1. Introduction
Sepsis is a syndrome of physiological, pathological, and biochemical abnormalities induced by infection [
1]. The conservative estimates indicate that sepsis is a leading cause of mortality and critical illness worldwide [
2,
3]. World Health Organization concerned that sepsis continues to cause approximately six million deaths worldwide every year, most of which are preventable [
4]. In their study, the Department of Health in Ireland reported that survival from sepsis-induced hypotension is over 75% if it is recognized promptly, but that every delay by an hour causes that figure to fall by over 7%, implying that the mortality increases by about 30%.
In this paper, we present our solution for the early detection of sepsis by joining the PhysioNet/Computing in Cardiology Challenge 2019 [
5]. Here, a detailed explanation of the Challenge data, participant evaluation metrics, and primary results are provided, and therefore, we will not explain it in this paper. However, a few important findings we should share in this paper in order to better explain the motivation to construct our algorithm in a particular way.
According to the requirements of the Challenge, our open-source algorithm works on clinical data provided on a real-time basis by giving a positive or negative prediction of sepsis for every single hour. The algorithm predicts sepsis development for the patient using a pre-trained mathematical model. Therefore, not only the appropriate model should be used but also the training should be performed in the right way.
Data used in the competition was collected from intensive care unit (ICU) patients in three separate hospital systems. However, data from two hospital systems only were publicly available for training (40,336 patients in total). Another set of records (24,819 patients in total), obtained from all three different hospital systems was hidden and used for official scoring only by challenges organizers. Such separation of the data prevented participants from over-fitting their models. Taking into account that the trained model may learn not only dependencies in the clinical records but also hospital system-related behavior, for our approach, we have tested different data selection strategies for training. Models trained on hospital system A data we tested on data from hospital system B and vice versa.
The most challenging issue in the available data records was a high number of unbalanced records. Only 2932 septic patients were included in the dataset, together with 37,404 non-septic patients. From the perspective of mathematical model training, the data balance is much worse. Since the sepsis prediction had to be made on an hourly basis, 6 h in advance to the onset time of sepsis, specified according to Sepsis-3 clinical criteria, a number of non-sepsis examples we also took from the septic patient early records. After such reorganization of training data, only 2% from 1,484,384 [1,424,171] events (16,933 from 752,946 [739,663] in set A and 10,557 from 73,438 [684,508] in set B) had to be classified as an early prediction of sepsis.
The imbalance of the data can be treated in different ways. Nemati et al. successfully used random subsampling to train deep cancer subtype classifier [
6]. Vicar et al. used special cost function—Generalized Dice Loss [
7]. Sweetly et al. created 54 datasets using the same sepsis data and different non-sepsis data records [
8]. He et al. have applied a random subsampling to this Challenge data [
9]. Although the rank of their solution was quite high in the Challenge, the model was highly overfitted on hospital systems A and B when comparing to the model performance on hidden hospital system C data. An interesting approach was proposed by Li et al., where they decided to divide data into three stages (1–9, 10–49 and above 50 h stay in ICU) [
10].
Dealing with missing values is another decision to be taken and it also may have an influence to the selected model training and overall performance. Forward-fill method [
8,
11,
12,
13,
14,
15]. Singh at al. found in their study, that mean imputation model gave worst results [
16]. Other authors successfully used mean calculation over whole dataset [
15,
17].
Our proposed algorithm was scored on a censored data set, dedicated for scoring and using utility function that rewards early predictions and penalizes late predictions as well as false alarms.
2. Materials and Methods
In this section, we address the challenges regarding the problem of early sepsis detection and propose a methodology to overcome them. A labeled clinical records dataset for training and verification of the algorithms was provided by the PhysioNet/Computing in Cardiology Challenge 2019 organizers [
5].
2.1. The Data
Data contained records of 40,336 ICU patients with up to 40 clinical variables divided into two datasets, based on hospital systems A and B. For each patient, the data were recorded at every hour during the stay in ICU. The records were labeled (on an hourly basis) according to Sepsis-3 clinical criteria. A total of 1,407,716 h of data was collected and labeled. Data labels included vital signs, laboratory values, and demographic values of the patients. Eight vital signs were a heart rate (HR), pulse oximetry (O2sat), temperature (Temp), systolic blood pressure (SBP), mean arterial pressure (MAP), diastolic blood pressure (DBP), respiration rate (Resp) and end-tidal carbon dioxide (EtCO2). A total of 26 laboratory values were included in the dataset. Demographic values include age, gender, hospital identifiers, the time between hospital and ICU admission (Hosp), and ICU length of stay (ICULOS). Data were labeled as positive 12 h before and 3 h after the onset time of sepsis. Positive labels of sepsis were found in 2932 of the 40,336 records, which is 7.27% of the data. Labels consisting of positive (sepsis) labels were found in 27,916 rows, which is only 1.98% of all data.
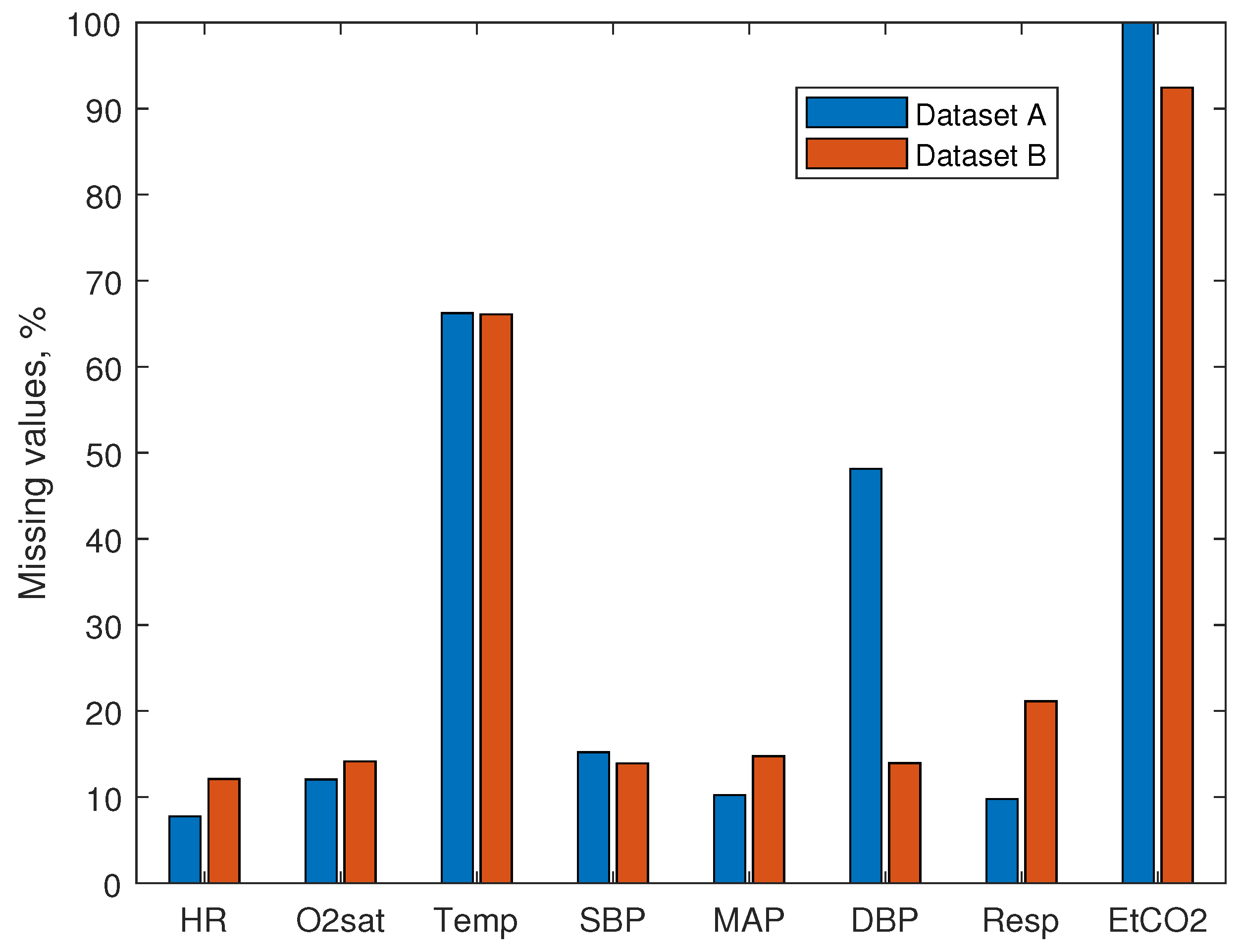
Investigation of the data showed large numbers of missing values. The percentage of missing rows of vital signs is shown in
Figure 1. Missing values of vital signs make about 10% of the data, with the exception of Temp ( 66% missing data) and EtCO2 (100% and 92% missing data, for dataset A and dataset B, respectively). Therefore, EtCO2 was not used as a feature for the model. The percentage of missing rows of laboratory measurements is shown in
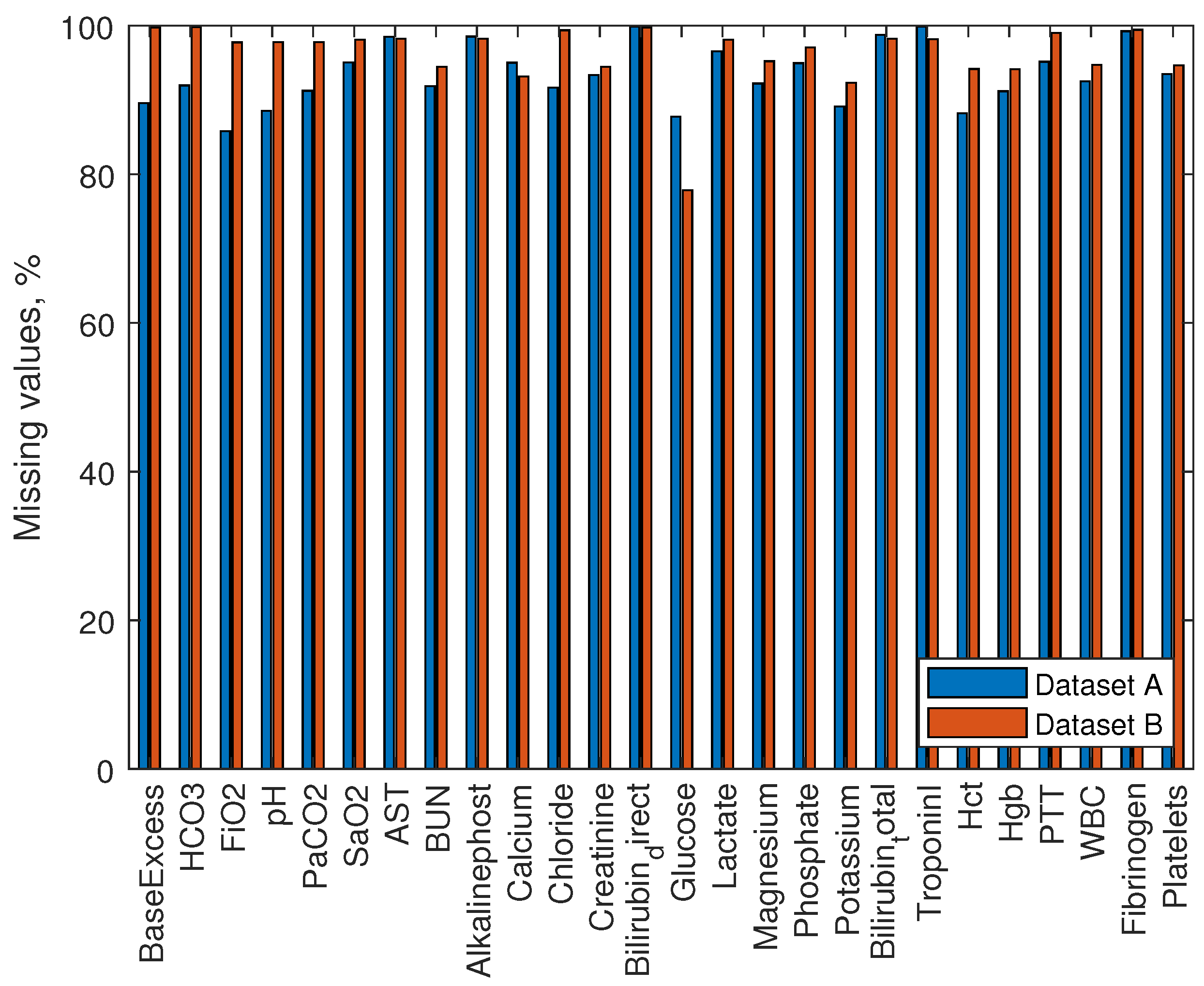
Figure 2. Missing data of laboratory values makes from 78% to 100% for all values. We did not use laboratory values to develop our model.
Average values of vitals are shown in
Table 1. Measured SBP, MAP and DBP values are higher in dataset B. Also, the measured HR is slightly lower in dataset B. Having two datasets collected in separate hospitals allowed us to develop models that are robust to measurement errors rising from the specificity of electronic medical record systems. Thus, the nature of data increases the difficulty of predicting sepsis. During the development of the model, we had to take into account the high unbalance of positive and negative cases, large amounts of missing values, and the fact that data was recorded using two different measurement systems.
2.2. Feature Extraction
A solution proposed in this paper to the early sepsis prediction problem employs information of the ICU length of stay, hospitalization time, age, and seven vital signs—HR, O2sat, Temp, SBP, MAP, DBP, and Resp. We did not use EtCO2 for feature extraction due to a large number of missing values.
We have calculated the mean, standard deviation, and the max-min difference for the vital sign data. We took those values from the whole duration of the record. Additionally, we have considered some other measures for our approach, such as kurtosis, entropy, and the standard error. However, after further analysis, we decided to discard these features. Kurtosis can only be calculated for four or more variables, not including the missing values. Additionally, kurtosis is not a representative statistic estimate for sample sizes less than 200 [
18]. The entropy value is proportional to the sample size. In the problem we have investigated in this paper, the sample size changes each hour of the patient stay and can be reduced with missing values for some patients [
19]. Therefore, in this case, the entropy just represents a number of samples used for its calculation. Thus, it is unlikely that entropy can carry useful information for the model training. The standard error is calculated by a division by the sample size, and it is inversely proportional to the sample size. Therefore, results can lead to a reduction of standard error for larger data sample sizes, which in its order increases unwanted load model training [
20].
After our theoretical investigation, we have calculated 21 features for each hour. Missing values of the data were removed when calculating features. In some cases, features could not be calculated due to a small set of available data (e.g., during the first few hours of ICU stay, or due to a large number of missing values). In such cases, we set the value of the feature to ‘−1’. Finally, we have assembled a feature set of 24 features for model training: 21 calculated featured from vital signs and three demographic values (Hosp, age, ICULOS). Obtained data had different measurement units, measurement errors, and scales. Therefore, we have applied data standardization to have zero mean and standard deviation ‘1’. The sample mean and sample standard deviation were used the same for each patient, obtained from all sample data of both datasets containing 40,336 patients in total.
2.3. Data Balancing
The data from the experiment were strongly unbalanced, as discussed in
Section 2.1. The balancing of the data can be performed using various oversampling or subsampling techniques that change the data in the dataset before the model is trained. In our investigation, we followed the alternative approach by setting different weights for the individual data points according to their class.
During our investigation of possible data balancing approaches, we surveyed various sampling methods, also used in the Challenge by other authors. The challengers applied undersampling, subsampling methods and some oversampling. The immensity of the imbalance problem is very great; the ratio of the labels is 1:50. Additionally, clinical data is very contextual. Oversampling methods haven’t shown good results in the Challenge. Undersampling methods possibly removed valuable information of non-septic patients. Most of the subsampling methods overtrained showed high Utility scores on known datasets and poor scores on hidden datasets. The variability of the clinical (ICU) data is very high. Non-septic cases have various other conditions and unknown prescribed medications. We think that adjustment of classification cost is a more robust and effective way to address this issue of data imbalance. The highest rank in the 2019 Challenge was received by a solution, where the classification cost was calculated using Utility function value differences between correct and incorrect classification [
13]. In our solution, we used simple weighting and investigated the behavior of the model training with differently selected weighs.
We have weighed the positive and negative predictions in accordance with the duration before or after the onset time of sepsis. Positive values, with various weights, make only 1.98% of the data. For this reason, the investigated models were trained with uneven classification costs. We have addressed the data balance issue by utilizing and modifying the classification cost function:
where
N is the number of observations,
is the target output for observation
i,
is the predicted value for observation
i, and
is the classification cost for the observation
i.
To investigate the influence of the selected weight on the model’s behavior during training, we trained our models using different classification costs. A misclassified non-septic observations were weighted by 1 (), and a misclassified septic observation weighted selecting different (1, 10, 20, 30, 100). Classification cost was selected based on the amount of “unbalance”. As it was expected, since the positive values make only 1.98% of the data, the most effective value should be found when the classification cost is between 20 and 30. Models with parameter lower than 20 tend to predict most of the data as non-septic, while models with higher tend to predict all data as septic.
2.4. Model Training
In our investigation, we have used models based on Decision trees, naive Gaussian Bayes, Support Vector Machines (SVM), and Ensemble learners. All models were trained using stratified 5-fold cross validation. Decision trees based models were important in this stage of the problem solution. The Decision tree models give insights about the relevance of selected features. However, they tend to overfit the data [
21]. Less over-fitting can be expected when using Ensemble learner models [
22]. We have trained the ensemble learning-based models using hyperparameter optimization among Bag, GentleBoost, LogitBoost, AdaBoost, and RUSBoost methods. Hyperparameter tuning was performed for the number of decision tree splits, number of learners used in the model, learning rate, number of features in the ensemble. The number of decision tree splits search scaled in the range from 1 to 500. Many branches tend to overfit the data, while simpler trees can be more robust, which is especially important for the clinical data. The number of learners used in the search was from 10 to 100. A high number of learners can produce higher accuracy but can be time-consuming to train. We changed the learning rate in the ranged from 0.0001 to 1, and the number of features in ensemble hyperparameter tuning scaled from 1 to 24.
Gaussian naive Bayes based models are known for their simplicity, high bias, and low overfit. Typically good results using Naive Bayes are achieved using low variance data. These models are not recommended for high variance data [
23]. Models trained using SVM tends to overfit data less. However, they are not very successful in problems with a high number of missing values in data [
24].
We have trained each model separately using features estimated on an hourly basis in random order. In order to avoid over-fitting and increase robustness, we have trained the models on records taken from a single hospital system (dataset A) and tested on records from another hospital (dataset B, hidden during the training). We trained models on the dataset A using 5-fold cross-validation. Using our selected approach, only half of the available data was used for training. However, as it was shown in the results of the challenge [
5], proposed solutions to the problem performed well on known datasets, even if scoring was done on a hidden part of the same set, and performed marginally worse on new hospital system C, hidden from challenge contestants. Additionally, the advantage of this approach to train and evaluate models is supported by Biglarbeigi [
25].
Based on the results of the trained models and insights into the data, we proposed a method that takes into account the amount of time the patients have already spent in the ICU. The first model used AdaBoost ensemble method with a decision tree to evaluate features of patients extracted during the first 56 h (11,146 sepsis labels and 654,866 non-sepsis labels; it makes approximate imbalance ratio of 1:59). The second model is applied for patients with ICULOS time greater than 56 (5990 sepsis labels and 118,213 non-sepsis labels; it makes an approximate imbalance ratio of 1:20). The second model was developed using the discriminant subspace-based ensemble method. The method generates decision trees using pseudorandomly selected feature components. Decisions of the trees are then combined by averaging the estimates. As this method is based on decision trees, it is fast to train and easy to interpret [
26]. Both models were using features described in
Section 2.2. Additionally, our proposed model was trained on both datasets, 75% of the data used for training. This allowed us to investigate how much model overtrains on known hospital systems.
2.5. Model Scoring
Models, proposed in this paper for Sepsis prediction, were evaluated using several different metrics. Traditional scoring metrics, such as the area under the receiver operating characteristics (AUROC), the area under the precision-recall curve (AUPRC), accuracy, F-measure, and Matthews correlation coefficient (MCC) were used. Additionally, investigated models were scored using a specific scoring function developed by the authors of the dataset, called Utility score. Performance of the investigated models was based on the Utility score metric. Additionally, using different scoring metrics allowed us a better comparison of investigated models. AUPRC is recommended for imbalanced data over the AUROC measure [
27,
28]. F-measure is a harmonic mean of precision and recall [
29]. Lately, MCC measure was shown to be more advantageous over F-measure in the binary classification of imbalanced data [
30].
The utility score metric was proposed by the authors of the dataset for the 2019 physionet Challenge [
5]. This metric was recently proposed. The utility score metric reward algorithms that facilitate early sepsis detection and treatment. Additionally, it addresses the problem of infrequent events and sequential prediction tasks. It is designed to capture the clinical utility of early sepsis detection by weighting early and late predictions. Moreover, decision threshold metrics (AUROC and AUPRC) have problems evaluating unbalanced datasets. Additionally, other challengers evaluated their models using the Utility score, therefore, it is easier to compare the results. However, the experimental results showed that Utility score correlates with two traditional metrics F-measure and MCC.
Utility score—a specifically designed scoring function rewards algorithms for early predictions and penalizes them for late or missed predictions and false alarms. Scoring was conducted by predicting each hourly label for each patient. Each positive label had a defined score depending on correct prediction time to sepsis. Scoring function awarded models for correct prediction at most 12 h before and 3 h after the onset time of sepsis. Scoring function penalized models who predicted septic state 12 h before onset time of sepsis and slightly penalized models with false-positive predictions. True negative predictions were not penalized or rewarded by the function. Best Utility score the most optimal model could achieve would be 1. Thus, a better model would have higher Utility score. A more detailed scoring description can be found in the original paper. The utility score was a reference metric. Using it, we evaluated the performance of our investigated models. AUROC, AUPRC, accuracy, F-measure, and MCC scores were used to gain insight into the models (e.g., if they correlate with any other parameter of the experiment, such as classification cost, feature reduction, model configuration, or Utility score).
Our investigated and proposed models were compared with five challengers who received the highest Utility score on hidden test C. Additionally, a result of three baseline models was generated. Baseline models were scored using all positive, all negative and random performance to clearly show the unbalance of the data and the difficulty of the challenge.
3. Results
As it was noted in
Section 2.4, decision trees, naive Gaussian Bayes, SVM and ensemble learners were investigated in our experiment. Various parameters of the models were adjusted. Additionally, the effect of classification cost and feature reduction was investigated. The performance of developed models was evaluated using Utility score as a reference metric. To complement, several other metrics, such as AUROC, AUPRC, accuracy, F-measure, and MCC were calculated to compare investigated models. Models were trained using dataset A, and scored using dataset B. Results of the experiment are given in
Table 2. Models, based on decision trees, are labeled from 1 to 14. Models, based on the Naive Bayes algorithm, are labeled from 21 to 25. SVM based models are labeled from 31 to 34. Models from 41 to 44 were using an optimizable ensemble method for searching the best model for the problem.
The random guess of the sepsis with an accuracy of 50% showed −0.529 Utility score and 0.125 AUROC. By labeling all cases as positive, the Utility score was reduced to −1.059 and AUROC was 0, F-measure—0.029. Labeling all cases as negative increased the Utility score to 0, AUROC was 0.5 and F-measure—0. AUROC was generally expected to be equal or above 0.5 when the dataset is balanced. In
Table 2, the third row indicates the AUROC value of a random performance, having an accuracy of 50%). The AUROC value in such a case is 0.125 (for balanced datasets it would be 0.5). It is a direct insight into the importance of the problem.
Based on the results of the Physionet challenge 2019 on the hidden dataset, several of the highest Utility scores were from 0.017 to 0.193. The highest Utility score (0.193) was achieved by Hong et al. using deep recurrent reinforcement learners [
31]. Murugesan et al. applied XGBoost algorithm and achieved 0.182 Utility score on hidden test set [
32]. Our proposed method was developed based on the results of further described investigations and results. The proposed method predicted 3753 true positive, 707,606 true negative, 7027 false positive and 43,309 false negative observations of dataset B. The precision of the method was
, recall—
. Our proposed method achieved
AUROC,
AUPROC,
accuracy,
F-measure,
MCC and
Utility scores. Additionally, this model was trained on both datasets (70% of the data used for training) achieved
Utility score.
Decision trees are fast to train and to evaluate. We started our investigation using these models. The baseline score of Model1, with default parameters, gave a Utility score of . Secondly, a feature reduction using principal component analysis (PCA) was applied. Six features to explain 95% variance was kept. Model2 gave Utility score of . For Model3, increasing feature set to 14 (out of 24) increased Utility score to . Forth model used 14 features and a modified classification cost ratio of 1:10. Model4 obtained Utility score of . Further increasing classification (Model5) ratio to 1:100 Utility score decreased to . Using all available features in the set (24 features) Utility score was slightly improved to , for Model6. Using 24 features and classification cost 1:10 obtained Utility score was for Model7. For Model8 we modified classification cost to 1:20, obtained Utility score was . Further increasing classification cost to 1:30 (Model9) decreased Utility score to . Model10, having a modified classification cost of 1:20, and a reduced feature set to 20 got Utility score decreased to . Next, we limited the tree split criterion to 50. Model11 achieved Utility score of . Higher Utility score was achieved by reducing split criterion to 4, it was (Model12). Reducing split criterion to 2 for Model13 got a similar Utility score of . Model14, with a further reduced split criterion to 1, achieved the highest Utility score of . Only 1 tree branch was used for this model. A feature that was used for this model was ICULOS. Features used for Model12 and Model14 also included ICULOS, and also mean SBP and Resp.
Models labeled from 21 to 25 were based on the Naive Bayes algorithm. Model21 without feature reduction and using classification cost 1:20 achieved Utility score of
. For Model23, using PCA, the number of features was reduced to 14, achieved Utility score was
. Further reducing the number of features to 6 (95% explained variance using PCA) improved Utility score to
, as shown in
Table 2 Model24 row. Adjusting the classification cost led to a reduced score—Model22 and Model25 used a reduced feature set and a modified classification cost of 1:10 and 1:30; they yielded Utility scores of
and
, respectively. SVM models were computed using the Gaussian kernel function. Results of the SVM models are shown in
Table 2, under Model31 to Model34. Model31, using a classification cost ratio of 1:20 and 24 features for training, achieved a
Utility score. Model32, using 6 features to explain 95% variance, achieved a
Utility score. Model33 and Model34, using classification costs 1:30 and 1:10, respectively, achieved
and
Utility scores.
Models labeled from 41 to 44 were trained using ensemble methods, searching between Bag, GentleBoost, LogitBoost, AdaBoost, and RUSBoost methods and other hyperparameters. The classification cost for all investigated models were set to 1:20. Model41 using a full feature set achieved Utility score of . Reducing the tree split criterion to 10 (Model42) gave an improved Utility score of—. Ensemble model (Model43) using bagged decision trees, having 29 learners, 4 splits and using 24 features achieved a Utility score of , further reducing split criterion to 1 did not improve the Utility score—. Using principal component analysis (95%) reduced the Utility score to (Model44).
High AUROC, AUPRC and accuracy scores using decision tree models were achieved when classification cost was 1:1, for example, , , AUROC score for Model1, Model2, Model3, respectively. Model12 and Model14 with high Utility scores gave low AUROC ( and , respectively), AUPRC (, both) and accuracy ( and , respectively) scores.
High AUROC, AUPRC, and accuracy scores using ensemble learners were achieved using Model41: , , and , respectively. However, this ensemble model achieved low Utility (). In the same manner, low AUROC (), AUPRC (), and accuracy () scores, and the highest Utility score () was achieved using Model43. Other investigated models performed similarly, high AUROC, AUPRC and accuracy, and low Utility score; or low AUROC, AUPRC and accuracy, and higher Utility score was observed in all investigated models, namely decision trees, SVM, naive Bayes, and ensemble-based models.
Highest F-measure and MCC scores using decision tree models were achieved for models that showed the highest Utility scores. F-measure score of Model14 was (Utility—), score of Model11 was (Utility—). MCC score of Model14 was ; the score of Model11 was . Lowest F-measure and MCC scores were achieved using Model2: F-measure—, MCC—. However, the lowest Utility score () using decision tree models was achieved using Model5. F-measure score of Model5 was , MCC—.
Naive Gaussian Bayes models achieved lowest F-measure () and MCC () scores when Model22 (Utility score—) was scored, highest F-measure () and MCC () scores using Model24 (Utility score—).
Model32 trained using SVM achieved F-measure () and MCC () score, and scored using Utility performance metric. However, Model31’s Utility score () was slightly higher, while F-measure () and MCC () score lower. Low F-measure () and MCC () scores also showed low Utility scores—, for Model33.
Comparably low F-measure () and MCC () scores using ensemble learners were achieved using Model44 (Utility score—). Model43 with a high Utility score () demonstrated a high F-measure () and MCC () scores.
4. Discussion
The highest Utility score was achieved using our proposed method, which divided patients based on their length of stay and then the appropriate model was applied. Additionally, decision trees with a low number of nodes achieved high Utility scores when ICU length of stay was included as a branch of decision tree. Therefore, we believe that future models should be developed based on ICU-stay time. For example, one model predicting recently hospitalized ICU patients, another would be used if a patient’s ICU length of stay reaches a certain length of time. Also, this approach can be implemented using three or more temporal divisions. This finding of our investigation is supported by Lauritsen [
36], Vincent [
37] and Shimabukuro [
38] papers. Each intervention, vital measurement, intravenous therapy, and duration of stay in general increases a chance of infection—a direct cause of sepsis.
Regarding the dataset, other papers tackled this problem and proposed methods, which were trained on both datasets, and officially scored on hidden set C. Dataset C is not available anymore. Therefore, one must find other means to compare the results with the challenge score. Most of the challengers performed well on known hospital systems, obtaining Utility scores of about
. However, Utility scores for the hidden hospital systems were low [
5]. One author suggested evaluating the proposed methodology using one dataset and testing it on another [
25]. Our achieved Utility score was for the known dataset, but the hidden percentage of data was
when trained on 75% of the records. This shows that our proposed model is robust to overtrain.
We assume that the Utility score can be improved a little by finding better value for classification cost, where a true positive prediction reward would be multiplied somewhere between 20 and 30. However, this would fit the data and would not solve the general problem of the Challenge. Therefore, we recommend using an arbitrary value between 20 and 30 to increases the robustness of the system.
MCC and F-measure scores gave similar results, which increases and decreases with the Utility score. However, the bounds of MCC are from −1 to 1, while the F-measure is from 0 to 1. The bounds of the Utility score are from −2 to 1. We support the idea of using the Utility score as a metric for this dataset. Moreover, we showed that the MCC and F-measure are effective metrics for this problem, while other traditional metrics AUROC, AURRC and Accuracy are misleading for a highly unbalanced dataset. Additionally, due to the nature of the Utility score, results can be difficult to interpret, as Roussel et al. pointed out in their work [
39].
Investigated decision trees achieved Utility score of
, AUROC score of
, and MCC score of
on hidden set. Models with such results are far from applicable to the clinical setting. Additionally, our investigation showed that increasing AUROC and accuracy usually leads to decreased Utility, F-measure, and MCC scores. Moreover, accuracy is high for all investigated models. Accuracy can be miss-leading when interpreting models, results for this kind of highly unbalanced data, and a large number of negatives [
22]. When developing methods for this kind of problem, one needs to be careful; the accuracy of 98.2% can be achieved just by guessing all rows as negative. We showed that balancing data reduces AUROC, accuracy scores and improves F-measure, MCC, and Utility scores.
There are many models to experiment with, for example, k-Nearest Neighbor (kNN) and Long Short-Term Memory (LSTM) models were not tested in our work. LSTM models are more difficult to configure to use them effectively. Additionally, LSTM tends to overfit the data. Moreover, even if one successfully tackles the overfitting problem, there is still another downside, which is more important in the current state of the early sepsis prediction problem. The developed model may be hard to interpret and would not reveal much insight into data [
40]. The clustering of unbalanced data (including Sepsis-related records) may give promising results for sepsis prediction. However, kNN overfits data with large variances [
41]. On the other hand, a trained kNN model having 1000 or 2000 clusters to represent the data can be expected to be robust. In general, we believe results using these models can be promising, and we encourage future works exploring LSTM and kNN model capabilities.
It is notable that investigated models do not differ significantly in Utility score if a number of features is reduced. This shows that some features are not useful for the model. On the other hand, our proposed features were relatively simple. We believe that more advanced features are needed to solve the early detection of the sepsis problem. Using advanced features should improve the score. However, feature engineering is a difficult, time-consuming process, which also requires understanding the nature of the data. In this paper, we provide many insights into the nature of the data, different scoring metrics, advantages of various models, and feature combinations.
We believe that the results of our investigation presented in this paper will benefit the fundamental need of early sepsis prediction and will answer some basic questions about the limits of early detection. Our results should benefit the search for advanced combinations of features, ease the use of machine learning tools. With meaningful insights peer researchers can apply advanced feature engineering techniques and develop more sophisticated and robust models in order to reach reliable results. Reaching better results is available through the use of combined models and handcrafted features [
42], thus, further contributing to this field. The main challenges of this problem, as we revealed, are—the highly unbalanced dataset, the high number of missing data, simple features calculated using vitals does not have enough predictive power, proposed solutions are prone to overtrain. Adjusting the classification cost function helps to address the latter problem. In addition, the insights and conclusions of our experiment may benefit not only machine learning specialists, researchers, but also ICU personnel and scientists in the medical field.