Identification and Molecular Binding Mechanism of Novel α-Glucosidase Inhibitory Peptides from Hot-Pressed Peanut Meal Protein Hydrolysates
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of PMHs
2.3. Determination of Degree of Hydrolysis
2.4. Determination of Trichloroacetic Acid-Nitrogen Soluble Index (TCA-NSI)
2.5. Determination of Protein Yield
2.6. Determination of α-Glucosidase Inhibitory Activity
2.7. Determination of Molecular Weight Distribution
2.8. Separation of α-Glucosidase Inhibitory Peptides from PMHs
2.8.1. Ultrafiltration
2.8.2. Ion Exchange Chromatography
2.9. Identification of α-Glucosidase Inhibitory Peptides Using Nano-HPLC-MS/MS
2.10. Molecular Docking Analysis
2.11. Stability of α-Glucosidase Inhibitory Peptides
2.12. Statistical Analysis
3. Results and Discussion
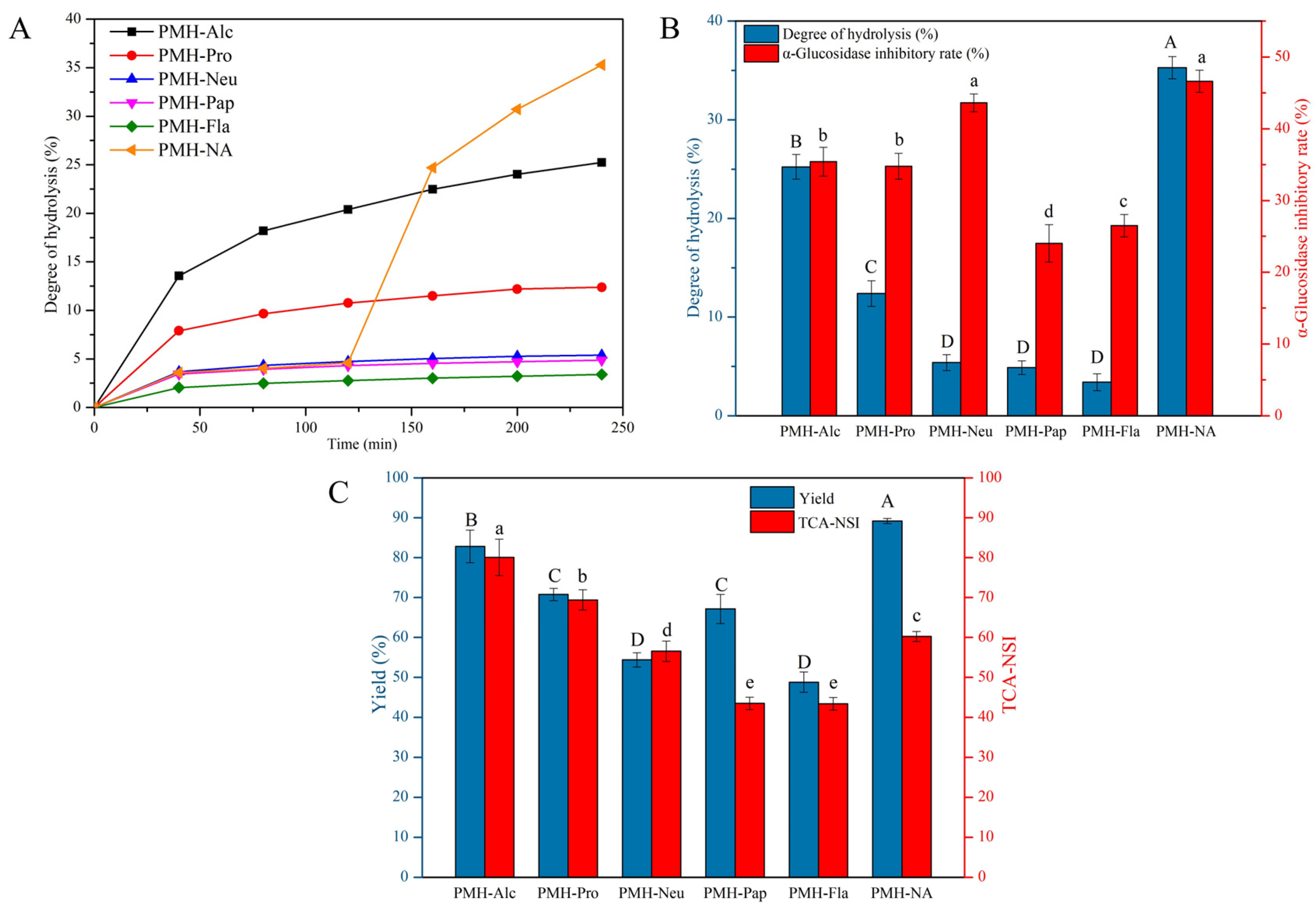
3.1. Degree of Hydrolysis and α-Glucosidase Inhibitory Activity of PMHs
3.2. Isolation and Purification of α-Glucosidase Inhibitory Peptide
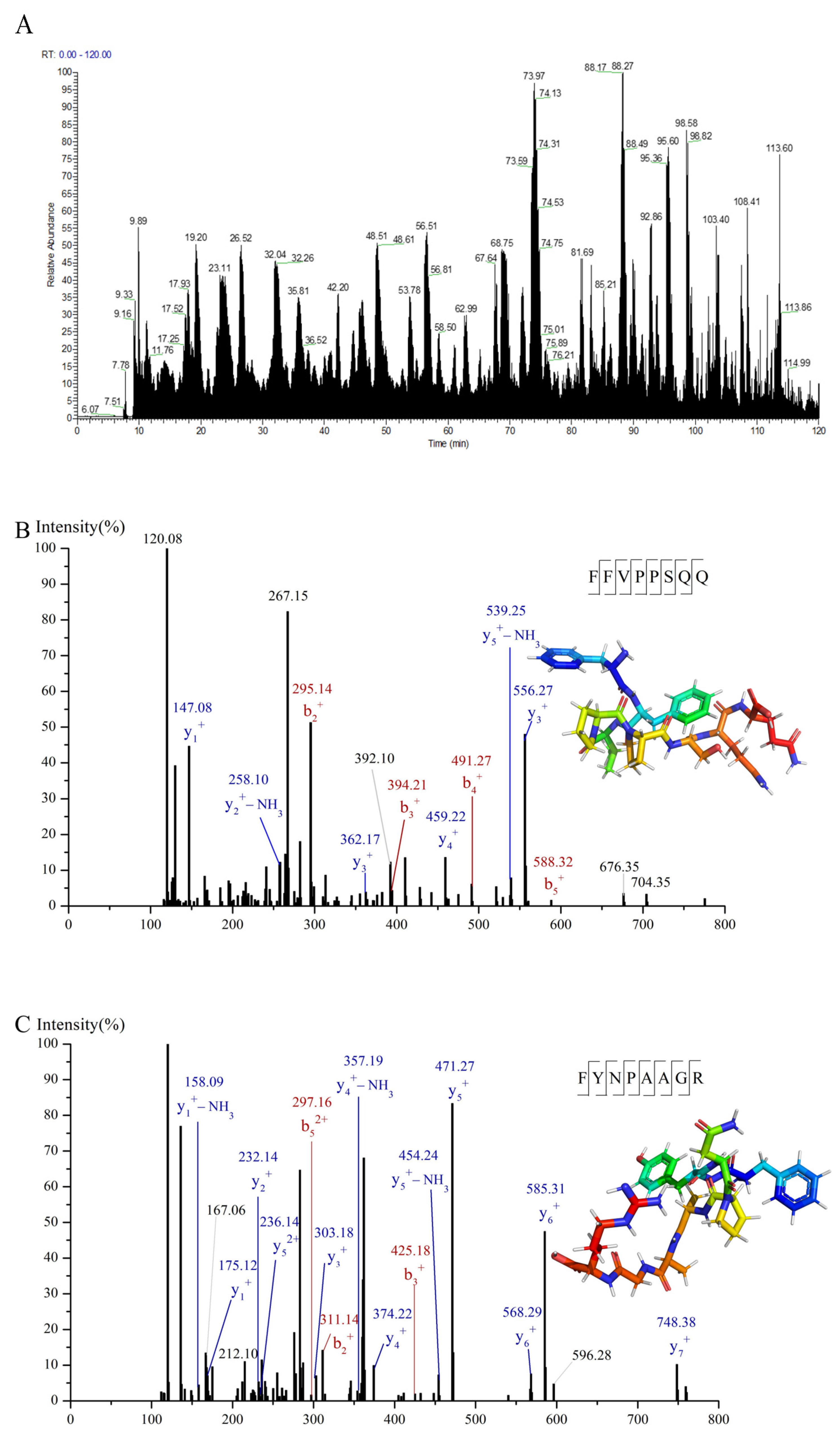
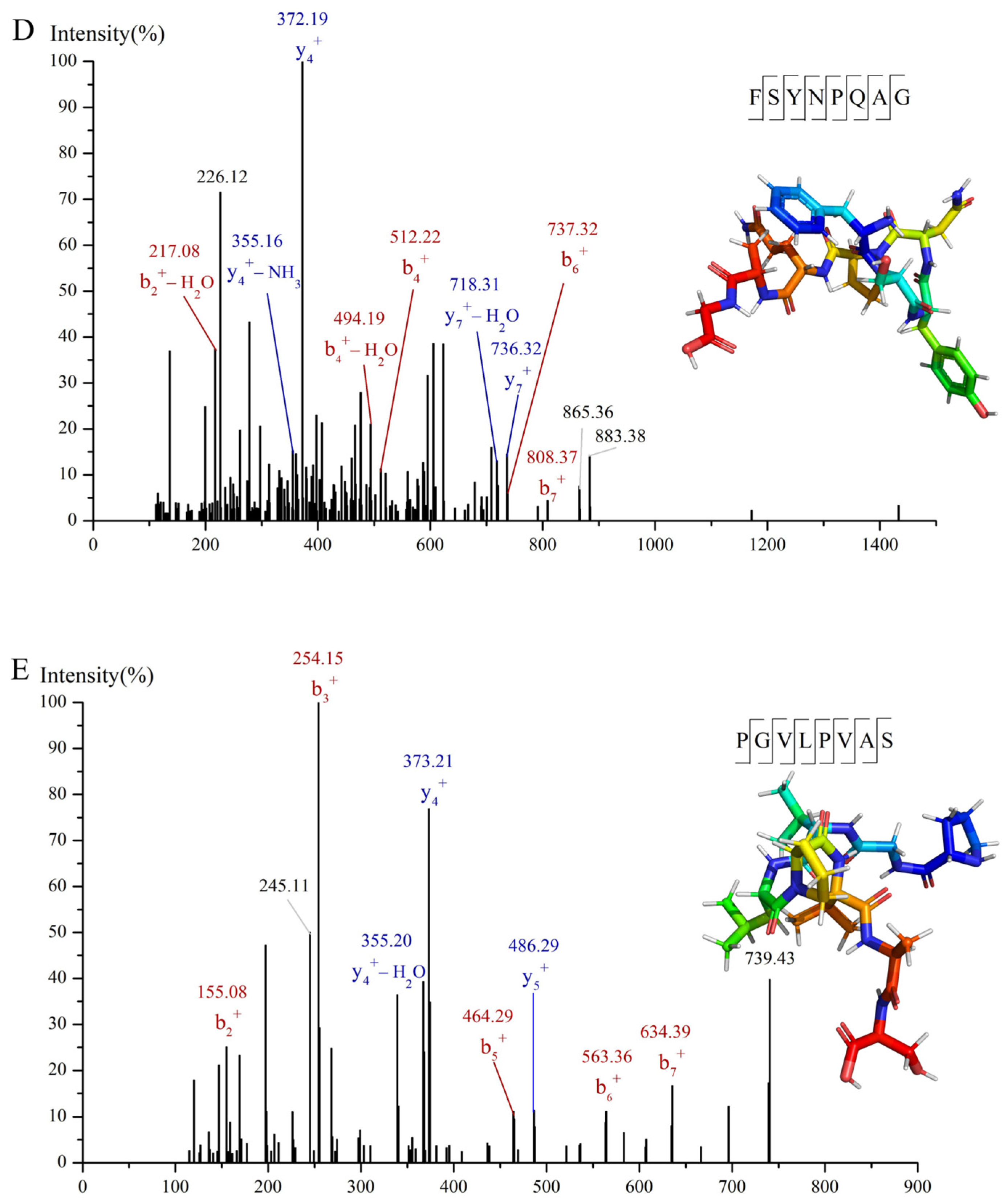
3.3. Identification and Screening of α-Glucosidase Inhibitory Peptides in C-I Fraction
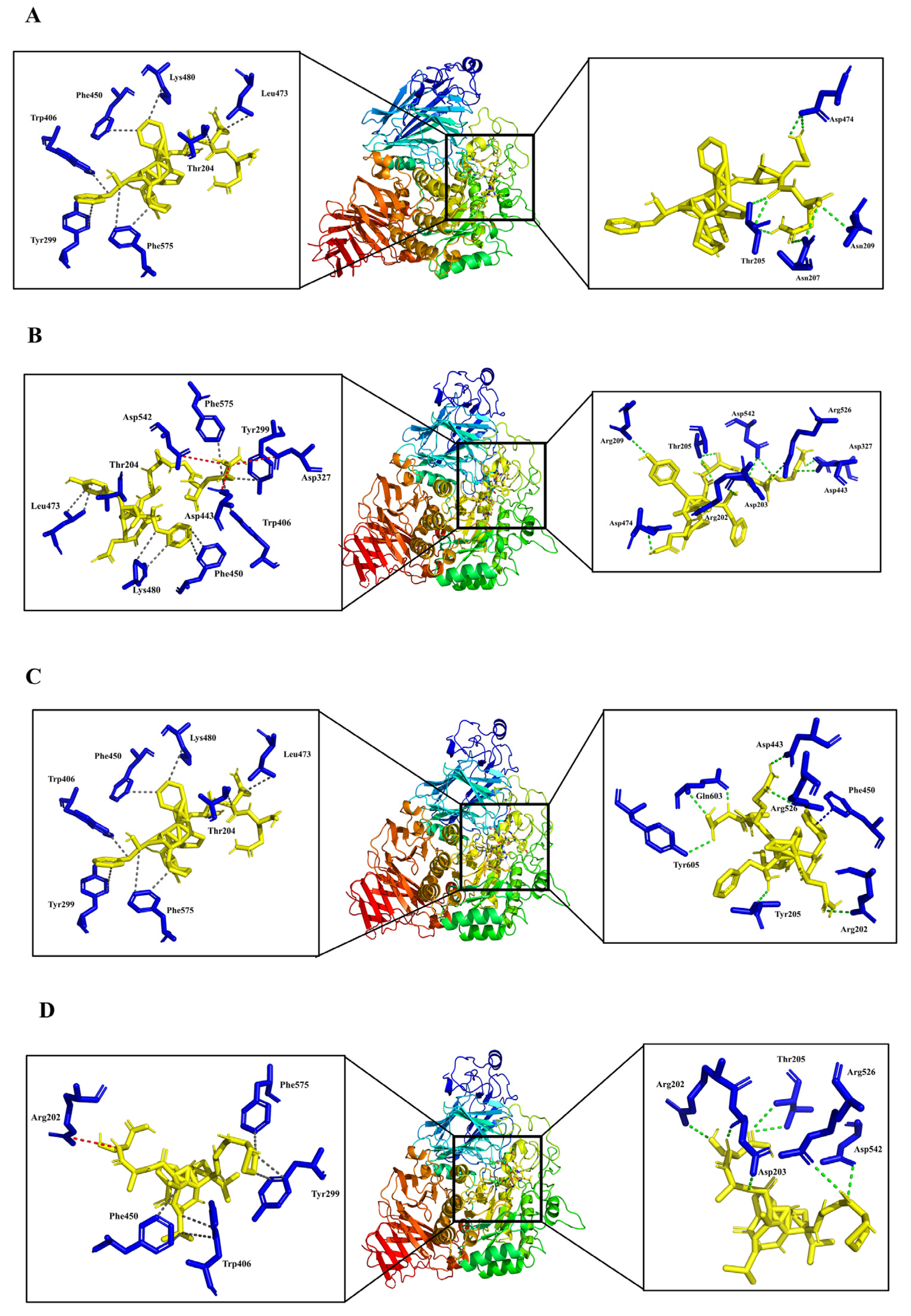
3.4. Analysis of Molecular Binding Mechanism
3.5. Chemical Stability of α-Glucosidase Inhibitory Peptides
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yagi, N.; Komiya, I.; Arai, K. Current status of oral antidiabetic drug prescribing patterns based on the body mass index for Japanese type 2 diabetes mellitus patients and yearly changes in diabetologists’ prescribing patterns from 2002 to 2019 (JDDM61). J. Diabetes Investig. 2022, 13, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Brand-Miller, J.; Buyken, A.E. The Relationship between Glycemic Index and Health. Nutrients 2020, 12, 536. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Wang, S.; Wu, J. The kinetics and mechanism of alpha-glucosidase inhibition by F5-SP, a novel compound derived from sericin peptides. Food Funct. 2017, 8, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Holman, R.; Coleman, R.; Chan, J. Effects of acarbose on cardiovascular and diabetes outcomes in patients with coronary heart disease and impaired glucose tolerance (ACE): A randomised, double-blind, placebo-controlled trial. Lancet Diabetes Endocrinol. 2017, 5, 877–886. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, N.; Wang, W. Molecular mechanisms of novel peptides from silkworm pupae that inhibit alpha-glucosidase. Peptides 2016, 76, 45–50. [Google Scholar] [CrossRef]
- Jiang, M.; Yan, H.; He, R. Purification and a molecular docking study of α-glucosidase-inhibitory peptides from a soybean protein hydrolysate with ultrasonic pretreatment. Eur. Food Res. Technol. 2018, 244, 1995–2005. [Google Scholar] [CrossRef]
- Liu, W.; Li, H.; Wen, Y. Molecular Mechanism for the alpha-Glucosidase Inhibitory Effect of Wheat Germ Peptides. J. Agric. Food Chem. 2021, 69, 15231–15239. [Google Scholar] [CrossRef]
- Abbasi, S.; Moslehishad, M.; Salami, M. Antioxidant and alpha-glucosidase enzyme inhibitory properties of hydrolyzed protein and bioactive peptides of quinoa. Int. J. Biol. Macromol. 2022, 213, 602–609. [Google Scholar] [CrossRef]
- Zhao, Q.; Wei, G.; Li, K. Identification and molecular docking of novel α-glucosidase inhibitory peptides from hydrolysates of Binglangjiang buffalo casein. LWT 2022, 156, 113062. [Google Scholar] [CrossRef]
- Lu, Y.; Ding, H.; Jiang, X. Effects of the extract from peanut meal fermented with Bacillus natto and Monascus on lipid metabolism and intestinal barrier function of hyperlipidemic mice. J. Sci. Food Agric. 2021, 101, 2561–2569. [Google Scholar] [CrossRef]
- Zhao, X.; Chen, J.; Du, F. Potential use of peanut by-products in food processing: A review. J. Food Sci Technol. 2012, 49, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, J.; Yu, L. Emulsification performance and interfacial properties of enzymically hydrolyzed peanut protein isolate pretreated by extrusion cooking. Food Hydrocoll. 2018, 77, 607–616. [Google Scholar] [CrossRef]
- Santos, F.S.D.; Signoretti, R.D.; Oliveira, J.S. Effect of replacing soybean meal with peanut meal on milk production and fat composition in lactating dairy cows. Trop. Anim. Health Prod. 2022, 54, 80. [Google Scholar] [CrossRef] [PubMed]
- Ye, M.J.; Xu, Q.L.; Tang, H.Y. Development and stability of novel selenium colloidal particles complex with peanut meal peptides. LWT 2020, 126, 109280. [Google Scholar] [CrossRef]
- Zen, Q.Z.; Cui, Y.L.; Sun, D.X. Process optimization and anti-oxidative activity of peanut meal Maillard reaction products. LWT 2018, 97, 573–580. [Google Scholar]
- Zhang, L.; Song, C.; Chang, J. Optimization of protein hydrolysates production from defatted peanut meal based on physicochemical characteristics and sensory analysis. LWT 2022, 163, 113572. [Google Scholar] [CrossRef]
- Zhang, P.; Chang, C.; Liu, H. Efficient enzymatic production of angiotensin I-converting enzyme inhibitory peptides from three protein-rich materials by electrolyzed water pretreatment. LWT 2022, 154, 112864. [Google Scholar] [CrossRef]
- Liu, J.; Chen, L.W.; Ji, K.M. An endurance-enhancing effect of peanut meal protein hydrolysate in mice: Possible involvement of a specific peanut peptide. J. Anim. Physiol. Anim. Nutr. 2014, 98, 830–837. [Google Scholar] [CrossRef]
- Rios-Herrera, G.D.; Salazar-Leyva, J.A.; Hernandez, C. Production of Protein Hydrolysates Using Marine Catfish Bagre panamensis Muscle or Casein as Substrates: Effect of Enzymatic Source and Degree of Hydrolysis on Antioxidant and Biochemical Properties. Appl. Biochem. Biotechnol. 2021, 193, 3214–3231. [Google Scholar] [CrossRef]
- Kong, X.; Zhang, L.; Song, W. Separation, identification and molecular binding mechanism of dipeptidyl peptidase IV inhibitory peptides derived from walnut (Juglans regia L.) protein. Food Chem. 2021, 347, 129062. [Google Scholar] [CrossRef]
- Mushtaq, M.; Gani, A.; Masoodi, F.A. Himalayan cheese (Kalari/Kradi) fermented with different probiotic strains: In vitro investigation of nutraceutical properties. LWT 2019, 104, 53–60. [Google Scholar] [CrossRef]
- Lee, H.J.; Lee, H.S.; Choi, J.W. Novel tripeptides with alpha-glucosidase inhibitory activity isolated from silk cocoon hydrolysate. J. Agric. Food Chem. 2011, 59, 11522–11525. [Google Scholar] [CrossRef] [PubMed]
- Awosika, T.O.; Aluko, R.E. Inhibition of the in vitro activities of α-amylase, α-glucosidase and pancreatic lipase by yellow field pea (Pisum sativum L.) protein hydrolysates. Int. J. Food Sci. Technol. 2019, 54, 2021–2034. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Ketnaw, A.S.; Benjakul, S.; Martinez-Alvarez, O. Fish skin gelatin hydrolysates produced by visceral peptidase and bovine trypsin: Bioactivity and stability. Food Chem. 2017, 215, 383–390. [Google Scholar] [CrossRef]
- Brodkorb, A.; Egger, L.; Alminger, M. INFOGEST static in vitro simulation of gastrointestinal food digestion. Nat. Protoc. 2019, 14, 991–1014. [Google Scholar] [CrossRef]
- Chen, N.; Yang, H.; Sun, Y. Purification and identification of antioxidant peptides from walnut (Juglans regia L.) protein hydrolysates. Peptides 2012, 38, 344–349. [Google Scholar] [CrossRef]
- Hu, F.; Ci, A.T.; Wang, H. Identification and hydrolysis kinetic of a novel antioxidant peptide from pecan meal using Alcalase. Food Chem. 2018, 261, 301–310. [Google Scholar] [CrossRef]
- Feng, J.; Ma, Y.L.; Sun, P. Purification and characterisation of α-glucosidase inhibitory peptides from defatted camellia seed cake. Int. J. Food Sci. Technol. 2020, 56, 138–147. [Google Scholar] [CrossRef]
- Luo, F.; Fu, Y.; Ma, L. Exploration of Dipeptidyl Peptidase-IV (DPP-IV) Inhibitory Peptides from Silkworm Pupae (Bombyx mori) Proteins Based on In Silico and In Vitro Assessments. J. Agric. Food Chem. 2022, 70, 3862–3871. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Wang, C.Y.; Wang, S.T. Physicochemical properties and antioxidant activities of tree peony (Paeonia suffruticosa Andr.) seed protein hydrolysates obtained with different proteases. Food Chem. 2021, 345, 128765. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.Y.; Li, M.Y.; Tian, G. Effects of ultrasonic pretreatment on the structure and functionality of chicken bone protein prepared by enzymatic method. Food Chem. 2019, 299, 125103. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Deng, Y.; Xie, P. Novel bioactive peptides from ginkgo biloba seed protein and evaluation of their alpha-glucosidase inhibition activity. Food Chem. 2023, 404, 134481. [Google Scholar] [CrossRef] [PubMed]
- De Matos, F.M.; De Lacerd, A.J.T.J.G.; Zanetti, G. Production of black cricket protein hydrolysates with α-amylase, α-glucosidase and angiotensin I-converting enzyme inhibitory activities using a mixture of proteases. Biocatal. Agric. Biotechnol. 2022, 39, 102276. [Google Scholar] [CrossRef]
- Yan, Z.F.; Yuan, S.; Qin, Q. Enhancement of rice protein hydrolysate quality using a novel dual enzyme system. LWT 2022, 158, 113110. [Google Scholar] [CrossRef]
- Cian, R.E.; Martínez-Augustin, O.; Drago, S.R. Bioactive properties of peptides obtained by enzymatic hydrolysis from protein byproducts of Porphyra columbina. Food Res. Int. 2012, 49, 364–372. [Google Scholar] [CrossRef]
- Agrawal, H.; Joshi, R.; Gupta, M. Purification, identification and characterization of two novel antioxidant peptides from finger millet (Eleusine coracana) protein hydrolysate. Food Res. Int. 2019, 120, 697–707. [Google Scholar] [CrossRef]
- Wang, R.; Zhao, H.; Pan, X. Preparation of bioactive peptides with antidiabetic, antihypertensive, and antioxidant activities and identification of alpha-glucosidase inhibitory peptides from soy protein. Food Sci. Nutr. 2019, 7, 1848–1856. [Google Scholar] [CrossRef]
- Wei, G.; Zhao, Q.; Wang, D. Novel ACE inhibitory, antioxidant and α-glucosidase inhibitory peptides identified from fermented rubing cheese through peptidomic and molecular docking. LWT 2022, 159, 113196. [Google Scholar] [CrossRef]
- Zambrowicz, A.; Pokora, M.; Setner, B. Multifunctional peptides derived from an egg yolk protein hydrolysate: Isolation and characterization. Amino Acids 2015, 47, 369–380. [Google Scholar] [CrossRef]
- Mudgil, P.; Kamal, H.; Priya Kilari, B. Simulated gastrointestinal digestion of camel and bovine casein hydrolysates: Identification and characterization of novel anti-diabetic bioactive peptides. Food Chem. 2021, 353, 129374. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.H.; Wen, J.Y.; Xu, X.M. Molecular Docking of Small-Molecule Monosaccharides and Their Analogues with α-Glucosidase. J. Food Sci. 2020, 41, 139–143. [Google Scholar]
- Sim, L.; Quezada-Calvillo, R.; Sterchi, E.E. Human intestinal maltase-glucoamylase: Crystal structure of the N-terminal catalytic subunit and basis of inhibition and substrate specificity. J. Mol. Biol. 2008, 375, 782–792. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.A.; Bester, M.J.; Neitz, A.W.H. Structural properties of bioactive peptides with alpha-glucosidase inhibitory activity. Chem. Biol. Drug Des. 2018, 91, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Young, J.F.; Dalsgaard, T.K. Separation of angiotensin I-converting enzyme inhibitory peptides from bovine connective tissue and their stability towards temperature, pH and digestive enzymes. Int. J. Food Sci. Technol. 2015, 50, 1234–1243. [Google Scholar] [CrossRef]


| Sequence | Molecular Weight (Da) | Binding Energy (kcal/mol) |
|---|---|---|
| MVDVT | 475.24 | −6.7 |
| PGVLPVAS | 739.43 | −8.6 |
| VEPTPVQ | 769.41 | −6.3 |
| LAFPGSGE | 777.38 | −7.3 |
| VIQQGQAT | 844.45 | −6.5 |
| ALRIPSGF | 860.49 | −6.6 |
| FSYNPQAG | 883.39 | −7.7 |
| FYNPAAGR | 895.44 | −8.1 |
| GALDPVQVT | 899.48 | −5.3 |
| VQVVDSNGN | 931.45 | −5.2 |
| TATDLVLTV | 932.52 | −7.0 |
| FFVPPSQQ | 949.47 | −8.0 |
| AEYTAPVAE | 950.44 | −6.8 |
| GDITNPINL | 956.50 | −5.3 |
| TISPLPVIK | 967.61 | −6.7 |
| ISPLPVIKE | 995.57 | −6.8 |
| LDLVPTLPQ | 995.61 | −6.5 |
| VNELDLPILG | 1082.61 | −6.3 |
| ADAYAATGLLQ | 1093.55 | −6.0 |
| LKNNNPFKF | 1121.61 | −5.1 |
| IAVGDVIPDGTL | 1169.64 | −4.4 |
| IYNPQAGSLKT | 1191.64 | −4.8 |
| NTGSPITVPVGR | 1197.66 | −4.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, X.; Wang, D.; Dai, Y.; Zhao, L.; Wang, W.; Ding, X. Identification and Molecular Binding Mechanism of Novel α-Glucosidase Inhibitory Peptides from Hot-Pressed Peanut Meal Protein Hydrolysates. Foods 2023, 12, 663. https://doi.org/10.3390/foods12030663
Yang X, Wang D, Dai Y, Zhao L, Wang W, Ding X. Identification and Molecular Binding Mechanism of Novel α-Glucosidase Inhibitory Peptides from Hot-Pressed Peanut Meal Protein Hydrolysates. Foods. 2023; 12(3):663. https://doi.org/10.3390/foods12030663
Chicago/Turabian StyleYang, Xinyu, Dan Wang, Yangyong Dai, Luping Zhao, Wentao Wang, and Xiuzhen Ding. 2023. "Identification and Molecular Binding Mechanism of Novel α-Glucosidase Inhibitory Peptides from Hot-Pressed Peanut Meal Protein Hydrolysates" Foods 12, no. 3: 663. https://doi.org/10.3390/foods12030663
APA StyleYang, X., Wang, D., Dai, Y., Zhao, L., Wang, W., & Ding, X. (2023). Identification and Molecular Binding Mechanism of Novel α-Glucosidase Inhibitory Peptides from Hot-Pressed Peanut Meal Protein Hydrolysates. Foods, 12(3), 663. https://doi.org/10.3390/foods12030663





