis spectrophotometry. Food Control 2021, 123, 107691. [Google Scholar] [CrossRef]
- Kim, S.; Kim, H. A new metric of absolute percentage error for intermittent demand forecasts. Int. J. Forecast. 2016, 32, 669–679. [Google Scholar] [CrossRef]
- Maricar, M.A.; Widiadnyana, P.; Arta Wijaya, I.W. Analysis of Data Mining for Forecasting Total Goods Delivery with Moving Average Method. Int. J. Eng. Emerg. Technol. 2017, 2, 7. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Liu, C.; Rubinato, M.; Guo, K.; Zhou, J.; Cui, M. An assessment of soil’s nutrient deficiencies and their influence on the restoration of degraded karst vegetation in Southwest China. Forests 2020, 11, 797. [Google Scholar] [CrossRef]
- Simatupang, N.F.; Pong-Masak, P.R.; Ratnawati, P.; Agusman; Paul, N.A.; Rimmer, M.A. Growth and product quality of the seaweed Kappaphycus alvarezii from different farming locations in Indonesia. Aquac. Rep. 2021, 20, 100685. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).