Abstract
In recent times, several machine learning models have been built to aid in the prediction of diverse diseases and to minimize diagnostic errors made by clinicians. However, since most medical datasets seem to be imbalanced, conventional machine learning algorithms tend to underperform when trained with such data, especially in the prediction of the minority class. To address this challenge and proffer a robust model for the prediction of diseases, this paper introduces an approach that comprises of feature learning and classification stages that integrate an enhanced sparse autoencoder (SAE) and Softmax regression, respectively. In the SAE network, sparsity is achieved by penalizing the weights of the network, unlike conventional SAEs that penalize the activations within the hidden layers. For the classification task, the Softmax classifier is further optimized to achieve excellent performance. Hence, the proposed approach has the advantage of effective feature learning and robust classification performance. When employed for the prediction of three diseases, the proposed method obtained test accuracies of 98%, 97%, and 91% for chronic kidney disease, cervical cancer, and heart disease, respectively, which shows superior performance compared to other machine learning algorithms. The proposed approach also achieves comparable performance with other methods available in the recent literature.
1. Introduction
Medical diagnosis is the process of deducing the disease affecting an individual [1]. This is usually done by clinicians, who analyze the patient’s medical record, conduct laboratory tests, and physical examinations, etc. Accurate diagnosis is essential and quite challenging, as certain diseases have similar symptoms. A good diagnosis should meet some requirements: it should be accurate, communicated, and timely. Misdiagnosis occurs regularly and can be life-threatening; in fact, over 12 million people get misdiagnosed every year in the United States alone [2]. Machine learning (ML) is progressively being applied in medical diagnosis and has achieved significant success so far.
In contrast to the shortfall of clinicians in most countries and expensive manual diagnosis, ML-based diagnosis can significantly improve the healthcare system and reduce misdiagnosis caused by clinicians, which can be due to stress, fatigue, and inexperience, etc. Machine learning models can also ensure that patient data are examined in more detail and results are obtained quickly [3]. Hence, several researchers and industry experts have developed numerous medical diagnosis models using machine learning [4]. However, some factors are hindering the growth of ML in the medical domain, i.e., the imbalanced nature of medical data and the high cost of labeling data. Imbalanced data are a classification problem in which the number of instances per class is not uniformly distributed. Recently, unsupervised feature learning methods have received massive attention since they do not entirely rely on labeled data [5], and are suitable for training models when the data are imbalanced.
There are various methods used to achieve feature learning, including supervised learning techniques such as dictionary learning and multilayer perceptron (MLP), and unsupervised learning techniques which include independent component analysis, matrix factorization, clustering, unsupervised dictionary learning, and autoencoders. An autoencoder is a neural network used for unsupervised feature learning. It is composed of input, hidden, and output layers [6]. The basic architecture of a three-layer autoencoder (AE) is shown in Figure 1. When given an input data, autoencoders (AEs) are helpful to automatically discover the features that lead to optimal classification [7]. There are diverse forms of autoencoders, including variational and regularized autoencoders. The regularized autoencoders have been mostly used in solving problems where optimal feature learning is needed for subsequent classification, which is the focus of this research. Examples of regularized autoencoders include denoising, contractive, and sparse autoencoders. We aim to implement a sparse autoencoder (SAE) to learn representations more efficiently from raw data in order to ease the classification process and ultimately, improve the prediction performance of the classifier.
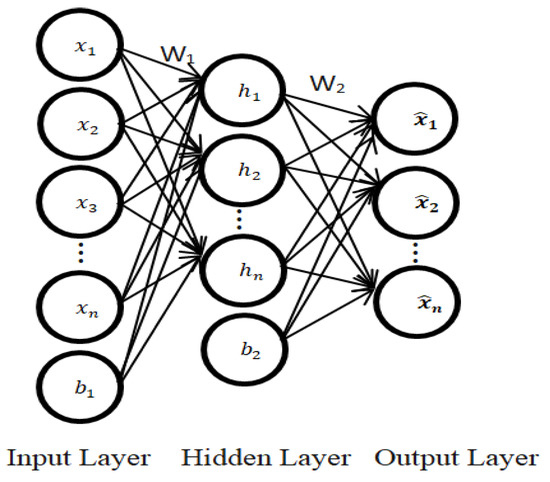
Figure 1.
The structure of an autoencoder.
Usually, the sparsity penalty in the sparse autoencoder network is achieved using either of these two methods: L1 regularization or Kullback–Leibler (KL) divergence. It is noteworthy that the SAE does not regularize the weights of the network; rather, the regularization is imposed on the activations. Consequently, suboptimal performances are obtained with this type of structure where the sparsity makes it challenging for the network to approximate a near-zero cost function [8]. Therefore, in this paper, we integrate an improved SAE and a Softmax classifier for application in medical diagnosis. The SAE imposes regularization on the weights, instead of the activations as in conventional SAE, and the Softmax classifier is used for performing the classification task.
To demonstrate the effectiveness of the approach, three publicly available medical datasets are used, i.e., the chronic kidney disease (CKD) dataset [9], cervical cancer risk factors dataset [10], and Framingham heart study dataset [11]. We also aim to use diverse performance evaluation metrics to assess the performance of the proposed method and compare it with some techniques available in the recent literature and other machine learning algorithms such as logistic regression (LR), classification and regression tree (CART), support vector machine (SVM), k-nearest neighbor (KNN), linear discriminant analysis (LDA), and conventional Softmax classifier. The rest of the paper is structured as follows: Section 2 reviews some related works, while Section 3 introduces the methodology and provides a detail background of the methods applied. The results are tabulated and discussed in Section 4, while Section 5 concludes the paper.
2. Related Works
This section discusses some recent applications of machine learning in medical diagnosis. Glaucoma is a vision condition that develops gradually and can lead to permanent vision loss. This condition destroys the optic nerve, the health of which is essential for good vision and is usually caused by too much pressure inside one or both eyes. There are diverse forms of glaucoma, and they have no warning signs; hence, early detection is difficult yet crucial. Recently, a method was developed for the early detection of glaucoma using a two-layer sparse autoencoder [7]. The SAE was trained using 1426 fundus images to identify salient features from the data and differentiate a normal eye from an affected eye. The structure of the network comprises of two cascaded autoencoders and a Softmax layer. The autoencoder network performed unsupervised feature learning, while the Softmax was trained in a supervised fashion. The proposed method obtained excellent performance with an F-measure of 0.95.
In another research, a two-stage approach was proposed for the prediction of heart disease using a sparse autoencoder and artificial neural network (ANN) [12]. Unsupervised feature learning was performed with the help of the sparse autoencoder, which was optimized using the adaptive moment estimation (Adam) algorithm, whereas the ANN was used as the classifier. The method achieved an accuracy of 90% on the Framingham heart disease dataset and 98% on the cervical cancer risk factors dataset, which outperformed some ML algorithms. In a similar research, Verma et al. [13] proposed a hybrid technique for the classification of heart disease, where optimal features were selected via the particle swarm optimization (PSO) search technique and k-means clustering. Several supervised learning methods, including decision tree, MLP, and Softmax regression, were then utilized for the classification task. The method was tested using a dataset containing 335 cases and 26 attributes, and the experimental results revealed that the hybrid model enhanced the accuracy of the various classifiers, with the Softmax regression model obtaining the best performance with 88.4% accuracy.
Tama et al. [14] implemented an ensemble learning method for the diagnosis of heart disease. The ensemble method was developed via a stacked structure, whereby the base learners were also ensembles. The base learners include gradient boosting, random forest (RF), and extreme gradient boosting (XGBoost). Additionally, feature ranking and selection were conducted using correlation-based feature selection and PSO, respectively. When tested on different heart disease datasets, the proposed method outperformed the conventional ensemble methods. Furthermore, Ahishakiye et al. [15] developed an ensemble learning classifier to detect cervical cancer risk. The model comprised of CART, KNN, SVM, and naïve Bayes (NB) as base learners, and the ensemble model achieved an accuracy of 87%.
The application of sparse autoencoders in the medical domain has been widely studied, especially for disease prediction [12]. Furthermore, sparse autoencoders have been utilized for classifying Parkinson’s disease (PD). Recently, Xiong and Lu [16] proposed an approach which involved a feature extraction step using a sparse autoencoder, to classify PD efficiently. Prior to the feature extraction, the data were preprocessed and an appropriate input subset was selected from the vocal features via the adaptive grey wolf optimization method. After feature extraction by the SAE, six ML classifiers were then applied to perform the classification task, and the experimental results signaled improved performance compared to other related works.
From the above-related works, we observed that most of the studies have some limitations: firstly, most of the authors utilized a single medical dataset to validate the performance of their models and not many studies experimented on more than two different diseases. By training and testing the model on two or more datasets, appropriate and more reliable conclusions can be drawn, and this can further validate the generalization ability of the ML method. Secondly, some recent research works have implemented sparse autoencoders for feature learning; however, most of these methods achieved sparsity by regularizing the activations [17], which is the norm. However, in this paper, sparsity is achieved via weight regularization. Additionally, poor generalization of ML algorithms resulting from imbalanced datasets, which is common in medical data, can be easily addressed using an effective feature learning method such as this.
3. Methodology
The sparse autoencoder (SAE) is an unsupervised learning method which is used to automatically learn features from unlabeled data [14]. In this type of autoencoder, the training criterion involves a sparsity penalty. Generally, the loss function of an SAE is constructed by penalizing activations within the hidden layers. For any particular sample, the network is encouraged to learn an encoding by activating only a small number of nodes. By introducing sparsity constraints on the network, such as limiting the number of hidden units, the algorithm can learn better relationships from the data [18]. An autoencoder consists of two functions: an encoder and decoder function. The encoder maps the d-dimensional input data to obtain a hidden representation. In contrast, the decoder maps the hidden representation back to a d-dimensional vector that is as close as possible to the encoder input [12,19]. Assuming denotes the input features and represents the neurons of the hidden layer, the encoding and decoding process can be represented with the following equations:
where and represent the weight matrices of the hidden layer and output layer, respectively; and denotes the bias matrices of the hidden layer and output layer, respectively; the vector denotes the inputs of the output layer; the vector represents the output of the sparse autoencoder, which is fed into the Softmax classifier for classification. The mean squared error function is used as the reconstruction error function between the input and reconstructed input . Additionally, we introduce a regularization function to the error function in order to achieve sparsity by penalizing the weights and . Therefore, the cost function of the sparse autoencoder can be represented as:
The mean squared error function and the regularization function can be expressed as:
Once the data have been transmitted from input to output of the sparse autoencoder, the next stage involves evaluating the cost function and fine-tuning the model parameters for optimal performance. Meanwhile, the cost function does not explicitly relate the weights and bias of the network; hence, it is necessary to define a sensitivity measure to sensitize the changes in and transmit the changes backwards via the backpropagation learning method [8]. To achieve this, and iteratively optimize the loss function, stochastic gradient descent is employed. The stochastic gradient descent to update the bias and weights of the output layer can be written as:
where represents the learning rate in relation to the output layer. The derivative of the loss function measures the sensitivity to change of the function value with respect to a change in its input value. Furthermore, the gradient indicates the extent to which the input parameter needs to change to minimize the loss function. Meanwhile, the gradients are computed using the chain rule. Therefore, (6) and (7) can be rewritten as:
The sensitivity at the output layer of the SAE is represented and defined as . Therefore, (8) and (9) can be rewritten as:
where
Using the same method for computing , the sensitivities can be transmitted back to the hidden layer
where denotes the learning rate with respect to the hidden layer, whereas is defined as:
Furthermore, the Softmax classifier is employed for the classification task. The learned features from the proposed SAE are used to train the classifier. Though, Softmax regression, otherwise called multinomial logistic regression (MLR), is a generalization of logistic regression that can be utilized for multi-class classification [20]. However, in the literature, the Softmax classifier has been applied for several binary classification tasks and has obtained excellent performance [21]. The Softmax function provides a method to interpret the outputs as probabilities and is expressed as:
where represent the input values and the output is the probability that the sample belongs to the label [22]. For input samples, the error at the Softmax layer is measured using the cross-entropy loss function:
where the true probability is the actual label and is the predicted value. is a measure of the dissimilarity between . Furthermore, neural networks can easily become stuck in local minima, whereby the algorithm assumes it has reached the global minima, thereby resulting in non-optimal performance. To prevent the local minima problem and further enhance classifier performance, the mini-batch gradient descent with momentum is applied to optimize the cross-entropy loss of the Softmax classifier. This optimization algorithm splits the training data into small batches which are then used to compute the model error and update the model parameters [23]. The momentum [24] ensures better convergence is obtained.
The flowchart to visualize the proposed methodology is shown in Figure 2. The initial dataset is preprocessed; then, it is divided into training and testing sets. The training set is utilized for training the sparse autoencoder in an unsupervised manner. Meanwhile, the testing set is transformed and inputted into the trained model to obtain the low-dimensional representation dataset. The low-dimensional training set is used to train the Softmax classifier, and its performance is tested using the low-dimensional test set. Hence, there is no possible data leakage since the classifier sees only the low-dimensional training set.
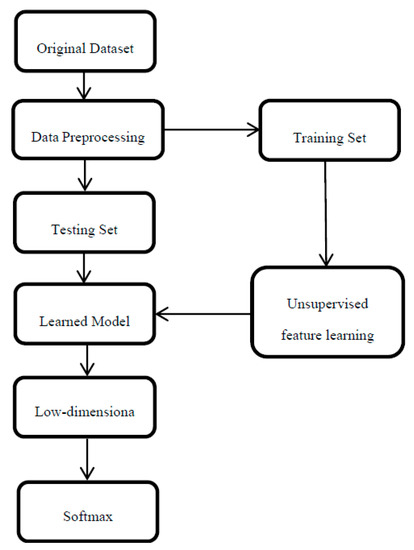
Figure 2.
Flowchart of the methodology.
4. Results and Discussion
The proposed method is applied for the prediction of three diseases in order to show its performance in diverse medical diagnosis situations. The datasets include the Framingham heart study [11], which was obtained from the Kaggle website, and it contains 4238 samples and 16 features. The second dataset is the cervical cancer risk factors dataset [10], which was obtained from the University of California, Irvine (UCI) ML repository, and it contains 858 instances and 36 attributes. Thirdly, the CKD dataset [9] was also obtained from the UCI ML repository, and it contains 400 samples and 25 features. We used mean imputation to handle missing variables in the datasets.
The training parameters of the SAE include: , and number of epochs = 200. The hyperparameters of the Softmax classifier include learning rate = 0.01, number of samples in mini batches = 32, momentum value = 0.9, and number of epochs = 200. These parameters were obtained from the literature [12,23], as they have led to optimal performance in diverse neural network applications.
The effectiveness of the proposed method is evaluated using the following performance metrics: accuracy, precision, recall, and F1 score. Accuracy is the ratio of the correctly classified instances to the total number of instances in the test set, and precision measures the fraction of correctly predicted instances among the ones predicted to have the disease, i.e., positive [25]. Meanwhile, recall measures the proportion of sick people that are predicted correctly, and F1 score is a measure of the balance between precision and recall [26]. The following equations are used to determine these metrics:
where
- True positive (TP): Sick people correctly predicted as sick.
- False-positive (FP): Healthy people wrongly predicted as sick.
- True negative (TN): Healthy people rightly predicted as healthy.
- False-negative (FN): Sick people wrongly predicted as healthy.
To demonstrate the efficacy of the proposed method, it is benchmarked with other algorithms, such as LR, CART, SVM, KNN, LDA, and conventional Softmax regression. In order to show the improved performance of the proposed method, no parameter tuning was performed on these algorithms; hence, their default parameter values in scikit-learn were used, which are adequate for most machine learning problems. The K-fold cross-validation technique was used to evaluate all the models. Table 1, Table 2 and Table 3 show the experimental results when the proposed method is tested on the Framingham heart study, cervical cancer risk factors, and CKD datasets, respectively. Meanwhile, Figure 3, Figure 4 and Figure 5 show the receiver operating characteristic (ROC) curves comparing the performance of the conventional Softmax classifier and the proposed approach for the various disease prediction models. The ROC curve illustrates the diagnostic ability of binary classifiers, and it is obtained by plotting the true positive rate (TPR) against the false positive rate (FPR).

Table 1.
Performance of the proposed method and other classifiers on the Framingham dataset.

Table 2.
Performance of the proposed method and other classifiers on the cervical cancer dataset.

Table 3.
Performance of the proposed method and other classifiers on the chronic kidney disease (CKD) dataset.
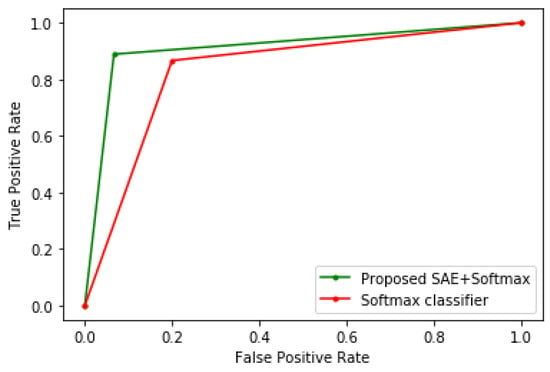
Figure 3.
Receiver operating characteristic (ROC) curve of the heart disease model.
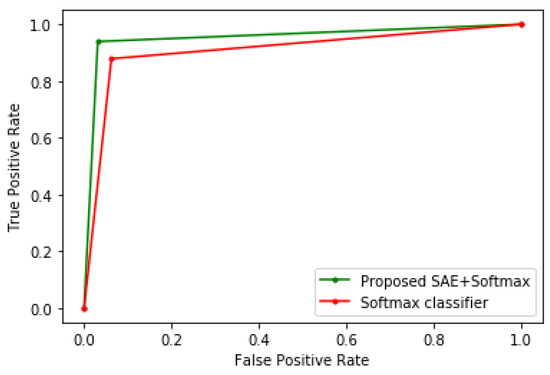
Figure 4.
ROC curve of the cervical cancer model.
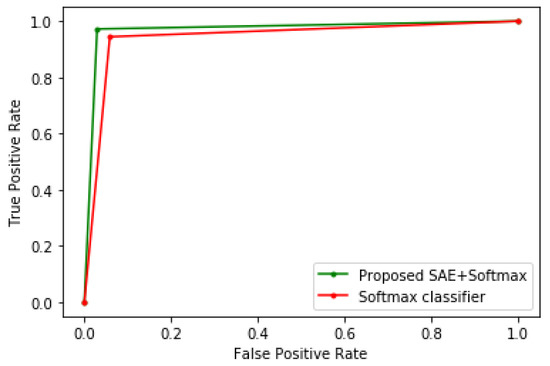
Figure 5.
ROC curve of the CKD model.
From the experimental results, it can be seen that the sparse autoencoder improves the performance of the Softmax classifier, which is further validated by the ROC curves of the various models. The proposed method also performed better than the other machine learning algorithms. Furthermore, the misclassifications obtained by the model in the various disease predictions are also considered. For the prediction of heart disease, the proposed method recorded an FPR of 7% and a false-negative rate (FNR) of 10%. In addition, the model specificity, which is the true negative rate (TNR), is 93%, and the TPR is 90%. For the cervical cancer dataset, the following were obtained: FPR = 3%, FNR = 5%, TNR = 97%, and TPR = 95%. For the CKD prediction: FPR = 0, FNR = 3%, TNR = 100%, and TPR = 97%.
Additionally, to further validate the performance of the proposed method, we compare it with some models for heart disease prediction available in the recent literature, including a feature selection method using PSO and Softmax regression [13], a two-tier ensemble method with PSO-based feature selection [14], an ensemble classifier comprising of the following base learners: NB, Bayes Net (BN), RF, and MLP [27], a hybrid method of NB and LR [28], and a hybrid RF with a linear model (HRFLM) [29]. The other techniques include a combination of LR and Lasso regression [30], an intelligent heart disease detection method based on NB and advanced encryption standard (AES) [31], a combination of ANN and Fuzzy analytic hierarchy method (Fuzzy-AHP) [32], and a sparse autoencoder feature learning method combined ANN classifier [12]. This comparison is tabulated in Table 4. Meanwhile, in order to give a fair comparison, only the accuracies of the various techniques were considered because some authors did not report the values for other performance metrics.

Table 4.
Comparison of the proposed method with the recent literature that used the heart disease dataset.
In Table 5, we compare the proposed approach with some recent scholarly works that used the cervical cancer dataset, including principal component analysis (PCA)-based SVM [33], a research work where the dataset was preprocessed and classified using numerous algorithms, in which LR and SVM had the best accuracy [34], a C5.0 decision tree [35]. The other methods include a multistage classification process which combined isolation forest (iForest), synthetic minority over-sampling technique (SMOTE), and RF [36], a sparse autoencoder feature learning method combined ANN classifier [12], and a feature selection method combined with C5.0 and RF [37].

Table 5.
Comparison of the proposed method with the recent literature that used the cervical cancer dataset.
In Table 6, we compare the proposed method with other recent CKD prediction research works, including an optimized XGBoost method [38], a probabilistic neural network (PNN) [39], and a method using adaptive boosting (AdaBoost) [40]. The other research works include a hybrid classifier of NB and decision tree (NBTree) [41], XGBoost [42], and a 7-7-1 MLP neural network [43].

Table 6.
Comparison of the proposed method with the recent literature that used the cervical CKD dataset.
From the tabulated comparisons, the proposed sparse autoencoder with Softmax regression obtained comparable performance with the state-of-the-art methods in various disease predictions. Additionally, the experimental results show an improved performance obtained due to efficient feature representation by the sparse autoencoder. This further demonstrates the importance of training classifiers with relevant data, since they can significantly affect the performance of the prediction model. Lastly, this research also showed that excellent classification performance could be obtained not only by performing hyperparameter tuning of algorithms but also by employing appropriate feature learning techniques.
5. Conclusions
In this paper, we developed an approach for improved prediction of diseases based on an enhanced sparse autoencoder and Softmax regression. Usually, autoencoders achieve sparsity by penalizing the activations within the hidden layers, but in the proposed method, the weights were penalized instead. This is necessary because by penalizing the activations, it makes approximating near-zero loss function challenging for the network. The proposed method was tested on three different diseases, including heart disease, cervical cancer, and chronic kidney disease, and it achieved accuracies of 91%, 97%, and 98%, respectively, which outperformed conventional Softmax regression and other algorithms. By experimenting with different datasets, we aimed to demonstrate the effectiveness of the method in diverse conditions. We also conducted a comparative study with some prediction models available in the recent literature, and the proposed approach obtained comparable performance in terms of accuracy. Thus, it can be concluded that the proposed approach is a promising method for the detection of diseases and can be further developed into a clinical decision support system to assist health professionals as in [44]. Meanwhile, future research will apply the method studied in this paper for the prediction of more diseases, and also employ other performance metrics such as training time, classification time, computational speed, and other metrics, which could be beneficial for the performance evaluation of the model.
Author Contributions
Conceptualization, S.A.E.-M.; methodology, S.A.E.-M., E.E., T.G.S.; software, S.A.E.-M., E.E.; validation, S.A.E.-M., E.E.; formal analysis, S.A.E.-M., E.E., T.G.S.; investigation, S.A.E.-M., E.E., T.G.S.; resources, E.E., T.G.S.; data curation, E.E., T.G.S.; writing—original draft preparation, S.A.E.-M., E.E.; writing—review and editing, E.E., T.G.S.; visualization, E.E.; supervision, E.E., T.G.S.; project administration, E.E., T.G.S.; funding acquisition, E.E., T.G.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding but will be funded by Research Center funds.
Acknowledgments
This work is supported by the Center of Telecommunications, University of Johannesburg, South Africa.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Stanley, D.E.; Campos, D.G. The Logic of Medical Diagnosis. Perspect. Biol. Med. 2013, 56, 300–315. [Google Scholar] [CrossRef] [PubMed]
- Epstein, H.M. The Most Important Medical Issue Ever: And Why You Need to Know More About It. Society to Improve Diagnosis in Medicine. 2019. Available online: https://www.improvediagnosis.org/dxiq-column/most-important-medical-issue-ever/ (accessed on 30 August 2020).
- Liu, N.; Li, X.; Qi, E.; Xu, M.; Li, L.; Gao, B. A novel Ensemble Learning Paradigm for Medical Diagnosis with Imbalanced Data. IEEE Access 2020, 8, 171263–171280. [Google Scholar] [CrossRef]
- Ma, Z.; Ma, J.; Miao, Y.; Liu, X.; Choo, K.K.R.; Yang, R.; Wang, X. Lightweight Privacy-preserving Medical Diagnosis in Edge Computing. IEEE Trans. Serv. Comput. 2020, 1. [Google Scholar] [CrossRef]
- Li, X.; Jia, M.; Islam, M.T.; Yu, L.; Xing, L. Self-supervised Feature Learning via Exploiting Multi-modal Data for Retinal Disease Diagnosis. IEEE Trans. Med. Imaging 2020, 1. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Guo, R.; Lin, Z.; Peng, T.; Peng, X. A data-driven health monitoring method using multi-objective optimization and stacked autoencoder based health indicator. IEEE Trans. Ind. Inform. 2020, 1. [Google Scholar] [CrossRef]
- Raghavendra, U.; Gudigar, A.; Bhandary, S.V.; Rao, T.N.; Ciaccio, E.J.; Acharya, U.R. A Two Layer Sparse Autoencoder for Glaucoma Identification with Fundus Images. J. Med. Syst. 2019, 43, 299. [Google Scholar] [CrossRef]
- Musafer, H.; Abuzneid, A.; Faezipour, M.; Mahmood, A. An Enhanced Design of Sparse Autoencoder for Latent Features Extraction Based on Trigonometric Simplexes for Network Intrusion Detection Systems. Electronics 2020, 9, 259. [Google Scholar] [CrossRef]
- Rubini, L.J.; Eswaran, P. UCI Machine Learning Repository: Chronic_Kidney_Disease Data Set. 2015. Available online: https://archive.ics.uci.edu/ml/datasets/chronic_kidney_disease (accessed on 26 June 2020).
- UCI Machine Learning Repository: Cervical cancer (Risk Factors) Data Set. Available online: https://archive.ics.uci.edu/ml/datasets/Cervical+cancer+%28Risk+Factors%29 (accessed on 27 January 2020).
- Framingham Heart Study Dataset. Available online: https://kaggle.com/amanajmera1/framingham-heart-study-dataset (accessed on 24 January 2020).
- Mienye, I.D.; Sun, Y.; Wang, Z. Improved sparse autoencoder based artificial neural network approach for prediction of heart disease. Inform. Med. Unlocked 2020, 18, 100307. [Google Scholar] [CrossRef]
- Verma, L.; Srivastava, S.; Negi, P.C. A Hybrid Data Mining Model to Predict Coronary Artery Disease Cases Using Non-Invasive Clinical Data. J. Med. Syst. 2016, 40, 178. [Google Scholar] [CrossRef]
- Tama, B.A.; Im, S.; Lee, S. Improving an Intelligent Detection System for Coronary Heart Disease Using a Two-Tier Classifier Ensemble. BioMed. Res. Int. 2020. Available online: https://www.hindawi.com/journals/bmri/2020/9816142/ (accessed on 28 August 2020). [CrossRef]
- Ahishakiye, E.; Wario, R.; Mwangi, W.; Taremwa, D. Prediction of Cervical Cancer Basing on Risk Factors using Ensemble Learning. In Proceedings of the 2020 IST-Africa Conference (IST-Africa), Kampala, Uganda, 6–8 May 2020; pp. 1–12. [Google Scholar]
- Xiong, Y.; Lu, Y. Deep Feature Extraction from the Vocal Vectors Using Sparse Autoencoders for Parkinson’s Classification. IEEE Access 2020, 8, 27821–27830. [Google Scholar] [CrossRef]
- Daoud, M.; Mayo, M.; Cunningham, S.J. RBFA: Radial Basis Function Autoencoders. In Proceedings of the 2019 IEEE Congress on Evolutionary Computation (CEC), Wellington, New Zealand, 10–13 June 2019; pp. 2966–2973. [Google Scholar] [CrossRef]
- Ng, A. Sparse Autoencoder. 2011. Available online: https://web.stanford.edu/class/cs294a/sparseAutoencoder.pdf (accessed on 6 June 2020).
- İrsoy, O.; Alpaydın, E. Unsupervised feature extraction with autoencoder trees. Neurocomputing 2017, 258, 63–73. [Google Scholar] [CrossRef]
- Kayabol, K. Approximate Sparse Multinomial Logistic Regression for Classification. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 42, 490–493. [Google Scholar] [CrossRef]
- Herrera, J.L.L.; Figueroa, H.V.R.; Ramírez, E.J.R. Deep fraud. A fraud intention recognition framework in public transport context using a deep-learning approach. In Proceedings of the 2018 International Conference on Electronics, Communications and Computers (CONIELECOMP), Cholula Puebla, Mexico, 21–23 February 2018; pp. 118–125. [Google Scholar] [CrossRef]
- Wang, M.; Lu, S.; Zhu, D.; Lin, J.; Wang, Z. A High-Speed and Low-Complexity Architecture for Softmax Function in Deep Learning. In Proceedings of the 2018 IEEE Asia Pacific Conference on Circuits and Systems (APCCAS), Chengdu, China, 26–28 October 2018; pp. 223–226. [Google Scholar] [CrossRef]
- Ruder, S. An overview of gradient descent optimization algorithms. arXiv 2017, arXiv:1609.04747. [Google Scholar]
- Qian, N. On the momentum term in gradient descent learning algorithms. Neural Netw. 1999, 12, 145–151. [Google Scholar] [CrossRef]
- Mienye, I.D.; Sun, Y.; Wang, Z. An improved ensemble learning approach for the prediction of heart disease risk. Inform. Med. Unlocked 2020, 20, 100402. [Google Scholar] [CrossRef]
- Abdulhammed, R.; Musafer, H.; Alessa, A.; Faezipour, M.; Abuzneid, A. Features Dimensionality Reduction Approaches for Machine Learning Based Network Intrusion Detection. Electronics 2019, 8, 322. [Google Scholar] [CrossRef]
- Latha, C.B.C.; Jeeva, S.C. Improving the accuracy of prediction of heart disease risk based on ensemble classification techniques. Inform. Med. Unlocked 2019, 16, 100203. [Google Scholar] [CrossRef]
- Amin, M.S.; Chiam, Y.K.; Varathan, K.D. Identification of significant features and data mining techniques in predicting heart disease. Telemat. Inform. 2019, 36, 82–93. [Google Scholar] [CrossRef]
- Mohan, S.; Thirumalai, C.; Srivastava, G. Effective Heart Disease Prediction Using Hybrid Machine Learning Techniques. IEEE Access 2019, 7, 81542–81554. [Google Scholar] [CrossRef]
- Haq, A.U.; Li, J.P.; Memon, M.H.; Nazir, S.; Sun, R. A Hybrid Intelligent System Framework for the Prediction of Heart Disease Using Machine Learning Algorithms. Mob. Inf. Syst. 2018, 2018, 3860146. [Google Scholar] [CrossRef]
- Repaka, A.N.; Ravikanti, S.D.; Franklin, R.G. Design and Implementing Heart Disease Prediction Using Naives Bayesian. In Proceedings of the 2019 3rd International Conference on Trends in Electronics and Informatics (ICOEI), Tirunelveli, India, 23–25 April 2019; pp. 292–297. [Google Scholar] [CrossRef]
- Samuel, O.W.; Asogbon, G.M.; Sangaiah, A.K.; Fang, P.; Li, G. An integrated decision support system based on ANN and Fuzzy_AHP for heart failure risk prediction. Expert Syst. Appl. 2017, 68, 163–172. [Google Scholar] [CrossRef]
- Wu, W.; Zhou, H. Data-Driven Diagnosis of Cervical Cancer with Support Vector Machine-Based Approaches. IEEE Access 2017, 5, 25189–25195. [Google Scholar] [CrossRef]
- Abdullah, F.B.; Momo, N.S. Comparative analysis on Prediction Models with various Data Preprocessings in the Prognosis of Cervical Cancer. In Proceedings of the 2019 10th International Conference on Computing, Communication and Networking Technologies (ICCCNT), Kanpur, India, 6–8 July 2019; pp. 1–6. [Google Scholar] [CrossRef]
- Chang, C.-C.; Cheng, S.-L.; Lu, C.-J.; Liao, K.-H. Prediction of Recurrence in Patients with Cervical Cancer Using MARS and Classification. Int. J. Mach. Learn. Comput. 2013, 3, 75–78. [Google Scholar] [CrossRef]
- Ijaz, M.F.; Attique, M.; Son, Y. Data-Driven Cervical Cancer Prediction Model with Outlier Detection and Over-Sampling Methods. Sensors 2020, 20, 2809. [Google Scholar] [CrossRef]
- Nithya, B.; Ilango, V. Evaluation of machine learning based optimized feature selection approaches and classification methods for cervical cancer prediction. SN Appl. Sci. 2019, 1, 641. [Google Scholar] [CrossRef]
- Ogunleye, A.A.; Qing-Guo, W. XGBoost Model for Chronic Kidney Disease Diagnosis. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019, 1. [Google Scholar] [CrossRef]
- Rady, E.-H.A.; Anwar, A.S. Prediction of kidney disease stages using data mining algorithms. Inform. Med. Unlocked 2019, 15, 100178. [Google Scholar] [CrossRef]
- Gupta, D.; Khare, S.; Aggarwal, A. A method to predict diagnostic codes for chronic diseases using machine learning techniques. In Proceedings of the 2016 International Conference on Computing, Communication and Automation (ICCCA), Greater Noida, India, 29–30 April 2016; pp. 281–287. [Google Scholar] [CrossRef]
- Khan, B.; Naseem, R.; Muhammad, F.; Abbas, G.; Kim, S. An Empirical Evaluation of Machine Learning Techniques for Chronic Kidney Disease Prophecy. IEEE Access 2020, 8, 55012–55022. [Google Scholar] [CrossRef]
- Raju, N.V.G.; Lakshmi, K.P.; Praharshitha, K.G.; Likhitha, C. Prediction of chronic kidney disease (CKD) using Data Science. In Proceedings of the 2019 International Conference on Intelligent Computing and Control Systems (ICCS), Madurai, India, 15–17 May 2019; pp. 642–647. [Google Scholar] [CrossRef]
- Aljaaf, A.J.; Al-Jumeily, D.; Haglan, H.M.; Alloghani, M.; Baker, T.; Hussain, A.J.; Mustafina, J. Early Prediction of Chronic Kidney Disease Using Machine Learning Supported by Predictive Analytics. In Proceedings of the 2018 IEEE Congress on Evolutionary Computation (CEC), Rio de Janeiro, Brazil, 8–13 July 2018; pp. 1–9. [Google Scholar] [CrossRef]
- Ebiaredoh-Mienye, S.A.; Esenogho, E.; Swart, T.G. Artificial Neural Network Technique for Improving Prediction of Credit Card Default: A Stacked Sparse Autoencoder Approach. Int. J. Electr. Comput. Eng. (IJECE) 2020. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).