A Systems Biology Approach on the Regulatory Footprint of Human Endogenous Retroviruses (HERVs)
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequence Analysis and Annotation
2.2. Analysis of Genomic Distribution Related to Human Genes
2.3. Network Analysis
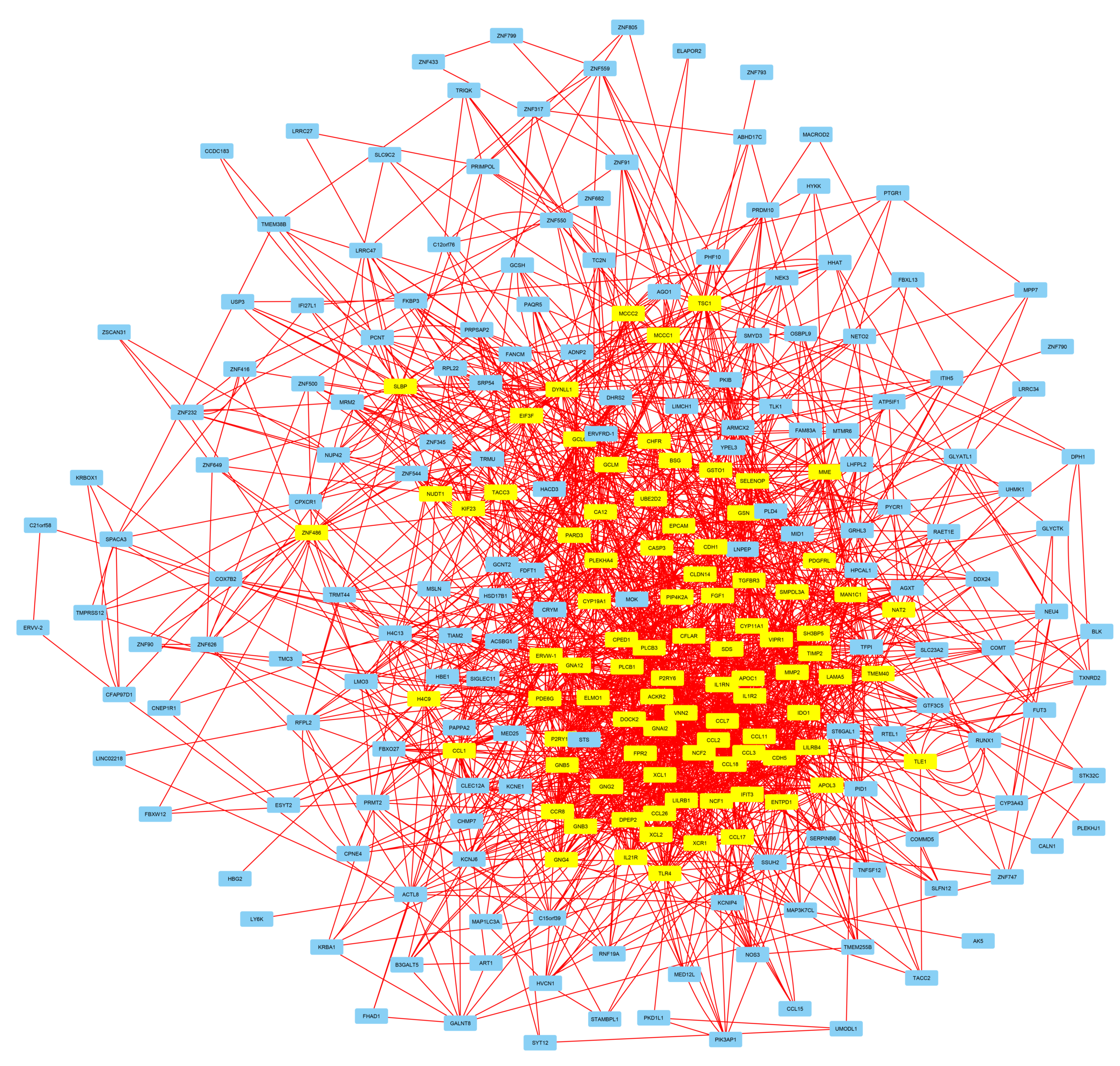
3. Results
3.1. Overview on HERV-Associated cis-Regulatory Elements
3.2. HERV-CREs Are Present in the Vicinity of the Majority of Human Genes
3.3. The Regulatory Footprint of Promoter-Specific HERV-CREs
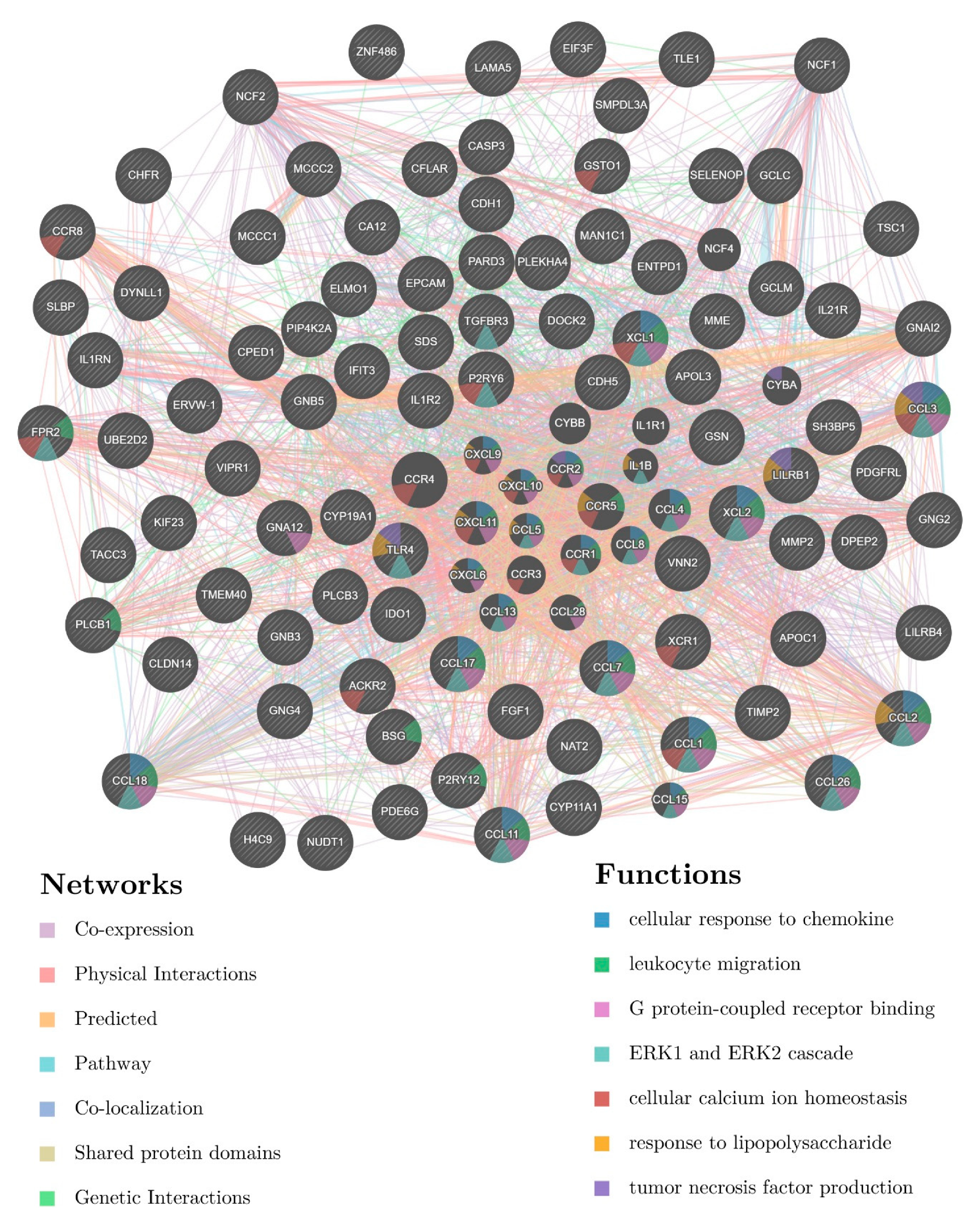
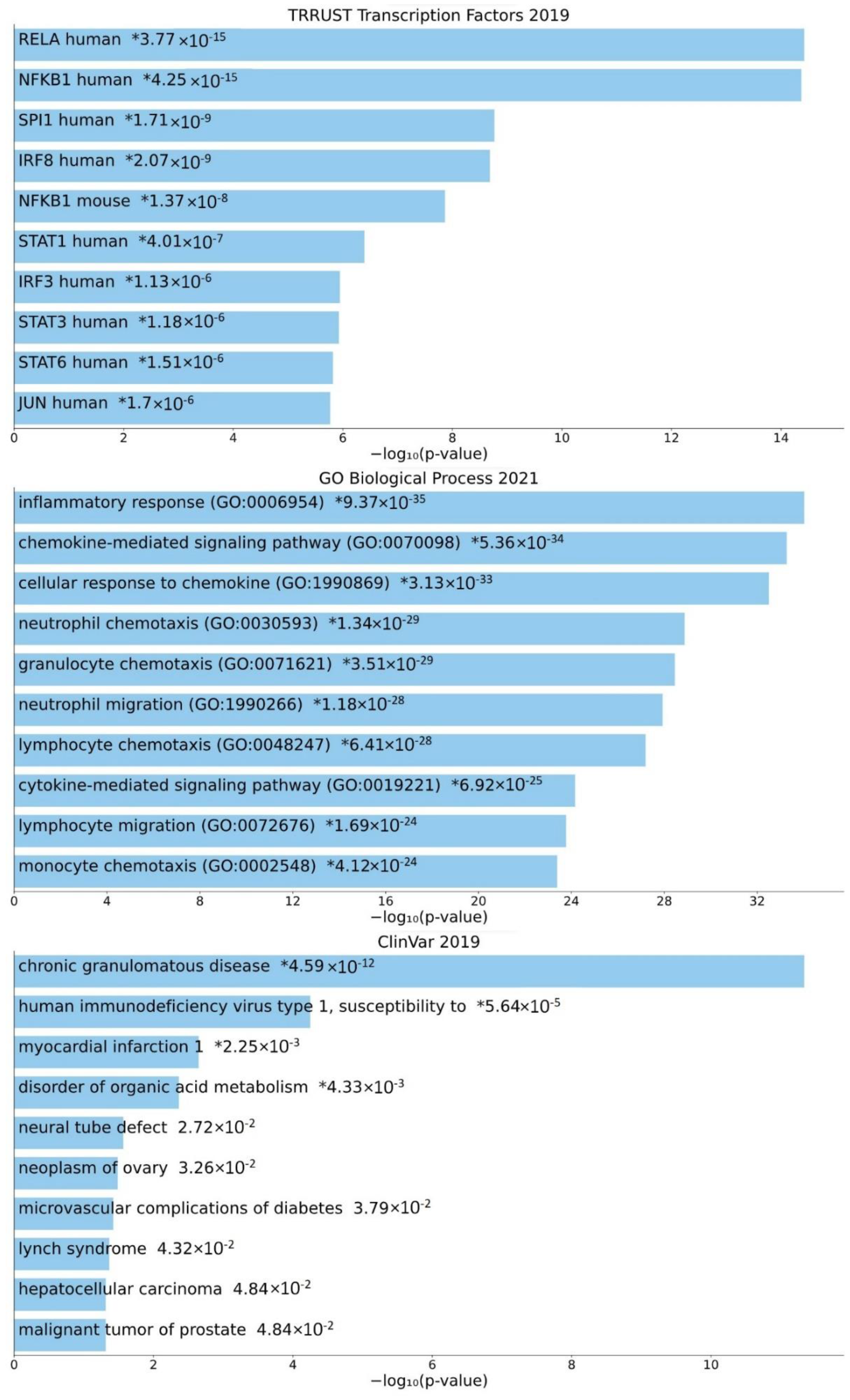
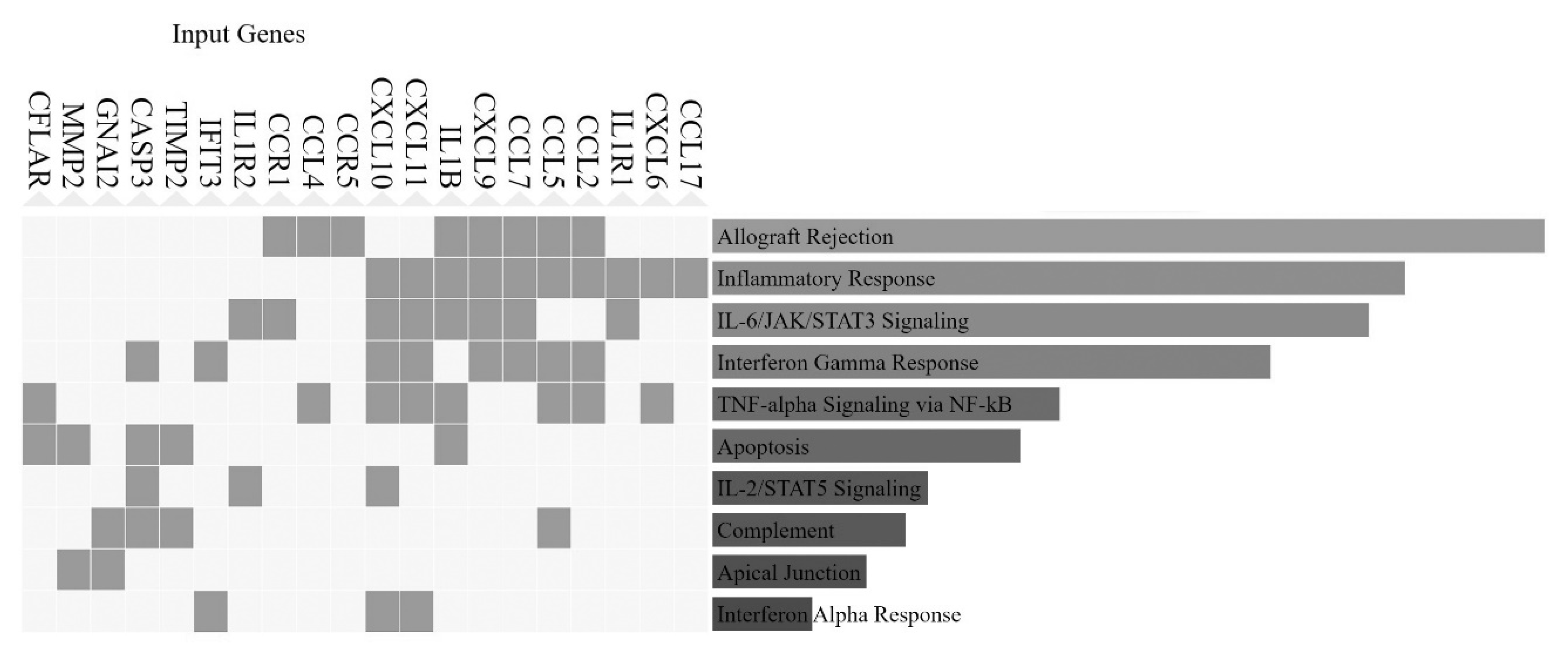
3.4. Involvement of Central Promoter-Specific HERV-CREs in Cellular Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921, Correction in Nature 2001, 412, 565–566; Erratum in Nature 2001, 7, 720. [Google Scholar] [CrossRef]
- Venter, J.C.; Adams, M.D.; Myers, E.W.; Li, P.W.; Mural, R.J.; Sutton, G.G.; Smith, H.O.; Yandell, M.; Evans, C.A.; Holt, R.A. The sequence of the human genome. Science 2001, 291, 1304–1351. [Google Scholar] [CrossRef] [PubMed]
- Kojima, K.K. Human transposable elements in Repbase: Genomic footprints from fish to humans. Mob. DNA 2018, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- de Koning, A.P.J.; Gu, W.; Castoe, T.A.; Batzer, M.A.; Pollock, D.D. Repetitive Elements May Comprise Over Two-Thirds of the Human Genome. PLoS Genet. 2011, 7, e1002384. [Google Scholar] [CrossRef] [PubMed]
- Goodier, J.L.; Kazazian, H.H., Jr. Retrotransposons Revisited: The Restraint and Rehabilitation of Parasites. Cell 2008, 135, 23–35. [Google Scholar] [CrossRef]
- Elbarbary, R.A.; Lucas, B.A.; Maquat, L.E. Retrotransposons as regulators of gene expression. Science 2016, 351, aac7247. [Google Scholar] [CrossRef]
- Dimitriadou, E.; Noutsopoulos, D.; Markopoulos, G.; Vlaikou, A.-M.; Mantziou, S.; Traeger-Synodinos, J.; Kanavakis, E.; Chrousos, G.P.; Tzavaras, T.; Syrrou, M. Abnormal DLK1/MEG3 imprinting correlates with decreased HERV-K methylation after assisted reproduction and preimplantation genetic diagnosis. Stress 2013, 16, 689–697. [Google Scholar] [CrossRef]
- Ladias, P.; Markopoulos, G.; Lazaros, L.; Markoula, S.; Tzavaras, T.; Georgiou, I. Holliday Junctions Are Associated with Transposable Element Sequences in the Human Genome. J. Mol. Biol. 2016, 428, 658–667. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, G.; Cui, J. Origin and Deep Evolution of Human Endogenous Retroviruses in Pan-Primates. Viruses 2022, 14, 1370. [Google Scholar] [CrossRef]
- Sverdlov, E.D. Retroviruses and Primate Genome Evolution; CRC Press: Boca Raton, FL, USA, 2005. [Google Scholar]
- Montesion, M.; Williams, Z.H.; Subramanian, R.P.; Kuperwasser, C.; Coffin, J.M. Promoter expression of HERV-K (HML-2) provirus-derived sequences is related to LTR sequence variation and polymorphic transcription factor binding sites. Retrovirology 2018, 15, 57. [Google Scholar] [CrossRef]
- Jia, L.; Liu, M.; Yang, C.; Li, H.; Liu, Y.; Han, J.; Zhai, X.; Wang, X.; Li, T.; Li, J.; et al. Comprehensive identification and characterization of the HERV-K (HML-9) group in the human genome. Retrovirology 2022, 19, 11. [Google Scholar] [CrossRef] [PubMed]
- Knössl, M.; Löwer, R.; Löwer, J. Expression of the Human Endogenous Retrovirus HTDV/HERV-K Is Enhanced by Cellular Transcription Factor YY1. J. Virol. 1999, 73, 1254–1261. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, N.V.; Kraft, M.; Tondera, C.; Hanschmann, K.-M.; Löwer, J.; Löwer, R. Expression of the human endogenous retrovirus (HERV) group HML-2/HERV-K does not depend on canonical promoter elements but is regulated by transcription factors Sp1 and Sp3. J. Virol. 2011, 85, 3436–3448. [Google Scholar] [CrossRef]
- De Parseval, N.; Alkabbani, H.; Heidmann, T. The long terminal repeats of the HERV-H human endogenous retrovirus contain binding sites for transcriptional regulation by the Myb protein. J. Gen. Virol. 1999, 80, 841–845. [Google Scholar] [CrossRef][Green Version]
- Matteucci, C.; Balestrieri, E.; Argaw-Denboba, A.; Sinibaldi-Vallebona, P. Human endogenous retroviruses role in cancer cell stemness. Semin. Cancer Biol. 2018, 53, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Mommert, M.; Tabone, O.; Oriol, G.; Cerrato, E.; Guichard, A.; Naville, M.; Fournier, P.; Volff, J.-N.; Pachot, A.; Monneret, G.; et al. LTR-retrotransposon transcriptome modulation in response to endotoxin-induced stress in PBMCs. BMC Genom. 2018, 19, 522. [Google Scholar] [CrossRef] [PubMed]
- Seifarth, W.; Frank, O.; Zeilfelder, U.; Spiess, B.; Greenwood, A.D.; Hehlmann, R.; Leib-Mösch, C. Comprehensive Analysis of Human Endogenous Retrovirus Transcriptional Activity in Human Tissues with a Retrovirus-Specific Microarray. J. Virol. 2005, 79, 341–352. [Google Scholar] [CrossRef]
- Georgiou, I.; Noutsopoulos, D.; Dimitriadou, E.; Markopoulos, G.; Apergi, A.; Lazaros, L.; Vaxevanoglou, T.; Pantos, K.; Syrrou, M.; Tzavaras, T. Retrotransposon RNA expression and evidence for retrotransposition events in human oocytes. Hum. Mol. Genet. 2009, 18, 1221–1228. [Google Scholar] [CrossRef]
- Lazaros, L.; Kitsou, C.; Kostoulas, C.; Bellou, S.; Hatzi, E.; Ladias, P.; Stefos, T.; Markoula, S.; Galani, V.; Vartholomatos, G.; et al. Retrotransposon expression and incorporation of cloned human and mouse retroelements in human spermatozoa. Fertil. Steril. 2017, 107, 821–830. [Google Scholar] [CrossRef]
- Smallwood, A.; Papageorghiou, A.; Nicolaides, K.; Alley, M.; Jim, A.; Nargund, G.; Ojha, K.; Campbell, S.; Banerjee, S. Temporal Regulation of the Expression of Syncytin (HERV-W), Maternally Imprinted PEG10, and SGCE in Human Placenta1. Biol. Reprod. 2003, 69, 286–293. [Google Scholar] [CrossRef]
- Sun, M.-A.; Wolf, G.; Wang, Y.; Senft, A.D.; Ralls, S.; Jin, J.; Dunn-Fletcher, C.E.; Muglia, L.J.; Macfarlan, T.S. Endogenous Retroviruses Drive Lineage-Specific Regulatory Evolution across Primate and Rodent Placentae. Mol. Biol. Evol. 2021, 38, 4992–5004. [Google Scholar] [CrossRef] [PubMed]
- Karamitros, T.; Paraskevis, D.; Hatzakis, A.; Psichogiou, M.; Elefsiniotis, I.; Hurst, T.; Geretti, A.-M.; Beloukas, A.; Frater, J.; Klenerman, P.; et al. A contaminant-free assessment of Endogenous Retroviral RNA in human plasma. Sci. Rep. 2016, 6, 33598. [Google Scholar] [CrossRef] [PubMed]
- Hurst, T.P.; Magiorkinis, G. Epigenetic Control of Human Endogenous Retrovirus Expression: Focus on Regulation of Long-Terminal Repeats (LTRs). Viruses 2017, 9, 130. [Google Scholar] [CrossRef]
- Wang, J.; Xie, G.; Singh, M.; Ghanbarian, A.T.; Raskó, T.; Szvetnik, A.; Cai, H.; Besser, D.; Prigione, A.; Fuchs, N.V.; et al. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature 2014, 516, 405–409. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, T.; Preissl, S.; Amaral, M.L.; Grinstein, J.D.; Farah, E.N.; Destici, E.; Qiu, Y.; Hu, R.; Lee, A.Y.; et al. Transcriptionally active HERV-H retrotransposons demarcate topologically associating domains in human pluripotent stem cells. Nat. Genet. 2019, 51, 1380–1388. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Sachs, F.; Ramsay, L.; Jacques, P.; Göke, J.; Bourque, G.; Ng, H.-H. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat. Struct. Mol. Biol. 2014, 21, 423–425. [Google Scholar] [CrossRef] [PubMed]
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57. [Google Scholar] [CrossRef]
- Markopoulos, G.; Noutsopoulos, D.; Mantziou, S.; Gerogiannis, D.; Thrasyvoulou, S.; Vartholomatos, G.; Kolettas, E.; Tzavaras, T. Genomic analysis of mouse VL30 retrotransposons. Mob. DNA 2016, 7, 10. [Google Scholar] [CrossRef]
- Mantziou, S.; Markopoulos, G.S. Origins and Function of VL30 lncRNA Packaging in Small Extracellular Vesicles: Implications for Cellular Physiology and Pathology. Biomedicines 2021, 9, 1742. [Google Scholar] [CrossRef]
- Vartholomatos, E.; Mantziou, S.; Alexiou, G.A.; Lazari, D.; Sioka, C.; Kyritsis, A.; Markopoulos, G.S. An NF-κB-and Therapy-Related Regulatory Network in Glioma: A Potential Mechanism of Action for Natural Antiglioma Agents. Biomedicines 2022, 10, 935. [Google Scholar] [CrossRef]
- Chen, N. Using Repeat Masker to Identify Repetitive Elements in Genomic Sequences. Curr. Protoc. Bioinform. 2004, 5, 4.10.1–4.10.14. [Google Scholar] [CrossRef] [PubMed]
- Gerstein, M.B.; Kundaje, A.; Hariharan, M.; Landt, S.G.; Yan, K.-K.; Cheng, C.; Mu, X.J.; Khurana, E.; Rozowsky, J.; Alexander, R.; et al. Architecture of the human regulatory network derived from ENCODE data. Nature 2012, 489, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhuang, J.; Iyer, S.; Lin, X.; Whitfield, T.W.; Greven, M.C.; Pierce, B.G.; Dong, X.; Kundaje, A.; Cheng, Y.; et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012, 22, 1798–1812. [Google Scholar] [CrossRef]
- Wang, J.; Zhuang, J.; Iyer, S.; Lin, X.-Y.; Greven, M.C.; Kim, B.-H.; Moore, J.; Pierce, B.; Dong, X.; Virgil, D.; et al. Factorbook.org: A Wiki-based database for transcription factor-binding data generated by the ENCODE consortium. Nucleic Acids Res. 2012, 41, D171–D176. [Google Scholar] [CrossRef]
- Landt, S.G.; Marinov, G.K.; Kundaje, A.; Kheradpour, P.; Pauli, F.; Batzoglou, S.; Bernstein, B.E.; Bickel, P.; Brown, J.B.; Cayting, P. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012, 22, 1813–1831. [Google Scholar] [CrossRef]
- The ENCODE Project Consortium; Moore, J.E.; Purcaro, M.J.; Pratt, H.E.; Epstein, C.B.; Shoresh, N.; Adrian, J.; Kawli, T.; Davis, C.A.; Dobin, A. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 2020, 583, 699–710. [Google Scholar] [CrossRef]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The Human Genome Browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef]
- Fujita, P.A.; Rhead, B.; Zweig, A.S.; Hinrichs, A.S.; Karolchik, D.; Cline, M.S.; Goldman, M.; Barber, G.P.; Clawson, H.; Coelho, A.; et al. The UCSC Genome Browser database: Update 2011. Nucleic Acids Res. 2010, 39, D876–D882. [Google Scholar] [CrossRef]
- Haeussler, M.; Zweig, A.S.; Tyner, C.; Speir, M.L.; Rosenbloom, K.R.; Raney, B.J.; Lee, C.M.; Lee, B.T.; Hinrichs, A.; Gonzalez, J.N.; et al. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res. 2018, 47, D853–D858. [Google Scholar] [CrossRef]
- Karolchik, D.; Hinrichs, A.S.; Furey, T.S.; Roskin, K.M.; Sugnet, C.W.; Haussler, D.; Kent, W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004, 32, D493–D496. [Google Scholar] [CrossRef]
- Frankish, A.; Diekhans, M.; Jungreis, I.; Lagarde, J.; Loveland, J.E.; Mudge, J.M.; Sisu, C.; Wright, J.C.; Armstrong, J.; Barnes, I. GENCODE 2021. Nucleic Acids Res. 2021, 49, D916–D923. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Loganantharaj, R.; Schroeder, B.; Fargo, D.; Li, L. Pavis: A tool for p eak a nnotation and vis ualization. Bioinformatics 2013, 29, 3097–3099. [Google Scholar] [CrossRef] [PubMed]
- McLean, C.Y.; Bristor, D.; Hiller, M.; Clarke, S.L.; Schaar, B.T.; Lowe, C.B.; Wenger, A.M.; Bejerano, G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010, 28, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Saito, R.; Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.-L.; Lotia, S.; Pico, A.R.; Bader, G.D.; Ideker, T. A travel guide to Cytoscape plugins. Nat. Methods 2012, 9, 1069–1076. [Google Scholar] [CrossRef]
- Su, G.; Morris, J.H.; Demchak, B.; Bader, G.D. Biological network exploration with Cytoscape 3. Curr. Protoc. Bioinform. 2014, 47, 8.13.1–8.13.24. [Google Scholar] [CrossRef]
- Scardoni, G.; Tosadori, G.; Faizan, M.; Spoto, F.; Fabbri, F.; Laudanna, C. Biological network analysis with CentiScaPe: Centralities and experimental dataset integration. F1000Research 2014, 3, 139. [Google Scholar] [CrossRef]
- Scardoni, G.; Petterlini, M.; Laudanna, C. Analyzing biological network parameters with CentiScaPe. Bioinformatics 2009, 25, 2857–2859. [Google Scholar] [CrossRef]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’Ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Clarke, D.J.; Jeon, M.; Stein, D.J.; Moiseyev, N.; Kropiwnicki, E.; Dai, C.; Xie, Z.; Wojciechowicz, M.L.; Litz, S.; Hom, J.; et al. Appyters: Turning Jupyter Notebooks into data-driven web apps. Gene Expr. Patterns 2021, 2, 100213. [Google Scholar] [CrossRef]
- Pratt, H.E.; Andrews, G.R.; Phalke, N.; Huey, J.D.; Purcaro, M.J.; van der Velde, A.; Moore, J.E.; Weng, Z. Factorbook: An updated catalog of transcription factor motifs and candidate regulatory motif sites. Nucleic Acids Res. 2021, 50, D141–D149. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, J.A.; Tjian, R. Unexpected roles for core promoter recognition factors in cell-type-specific transcription and gene regulation. Nat. Rev. Genet. 2010, 11, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Haberle, V.; Stark, A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol. 2018, 19, 621–637. [Google Scholar] [CrossRef]
- Grandi, N.; Pisano, M.; Pessiu, E.; Scognamiglio, S.; Tramontano, E. HERV-K(HML7) Integrations in the Human Genome: Comprehensive Characterization and Comparative Analysis in Non-Human Primates. Biology 2021, 10, 439. [Google Scholar] [CrossRef]
- Li, W.; Lin, L.; Malhotra, R.; Yang, L.; Acharya, R.; Poss, M. A computational framework to assess genome-wide distribution of polymorphic human endogenous retrovirus-K In human populations. PLoS Comput. Biol. 2019, 15, e1006564. [Google Scholar] [CrossRef]
- Thomas, J.; Perron, H.; Feschotte, C. Variation in proviral content among human genomes mediated by LTR recombination. Mob. DNA 2018, 9, 1–15. [Google Scholar] [CrossRef]
- Heidari, N.; Phanstiel, D.H.; He, C.; Grubert, F.; Jahanbani, F.; Kasowski, M.; Zhang, M.Q.; Snyder, M.P. Genome-wide map of regulatory interactions in the human genome. Genome Res. 2014, 24, 1905–1917. [Google Scholar] [CrossRef]
- Chua, E.H.Z.; Yasar, S.; Harmston, N. The importance of considering regulatory domains in genome-wide analyses–the nearest gene is often wrong! Biol. Open 2022, 11, bio059091. [Google Scholar] [CrossRef] [PubMed]
- Nasser, J.; Bergman, D.T.; Fulco, C.P.; Guckelberger, P.; Doughty, B.R.; Patwardhan, T.A.; Jones, T.R.; Nguyen, T.H.; Ulirsch, J.C.; Lekschas, F.; et al. Genome-wide enhancer maps link risk variants to disease genes. Nature 2021, 593, 238–243. [Google Scholar] [CrossRef] [PubMed]
- Lappalainen, T.; Scott, A.J.; Brandt, M.; Hall, I.M. Genomic Analysis in the Age of Human Genome Sequencing. Cell 2019, 177, 70–84. [Google Scholar] [CrossRef]
- Jung, I.; Schmitt, A.; Diao, Y.; Lee, A.J.; Liu, T.; Yang, D.; Tan, C.; Eom, J.; Chan, M.; Chee, S.; et al. A compendium of promoter-centered long-range chromatin interactions in the human genome. Nat. Genet. 2019, 51, 1442–1449. [Google Scholar] [CrossRef]
- Fu, B.; Ma, H.; Liu, D. Endogenous Retroviruses Function as Gene Expression Regulatory Elements During Mammalian Pre-implantation Embryo Development. Int. J. Mol. Sci. 2019, 20, 790. [Google Scholar] [CrossRef]
- Buttler, C.A.; Chuong, E.B. Emerging roles for endogenous retroviruses in immune epigenetic regulation*. Immunol. Rev. 2021, 305, 165–178. [Google Scholar] [CrossRef]
- Jansz, N.; Faulkner, G.J. Endogenous retroviruses in the origins and treatment of cancer. Genome Biol. 2021, 22, 147. [Google Scholar] [CrossRef] [PubMed]
- Tokuyama, M.; Kong, Y.; Song, E.; Jayewickreme, T.; Kang, I.; Iwasaki, A. ERVmap analysis reveals genome-wide transcription of human endogenous retro-viruses. Proc. Natl. Acad. Sci. USA 2018, 115, 12565–12572. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Foroozesh, M.; Qin, Z. Transactivation of human endogenous retroviruses by tumor viruses and their functions in virus-associated malignancies. Oncogenesis 2019, 8, 6. [Google Scholar] [CrossRef]
- Dai, L.; Del Valle, L.; Miley, W.; Whitby, D.; Ochoa, A.C.; Flemington, E.K.; Qin, Z. Transactivation of human endogenous retrovirus K (HERV-K) by KSHV promotes Kaposi’s sarcoma development. Oncogene 2018, 37, 4534–4545. [Google Scholar] [CrossRef]
- Vincendeau, M.; Göttesdorfer, I.; Schreml, J.M.H.; Wetie, A.G.N.; Mayer, J.; Greenwood, A.D.; Helfer, M.; Kramer, S.; Seifarth, W.; Hadian, K.; et al. Modulation of human endogenous retrovirus (HERV) transcription during persistent and de novo HIV-1 infection. Retrovirology 2015, 12, 27. [Google Scholar] [CrossRef] [PubMed]
- van der Kuyl, A.C. HIV infection and HERV expression: A review. Retrovirology 2012, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Hernandez, M.J.; Swanson, M.D.; Contreras-Galindo, R.; Cookinham, S.; King, S.R.; Noel, R.J.; Kaplan, M.H.; Markovitz, D.M. Expression of Human Endogenous Retrovirus Type K (HML-2) Is Activated by the Tat Protein of HIV-1. J. Virol. 2012, 86, 7790–7805. [Google Scholar] [CrossRef]
- Delaneau, O.; Zazhytska, M.; Borel, C.; Giannuzzi, G.; Rey, G.; Howald, C.; Kumar, S.; Ongen, H.; Popadin, K.; Marbach, D. Chromatin three-dimensional interactions mediate genetic effects on gene ex-pression. Science 2019, 364, eaat8266. [Google Scholar] [CrossRef]
- Wang, T.; Medynets, M.; Johnson, K.R.; Doucet-O’Hare, T.T.; DiSanza, B.; Li, W.; Xu, Y.; Bagnell, A.; Tyagi, R.; Sampson, K.; et al. Regulation of stem cell function and neuronal differentiation by HERV-K via mTOR pathway. Proc. Natl. Acad. Sci. USA 2020, 117, 17842–17853. [Google Scholar] [CrossRef] [PubMed]
- Tamouza, R.; Meyer, U.; Foiselle, M.; Richard, J.-R.; Wu, C.-L.; Boukouaci, W.; Le Corvoisier, P.; Barrau, C.; Lucas, A.; Perron, H. Identification of inflammatory subgroups of schizophrenia and bipolar disorder patients with HERV-W ENV antigenemia by unsupervised cluster analysis. Transl. Psychiatry 2021, 11, 377. [Google Scholar] [CrossRef] [PubMed]
- Johnston, J.B.; Silva, C.; Holden, J.; Warren, K.G.; Clark, A.W.; Power, C. Monocyte activation and differentiation augment human endogenous retrovirus expression: Implications for inflammatory brain diseases. Ann. Neurol. 2001, 50, 434–442. [Google Scholar] [CrossRef] [PubMed]
- Ko, E.-J.; Cha, H.-J. The Roles of Human Endogenous Retroviruses (HERVs) in Inflammation. Kosin Med. J. 2021, 36, 69–78. [Google Scholar] [CrossRef]
- Mantovani, A.; Dinarello, C.A.; Molgora, M.; Garlanda, C. Interleukin-1 and related cytokines in the regulation of inflam-mation and immunity. Immunity 2019, 50, 778–795. [Google Scholar] [CrossRef]
- Kany, S.; Vollrath, J.T.; Relja, B. Cytokines in Inflammatory Disease. Int. J. Mol. Sci. 2019, 20, 6008. [Google Scholar] [CrossRef]
- Chen, K.; Bao, Z.; Tang, P.; Gong, W.; Yoshimura, T.; Wang, J.M. Chemokines in homeostasis and diseases. Cell. Mol. Immunol. 2018, 15, 324–334. [Google Scholar] [CrossRef] [PubMed]
- Eckardt, V.; Miller, M.C.; Blanchet, X.; Duan, R.; Leberzammer, J.; Duchene, J.; Soehnlein, O.; Megens, R.T.; Ludwig, A.; Dregni, A.; et al. Chemokines and galectins form heterodimers to modulate inflammation. EMBO Rep. 2020, 21, e47852. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, L.M. The Role of Nuclear Factor κB in the Interferon Response. J. Interf. Cytokine Res. 2011, 31, 553–559. [Google Scholar] [CrossRef]
- Pires, B.R.; Silva, R.C.; Ferreira, G.M.; Abdelhay, E. NF-kappaB: Two sides of the same coin. Genes 2018, 9, 24. [Google Scholar] [CrossRef] [PubMed]
- Sehnert, B.; Burkhardt, H.; Dübel, S.; Voll, R.E. Cell-type targeted NF-kappaB inhibition for the treatment of inflammatory diseases. Cells 2020, 9, 1627. [Google Scholar] [CrossRef] [PubMed]
- Schuliga, M. NF-kappaB signaling in chronic inflammatory airway disease. Biomolecules 2015, 5, 1266–1283. [Google Scholar] [CrossRef]
- Dev, A.; Iyer, S.; Razani, B.; Cheng, G. NF-κB and innate immunity. In NF-Kb Health and Disease; Springer: Berlin/Heidelberg, Germany, 2010; pp. 115–143. [Google Scholar]
- Antonaki, A.; Demetriades, C.; Polyzos, A.; Banos, A.; Vatsellas, G.; Lavigne, M.D.; Apostolou, E.; Mantouvalou, E.; Papadopoulou, D.; Mosialos, G. Genomic analysis reveals a novel nuclear factor-κB (NF-κB)-binding site in Alu-repetitive elements. J. Biol. Chem. 2011, 286, 38768–38782. [Google Scholar] [CrossRef]
- Apostolou, E.; Thanos, D. Virus infection induces NF-κB-dependent interchromosomal associations mediating monoallelic IFN-β gene expression. Cell 2008, 134, 85–96. [Google Scholar] [CrossRef]
- Markopoulos, G.S.; Roupakia, E.; Marcu, K.B.; Kolettas, E. Epigenetic regulation of inflammatory cytokine-induced epithe-lial-to-mesenchymal cell transition and cancer stem cell generation. Cells 2019, 8, 1143. [Google Scholar] [CrossRef]
- Markopoulos, G.S.; Roupakia, E.; Tokamani, M.; Chavdoula, E.; Hatziapostolou, M.; Polytarchou, C.; Marcu, K.B.; Papavassiliou, A.G.; Sandaltzopoulos, R.; Kolettas, E. A step-by-step microRNA guide to cancer development and metastasis. Cell. Oncol. 2017, 40, 303–339. [Google Scholar] [CrossRef]
- Markopoulos, G.S.; Roupakia, E.; Tokamani, M.; Alabasi, G.; Sandaltzopoulos, R.; Marcu, K.B.; Kolettas, E. Roles of NF-κB signaling in the regulation of miRNAs impacting on inflammation in cancer. Biomedicines 2018, 6, 40. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, H.A.; Iliopoulos, D.; Joshi, A.; Zhang, Y.; Jaeger, S.A.; Bulyk, M.; Tsichlis, P.N.; Liu, X.S.; Struhl, K. A Transcriptional Signature and Common Gene Networks Link Cancer with Lipid Metabolism and Diverse Human Diseases. Cancer Cell 2010, 17, 348–361. [Google Scholar] [CrossRef]
- Ditsworth, D.; Zong, W.-X. NF-kappaB: Key mediator of inflammation-associated cancer. Cancer Biol. Ther. 2004, 3, 1214–1216. [Google Scholar] [CrossRef] [PubMed]
- Salas, A.; Hernandez-Rocha, C.; Duijvestein, M.; Faubion, W.; McGovern, D.; Vermeire, S.; Vetrano, S.; Casteele, N.V. JAK–STAT pathway targeting for the treatment of inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Pfitzner, E.; Kliem, S.; Baus, D.; Litterst, M.C. The Role of STATs in Inflammation and Inflammatory Diseases. Curr. Pharm. Des. 2004, 10, 2839–2850. [Google Scholar] [CrossRef] [PubMed]
- Stat, C.J.K.; Egf, P. The Jak-STAT pathway in rheumatoid arthritis. J. Rheumatol. 2005, 32, 1650–1653. [Google Scholar]
- Gonzalez-Cao, M.; Iduma, P.; Karachaliou, N.; Santarpia, M.; Blanco, J.; Rosell, R. Human endogenous retroviruses and cancer. Cancer Biol. Med. 2016, 13, 483. [Google Scholar]
- Kitsou, K.; Iliopoulou, M.; Spoulou, V.; Lagiou, P.; Magiorkinis, G. Viral Causality of Human Cancer and Potential Roles of Human Endogenous Retroviruses in the Multi-Omics Era: An Evolutionary Epidemiology Review. Front. Oncol. 2021, 11. [Google Scholar] [CrossRef]
- Agoni, L.; Guha, C.; Lenz, J. Detection of Human Endogenous Retrovirus K (HERV-K) Transcripts in Human Prostate Cancer Cell Lines. Front. Oncol. 2013, 3, 180. [Google Scholar] [CrossRef]
- Bonaventura, P.; Alcazer, V.; Mutez, V.; Tonon, L.; Martin, J.; Chuvin, N.; Michel, E.; Boulos, R.E.; Estornes, Y.; Valladeau-Guilemond, J.; et al. Identification of shared tumor epitopes from endogenous retroviruses inducing high-avidity cytotoxic T cells for cancer immunotherapy. Sci. Adv. 2022, 8, eabj3671. [Google Scholar] [CrossRef]
- Wang-Johanning, F.; Frost, A.R.; Jian, B.; Epp, L.; Lu, D.W.; Johanning, G.L. Quantitation of HERV-K env gene expression and splicing in human breast cancer. Oncogene 2003, 22, 1528–1535. [Google Scholar] [CrossRef]
- Golan, M.; Hizi, A.; Resau, J.H.; Yaal-Hahoshen, N.; Reichman, H.; Keydar, I.; Tsarfaty, I. Human Endogenous Retrovirus (HERV-K) Reverse Transcriptase as a Breast Cancer Prognostic Marker. Neoplasia 2008, 10, 521–533, IN1–IN2. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Li, M.; Wei, Y.; Lin, K.; Lu, Y.; Shen, J.; Johanning, G.L.; Wang-Johanning, F. Activation of HERV-K Env protein is essential for tumorigenesis and metastasis of breast cancer cells. Oncotarget 2016, 7, 84093–84117. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Hong, Z.; Liu, H.; Chen, X.; Ding, L.; Liu, Z.; Zhou, F.; Yuan, Y. Human Endogenous Retroviruses-K (HML-2) Expression Is Correlated with Prognosis and Progress of Hepatocellular Carcinoma. BioMed Res. Int. 2016, 2016, 8201642. [Google Scholar] [CrossRef] [PubMed]
- Wang-Johanning, F.; Liu, J.; Rycaj, K.; Huang, M.; Tsai, K.; Rosen, D.G.; Chen, D.-T.; Lu, D.W.; Barnhart, K.F.; Johanning, G.L. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int. J. Cancer 2006, 120, 81–90. [Google Scholar] [CrossRef]
- Golkaram, M.; Salmans, M.L.; Kaplan, S.; Vijayaraghavan, R.; Martins, M.; Khan, N.; Garbutt, C.; Wise, A.; Yao, J.; Casimiro, S.; et al. HERVs establish a distinct molecular subtype in stage II/III colorectal cancer with poor outcome. npj Genom. Med. 2021, 6, 13. [Google Scholar] [CrossRef]
- Ko, E.-J.; Ock, M.-S.; Choi, Y.-H.; Iovanna, J.; Mun, S.; Han, K.; Kim, H.-S.; Cha, H.-J. Human Endogenous Retrovirus (HERV)-K env Gene Knockout Affects Tumorigenic Characteristics of nupr1 Gene in DLD-1 Colorectal Cancer Cells. Int. J. Mol. Sci. 2021, 22, 3941. [Google Scholar] [CrossRef]
- Giebler, M.; Staege, M.S.; Blauschmidt, S.; Ohm, L.I.; Kraus, M.; Würl, P.; Taubert, H.; Greither, T. Elevated HERV-K Expression in Soft Tissue Sarcoma Is Associated with Worsened Relapse-Free Survival. Front. Microbiol. 2018, 9, 211. [Google Scholar] [CrossRef]
- Alcazer, V.; Bonaventura, P.; Tonon, L.; Michel, E.; Mutez, V.; Fabres, C.; Chuvin, N.; Boulos, R.; Estornes, Y.; Maguer-Satta, V.; et al. HERVs characterize normal and leukemia stem cells and represent a source of shared epitopes for cancer immunotherapy. Am. J. Hematol. 2022, 97, 1200–1214. [Google Scholar] [CrossRef]
- Rivas, S.R.; Valdez, M.J.M.; Govindarajan, V.; Seetharam, D.; Doucet-O’Hare, T.T.; Heiss, J.D.; Shah, A.H. The Role of HERV-K in Cancer Stemness. Viruses 2022, 14, 2019. [Google Scholar] [CrossRef]
- Larouche, J.-D.; Trofimov, A.; Hesnard, L.; Ehx, G.; Zhao, Q.; Vincent, K.; Durette, C.; Gendron, P.; Laverdure, J.-P.; Bonneil, É.; et al. Widespread and tissue-specific expression of endogenous retroelements in human somatic tissues. Genome Med. 2020, 12, 40. [Google Scholar] [CrossRef] [PubMed]
- Mantziou, S.; Markopoulos, G.; Thrasyvoulou, S.; Noutsopoulos, D.; Gkartziou, F.; Artholomatos, G.; Tzavaras, T. Tinzaparin inhibits VL30 retrotransposition induced by oxidative stress and/or VEGF in HC11 mouse progenitor mammary cells: Association between inhibition of cancer stem cell proliferation and mammosphere disaggregation. Oncol. Rep. 2021, 46, 241. [Google Scholar] [CrossRef] [PubMed]
- Noutsopoulos, D.; Markopoulos, G.; Vartholomatos, G.; Kolettas, E.; Kolaitis, N.; Tzavaras, T. VL30 retrotransposition signals activation of a caspa-se-independent and p53-dependent death pathway associated with mitochondrial and lysosomal damage. Cell Res. 2010, 20, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Noutsopoulos, D.; Markopoulos, G.; Koliou, M.; Dova, L.; Vartholomatos, G.; Kolettas, E.; Tzavaras, T. Vanadium Induces VL30 Retrotransposition at an Unusually High Level: A Possible Carcinogenesis Mechanism. J. Mol. Biol. 2007, 374, 80–90. [Google Scholar] [CrossRef]
- Markopoulos, G.; Noutsopoulos, D.; Mantziou, S.; Vartholomatos, G.; Monokrousos, N.; Angelidis, C.; Tzavaras, T. Arsenic Induces VL30 Retrotransposition: The Involvement of Oxidative Stress and Heat-Shock Protein 70. Toxicol. Sci. 2013, 134, 312–322. [Google Scholar] [CrossRef][Green Version]
- Thrasyvoulou, S.; Vartholomatos, G.; Markopoulos, G.; Noutsopoulos, D.; Mantziou, S.; Gkartziou, F.; Papageorgis, P.; Charchanti, A.; Kouklis, P.; Constantinou, A.I.; et al. VL30 retrotransposition is associated with induced EMT, CSC generation and tumorigenesis in HC11 mouse mammary stem-like epithelial cells. Oncol. Rep. 2020, 44, 126–138. [Google Scholar] [CrossRef]
- Nellåker, C.; Yao, Y.; Jones-Brando, L.; Mallet, F.; Yolken, R.H.; Karlsson, H. Transactivation of elements in the human endogenous retrovirus W family by viral infection. Retrovirology 2006, 3, 44. [Google Scholar] [CrossRef]
- Badarinarayan, S.S.; Shcherbakova, I.; Langer, S.; Koepke, L.; Preising, A.; Hotter, D.; Kirchhoff, F.; Sparrer, K.M.J.; Schotta, G.; Sauter, D. HIV-1 infection activates endogenous retroviral promoters regulating antiviral gene expression. Nucleic Acids Res. 2020, 48, 10890–10908. [Google Scholar] [CrossRef]
- Monde, K.; Terasawa, H.; Nakano, Y.; Soheilian, F.; Nagashima, K.; Maeda, Y.; Ono, A. Molecular mechanisms by which HERV-K Gag interferes with HIV-1 Gag assembly and particle infectivity. Retrovirology 2017, 14, 27. [Google Scholar] [CrossRef]
- Monde, K.; Contreras-Galindo, R.; Kaplan, M.H.; Markovitz, D.M.; Ono, A. Human Endogenous Retrovirus K Gag Coassembles with HIV-1 Gag and Reduces the Release Efficiency and Infectivity of HIV-1. J. Virol. 2012, 86, 11194–11208. [Google Scholar] [CrossRef]
- Hurst, T.P.; Magiorkinis, G. Activat ion of the innate immune response by endogenous retroviruses. J. Gen. Virol. 2015, 96, 1207–1218. [Google Scholar] [CrossRef] [PubMed]
- Curty, G.; Iniguez, L.P.; Soares, M.A.; Nixon, D.F.; de Mulder Rougvie, M. Off-Target Effect of Activation of NF-κB by HIV Latency Reversal Agents on Transposable Elements Expression. Viruses 2022, 14, 1571. [Google Scholar] [CrossRef] [PubMed]
- McClintock, B. The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. USA 1950, 36, 344–355. [Google Scholar] [CrossRef]
- McClintock, B. Controlling elements and the gene. Proc. Cold Spring Harb. Symp. Quant. Biol. 1956, 21, 197–216. [Google Scholar] [CrossRef]
- Feschotte, C. Transposable elements and the evolution of regulatory networks. Nat. Rev. Genet. 2008, 9, 397–405. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Markopoulos, G.S. A Systems Biology Approach on the Regulatory Footprint of Human Endogenous Retroviruses (HERVs). Diseases 2022, 10, 98. https://doi.org/10.3390/diseases10040098
Markopoulos GS. A Systems Biology Approach on the Regulatory Footprint of Human Endogenous Retroviruses (HERVs). Diseases. 2022; 10(4):98. https://doi.org/10.3390/diseases10040098
Chicago/Turabian StyleMarkopoulos, Georgios S. 2022. "A Systems Biology Approach on the Regulatory Footprint of Human Endogenous Retroviruses (HERVs)" Diseases 10, no. 4: 98. https://doi.org/10.3390/diseases10040098
APA StyleMarkopoulos, G. S. (2022). A Systems Biology Approach on the Regulatory Footprint of Human Endogenous Retroviruses (HERVs). Diseases, 10(4), 98. https://doi.org/10.3390/diseases10040098





