Automatic Detection of Atrial Fibrillation and Other Arrhythmias in ECG Recordings Acquired by a Smartphone Device
Abstract
1. Introduction
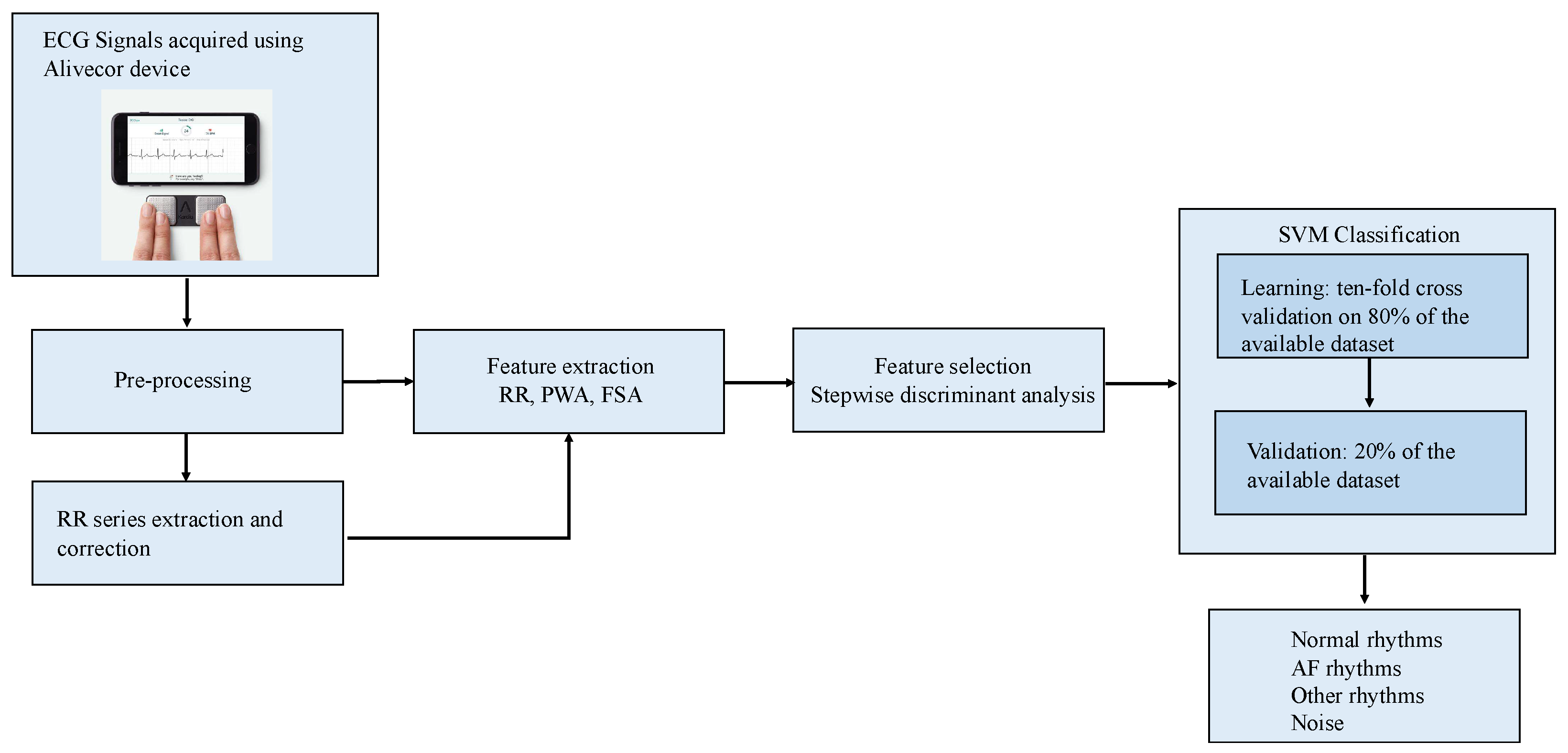
2. Materials and Methods
2.1. Datasets
2.2. ECG Pre-Processing and RR Series Construction
2.3. Feature Extraction and Selection
2.3.1. RR Analysis
2.3.2. P Wave Analysis
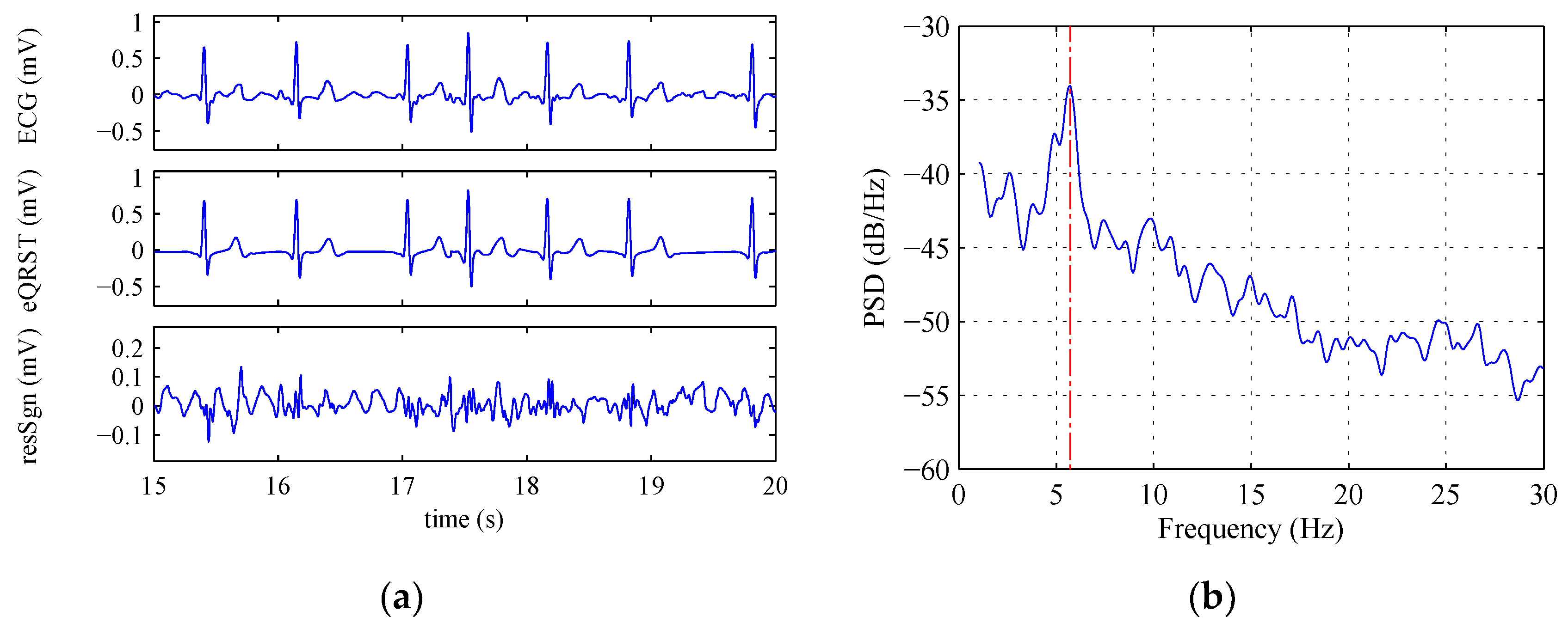
2.3.3. Frequency Spectrum Analysis
2.3.4. Other Features
2.4. Classification
2.5. Scoring
3. Results
3.1. Physionet Challenge 2017 Database
3.2. MIT-BH AF Database
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Chugh, S.S.; Havmoeller, R.; Narayanan, K.; Singh, D.; Rienstra, M.; Benjamin, E.J.; Gillum, R.F.; Kim, Y.H.; McAnulty, J.H., Jr.; Zheng, Z.J.; et al. Worldwide epidemiology of atrial fibrillation: A Global Burden of Disease 2010 Study. Circulation 2014, 129, 837–847. [Google Scholar] [CrossRef] [PubMed]
- Krijthe, B.P.; Kunst, A.; Benjamin, E.J.; Lip, G.Y.; Franco, O.H.; Hofman, A.; Witteman, J.C.; Stricker, B.H.; Heeringa, J. Projections on the number of individuals with atrial fibrillation in the European Union, from 2000 to 2060. Eur. Heart J. 2013, 34, 2746–2751. [Google Scholar] [CrossRef] [PubMed]
- Chugh, S.S.; Blackshear, J.L.; Shen, W.K.; Hammill, S.C.; Gersh, B.J. Epidemiology and natural history of atrial fibrillation: Clinical implications. J. Am. Coll. Cardiol. 2001, 37, 371–378. [Google Scholar] [CrossRef]
- Miyasaka, Y.; Barnes, M.E.; Gersh, B.J.; Cha, S.S.; Bailey, K.R.; Abhayaratna, W.P.; Seward, J.B.; Tsang, T.S. Secular trends in incidence of atrial fibrillation in olmsted county, minnesota, 1980 to 2000, and implications on the projections for future prevalence. Circulation 2006, 114, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.J.; Larson, M.G.; Levy, D.; Vasan, R.S.; Leip, E.P.; Wolf, P.A.; D’Agostino, R.B.; Murabito, J.M.; Kannel, W.B.; Benjamin, E.J. Temporal relations of atrial fibrillation and congestive heart failure and their joint influence on mortality: The framingham heart study. Circulation 2003, 107, 2920–2925. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Chun, S.; Hadley, D.; Froelicher, V. Clinical Implications of Technological Advances in Screening for Atrial Fibrillation. Prog. Cardiovasc. Dis. 2018, 60, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Tateno, K.; Glass, L. A method for detection of atrial fibrillation using rr intervals. Comput. Cardiol. 2000, 27, 391–394. [Google Scholar]
- Dash, S.; Chon, K.; Lu, S.; Raeder, A. Automatic real time detection of atrial fibrillation. Ann. Biomed. Eng. 2009, 37, 1701–1709. [Google Scholar] [CrossRef] [PubMed]
- Chiarugi, F.; Varanini, M.; Cantini, F.; Conforti, F.; Vrouchos, G. Noninvasive ecg as a tool for predicting termination of paroxysmal atrial fibrillation. IEEE Trans. Biomed. Eng. 2007, 54, 1399–1406. [Google Scholar] [CrossRef] [PubMed]
- Stridth, M.; Sornmo, L. Shape characterization of atrial fibrillation using time-frequency analysis. Comput. Cardiol. 2002, 29, 17–20. [Google Scholar]
- Slocum, J.; Sahakian, A.; Swiryn, S. Diagnosis of Atrial Fibrillation from Surface Electrocardiograms Based on Computer-detected Atrial Activity. J. Electrocardiol. 1992, 25, 1–8. [Google Scholar] [CrossRef]
- Clavier, L.; Boucher, J.; Lepage, R.; Blanc, J.; Cornily, J. Automatic p-wave analysis of patients prone to atrial fibrillation. Med. Biol. Eng. Comput. 2002, 40, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Larburu, N.; Lopetegi, T.; Romero, I. Comparative study of algorithms for atrial fibrillation detection. Comput. Cardiol. 2011, 38, 265–268. [Google Scholar]
- Couceiro, R.; Carvalho, P.; Henriques, J.; Antunes, M.; Harris, M.; Habetha, J. Detection of Atrial Fibrillation using model-based ECG analysis. In Proceedings of the 19th International Conference on Pattern Recognition, Tampa, FL, USA, 8–11 December 2008; pp. 1–5. [Google Scholar]
- Schmidt, R.; Harris, M.; Novac, D.; Perkhun, M. Atrial Fibrillation Detection; Patent Cooperation Treaty: Eindhoven, The Netherlands, 2008. [Google Scholar]
- Babaeizadeh, S.; Gregg, R.E.; Helfenbein, E.D.; Lindauer, J.M.; Zhou, S.H. Improvements in atrial fibrillation detection for real-time monitoring. J. Electrocardiol. 2009, 42, 522–526. [Google Scholar] [CrossRef] [PubMed]
- Kara, S.; Okandan, M. Atrial fibrillation classification with artificial neural networks. Pattern Recogn. 2007, 40, 2967–2973. [Google Scholar] [CrossRef]
- Kennedy, A.; Finlay, D.; Guldenring, D.; Bond, R.; Moran, K.; McLaughlin, J. Automated detection of atrial fibrillation using R-R intervals and multivariate-based classification. J. Electrocardiol. 2016, 49, 871–876. [Google Scholar] [CrossRef] [PubMed]
- Nayak, J.; Naik, B.; Behera, H. A comprehensive survey on support vector machine in data mining tasks: Applications & challenges. Int. J. Database Theory Appl. 2015, 8, 169–186. [Google Scholar]
- Mohebbi, M.; Ghassemian, H. Detection of atrial fibrillation episodes using SVM. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2008, 2008, 177–180. [Google Scholar] [PubMed]
- Asgari, S.; Mehrnia, A.; Moussavi, M. Automatic detection of atrial fibrillation using stationary wavelet transform and support vector machine. Comput. Biol. Med. 2015, 60, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Carrara, M.; Carozzi, L.; Moss, T.; De Pasquale, M.; Cerutti, S.; Ferrario, M. Heart rate dynamics distinguish among atrial fibrillation, normal sinus rhythm and sinus rhythm with frequent ectopy. Physiol. Meas. 2015, 36, 1873–1888. [Google Scholar] [CrossRef] [PubMed]
- Carrara, M.; Carozzi, L.; Moss, T.; De Pasquale, M.; Cerutti, S.; Lake, D.E.; Moorman, J.R.; Ferrario, M. Classification of cardiac rhythm using heart rate dynamical measures: Validation in MIT-BIH databases. J. Electrocardiol. 2015, 48, 943–946. [Google Scholar] [CrossRef] [PubMed]
- DeMazumder, D.; Lake, D.E.; Cheng, A.; Moss, T.J.; Guallar, E.; Weiss, R.G.; Jones, S.R.; Tomaselli, G.F.; Moorman, J.R. Dynamic analysis of cardiac rhythms for discriminating atrial fibrillation from lethal ventricular arrhythmias. Circ. Arrhythm. Electrophysiol. 2013, 6, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Clifford, G.; Liu, C.; Moody, B.; Lehman, L.H.; Silva, I.; Li, Q.; Johnson, A.; Mark, R.G. AF classification from a short single lead ecg recording: The PhysioNet/Computing in Cardiology Challenge 2017. Comput. Cardiol. 2017, 44, 1–4. [Google Scholar]
- Lau, J.K.; Lowres, N.; Neubeck, L.; Brieger, D.B.; Sy, R.W.; Galloway, C.D.; Albert, D.E.; Freedman, S.B. iPhone ECG application for community screening to detect silent atrial fibrillation: A novel technology to prevent stroke. Int. J. Cardiol. 2013, 165, 193–194. [Google Scholar] [CrossRef] [PubMed]
- Tu, H.T.; Chen, Z.; Swift, C.; Churilov, L.; Guo, R.; Liu, X.; Jannes, J.; Mok, V.; Freedman, B.; Davis, S.M.; et al. Smartphone electrographic monitoring for atrial fibrillation in acute ischemic stroke and transient ischemic attack. Int. J. Stroke 2017, 12, 786–789. [Google Scholar] [CrossRef] [PubMed]
- Billeci, L.; Chiarugi, F.; Costi, M.; Lombardi, D.; Varanini, M. Detection of AF and Other Rhythms Using RR Variability and ECG Spectral Measures. Comput. Cardiol. 2017, 44, 44. [Google Scholar] [CrossRef]
- The Physionet ECG Database. Available online: http://physionet.org/physiobank/database/.
- Varanini, M.; Tartarisco, G.; Billeci, L.; Macerata, A.; Pioggia, G.; Balocchi, R. An efficient unsupervised fetal qrs complex detection from abdominal maternal ecg. Physiol. Meas. 2014, 35, 1607–1619. [Google Scholar] [CrossRef] [PubMed]
- Taddei, A.; Marchesi, C.; Landucci, L. Performance comparison of fast QRS detection algorithms. In Ambulatory Monitoring; Marchesi, C., Ed.; Martinus Nijhoff: Boston, MA, USA, 1984; pp. 189–207. [Google Scholar]
- Lake, D.E.; Moorman, J.R. Accurate estimation of entropy in very short physiological time series: The problem of atrial fibrillation detection in implanted ventricular devices. Am. J. Physiol. Heat Circ. Physiol. 2011, 300, H319–H325. [Google Scholar] [CrossRef] [PubMed]
- Katz, M. Fractals and the analysis of waveforms. Comput. Biol. Med. 1988, 18, 45–156. [Google Scholar] [CrossRef]
- Le Digabel, S. Algorithm 909: NOMAD: Nonlinear Optimization with the MADS algorithm. ACM Trans. Math. Softw. 2011, 37, 1–15. [Google Scholar] [CrossRef]
- Elgendi, M.; Meo, M.; Abbott, D. A Proof-of-Concept Study: Simple and Effective Detection of P and T Waves in Arrhythmic ECG Signals. Bioengineering 2016, 3, 26. [Google Scholar] [CrossRef] [PubMed]
- Suykens, J.; Vandewalle, J. Least Squares Support Vector Machine Classifier. Neural Process. Lett. 1999, 9, 293–300. [Google Scholar] [CrossRef]
- Vapnik, V. Pattern recognition using generalized portrait method. Autom. Remote Control 1963, 24, 774–780. [Google Scholar]
- Hastie, T.; Tibshirani, R. Classification by pairwise coupling. Ann. Stat. 1998, 26, 451–471. [Google Scholar] [CrossRef]
- Suykens, J.A.K.; Van Gestel, T.; De Brabanter, J.; De Moor, B.; Vandewalle, J. Least Squares Support Vector Machines; World Scientific: Singapore, 1984. [Google Scholar]
- Teijeiro, T.; García, C.A.; Castro, D.; Félix, P. Abductive reasoning as the basis to reproduce expert criteria in ECG Atrial Fibrillation identification. Physiol. Meas. 2018, 39, 084006. [Google Scholar] [CrossRef]
- Kropf, M.; Hayn, D.; Schreier, G. ECG Classification Based on Time and Frequency Domain Features Using Random Forests. Comput. Cardiol. 2017, 44. [Google Scholar] [CrossRef]
- Datta, S.; Chetanya, P.; Mukherjee, A.; Banerjee, R.; Choudhury, A.D.; Singh, R.; Ukil, A.; Bandyopadhyay, S.; Pal, A.; Khandelwal, S. Identifying Normal, AF and other Abnormal ECG Rhythms using a Cascaded Binary Classifier. Comput. Cardiol. 2017, 44. [Google Scholar] [CrossRef]
- Plesinger, F.; Nejedly, P.; Viscor, I.; Halamek, J.; Jurak, P. Parallel use of a convolutional neural network and bagged tree ensemble for the classification of Holter ECG. Physiol. Meas. 2018, 39, 094002. [Google Scholar] [CrossRef]
- Langley, P.; Dewhurst, M.; Di Marco, L.Y.; Adams, P.; Dewhurst, F.; Mwita, J.C.; Walker, R.; Murray, A. Accuracy of algorithms for detection of atrial fibrillation from short duration beat interval recordings. Med. Eng. Phys. 2012, 34, 1441–1447. [Google Scholar] [CrossRef] [PubMed]
- Saeed, M. Fractals analysis of cardiac arrhythmias. Sci. World J. 2005, 5, 691–701. [Google Scholar] [CrossRef] [PubMed]
- Stridh, M.; Sörnmo, L. Spatiotemporal QRST cancellation techniques for analysis of atrial fibrillation. IEEE Trans. Biomed. Eng. 2001, 48, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Senhaji, L.; Wang, F.; Hernandez, A.I.; Carrault, G. Wavelets Extrema Representation for QRS-T Cancellation and P Wave Detection. Comput. Cardiol. 2002, 29, 37–40. [Google Scholar]
- Rieta, J.J.; Castells, F.; Sánchez, C.; Zarzoso, V.; Millet, J. Atrial activity extraction for atrial fibrillation analysis using blind source separation. IEEE Trans. Biomed. Eng. 2004, 51, 1176–1186. [Google Scholar] [CrossRef] [PubMed]
- Alcaraz, R.; Rieta, J.J. Adaptive singular value cancelation of ventricular activity in single-lead atrial fibrillation electrocardiograms. Physiol. Meas. 2008, 29, 1351–1369. [Google Scholar] [CrossRef] [PubMed]
- Dewland, T.A.; Vittinghoff, E.; Mandyam, M.C.; Heckbert, A.R.; Siscovick, D.S.; Stein, P.K.; Psaty, B.M.; Sotoodehnia, N.; Gottdiener, J.S.; Marcus, G.M. Atrial ectopy as a predictor of incident atrial fibrillation: A cohort study. Ann. Intern. Med. 2013, 159, 721–728. [Google Scholar] [CrossRef] [PubMed]
- Lee, V.; Hemingway, H.; Harb, R.; Crake, T.; Lambiase, P. The prognostic significance of premature ventricular complexes in adults without clinically apparent heart disease: A meta-analysis and systematic review. Heart 2012, 98, 1290–1298. [Google Scholar] [CrossRef] [PubMed]
| Feature | F | Description |
|---|---|---|
| Features extracted from the RR series | ||
| bigem | 910 | Bigemy rhythm index |
| trigem | 440 | Trigemy rhythm index |
| RRmean | 250 | Mean value of the RR intervals |
| RRmax | 168 | Max value of the RR intervals |
| RRbrady | 125 | Mean value of the RR intervals above the threshold of 1.2 s |
| RRtachy | 659 | Mean value of the RR intervals below the threshold of 0.7 s |
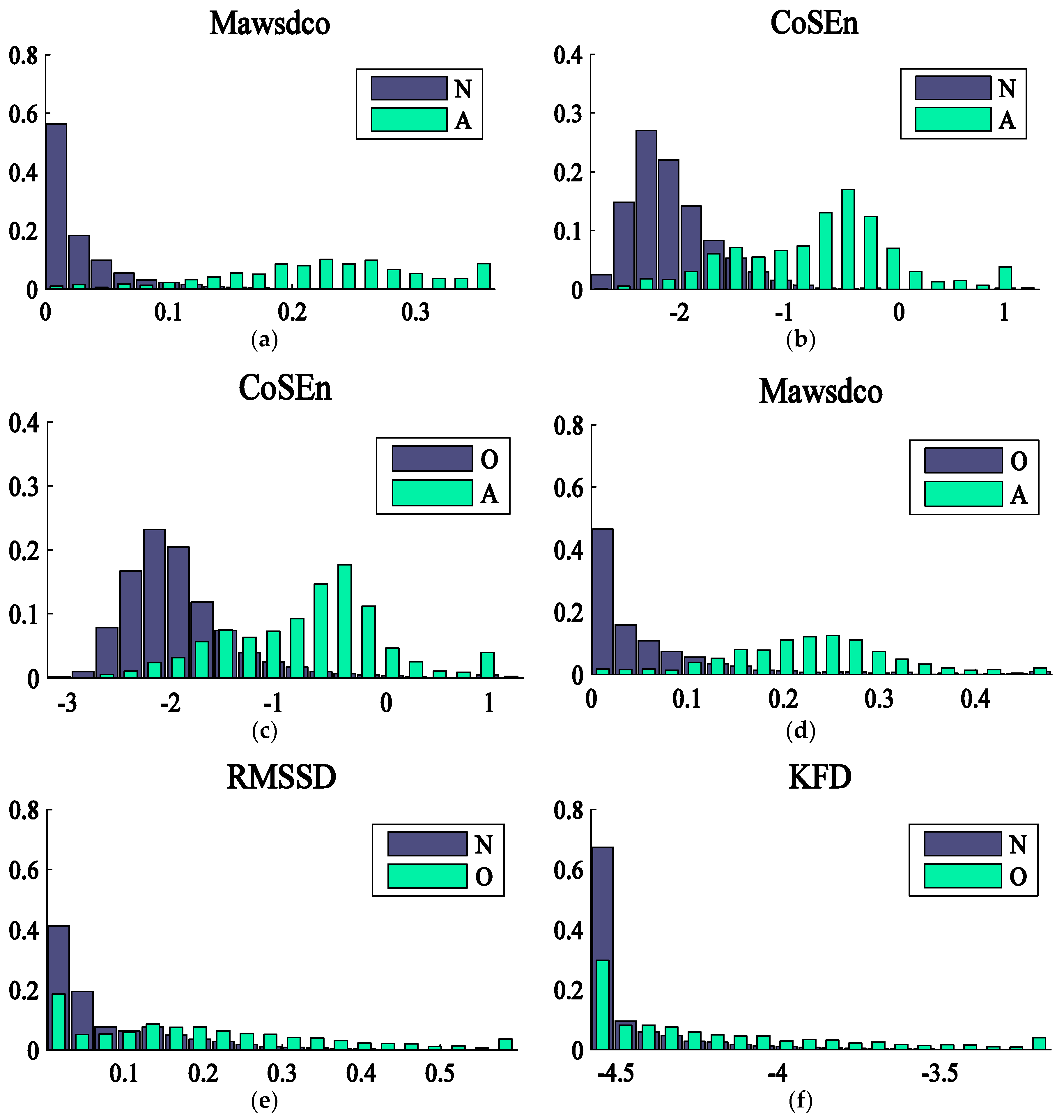
| CoSEn | 1913 | Coefficient of sample entropy |
| Mawsdo | 1771 | Mean of the absolute weighted successive difference with weight optimization |
| Mawsdco | 2114 | Mean of the absolute weighted successive difference controlling for premature beats with compensatory pause (weight optimization) |
| KFD | 993 | Katz Fractal Dimension |
| Frequency Spectrum Analysis (FSA) | ||
| mPdAF | 357 | Power around the peak of the weighted spectral density in the 3–20 Hz band. The power of the three harmonics was considered |
| mPdAFr | 449 | Ratio between the power of the peak and the power in the around |
| P wave analysis (PWA) | ||
| P_numr | 88 | Index of P wave existence based on the comparison between a moving average of 0.06 s width and a moving average of 0.12 s width |
| Pexist | 67 | Index of P wave existence obtained comparing a moving average of 0.06 s width with the output of a comb filter consisting of two moving average of 0.04 s width, delayed by 0.04 s |
| PintNr | 240 | Index of P wave existence obtained calculating the power peaks within intervals before the QRS onset and computing the mode of the distribution of the positions of such peaks |
| Other features extracted from the ECG and the QRS complex | ||
| pRhf | 62 | Power of the signal obtained as the difference between the original and the low-pass filtered (30 Hz) signal (index of noise) |
| QRSwidthMeanQ | 1101 | Trimmed mean (2% tail discard) of the QRS width values |
| QRSwidthStdQ | 545 | Trimmed standard deviation (2% tail discard) of the QRS width values |
| QRSwidthMax3Q | 619 | Mean of the three max values of the QRS width |
| QRSwidthMaxQs | 292 | Max value of the moving average on 3 values of the QRS width |
| QRSppAmplMea | 44 | Trimmed mean (2% tail discard) of the QRS peak-peak amplitude |
| QRSpprAmplMea | 311 | Ratio between the QRSppAmplMea and the peak-peak amplitude of the signal outside the QRST interval |
| QRSpolMea | 26 | Trimmed mean (3% tail discard) of the QRS polarity computed as the sum of the maximum and the minimum of the QRS values |
| QRSwidthMin | 183 | Min value of the QRS equivalent widths, defined as the QRS absolute area divided by the peak-peak amplitude |
| QRSwidthNmea | 508 | Trimmed mean (discarding 40% of high values) of the QRS equivalent widths |
| PnWideQRS | 149 | Ratio between the number of QRS with equivalent width >1.25 × QRSwidthNmea |
| morphDiffr | 550 | Mean of the sum of absolute differences between the QRSs and the QRS mean |
| QRSsadV | 19 | Sum of the absolute differences between the two first right-singular-vectors |
| nPVCp | 353 | Fraction of the premature beats that are followed by a compensatory pause |
| AtypBeatPrk | 450 | Ratio between the relative QRS width and the beat prematurity |
| n | a | o | p | |
|---|---|---|---|---|
| N | 4923 | 16 | 129 | 8 |
| A | 23 | 635 | 95 | 5 |
| O | 459 | 74 | 1875 | 6 |
| P | 58 | 7 | 3 | 108 |
| Reference | Number of Features | Classification Method | Overall Performance |
|---|---|---|---|
| Teijeiro et al. [40] | 42 | Gradient Boosting classifier and Recurrent Neural Network | F1 = 0.848 |
| Kropf et al. [41] | 380 | Random Forest | F1 = 0.832 |
| Billeci et al. (proposed method) | 30 | Least Square-Support Vector Machine | F1 = 0.830 |
| Datta et al. [42] | 150 | Multi-layer Cascaded Binary | F1 = 0.8294 |
| Plesinger [43] | 60 | Neural Netwok + Bagged Tree Ensemble | F1 = 0.8278 |
| n | a | |
|---|---|---|
| N | 5393 | 75 |
| A | 80 | 10,937 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Billeci, L.; Costi, M.; Lombardi, D.; Chiarugi, F.; Varanini, M. Automatic Detection of Atrial Fibrillation and Other Arrhythmias in ECG Recordings Acquired by a Smartphone Device. Electronics 2018, 7, 199. https://doi.org/10.3390/electronics7090199
Billeci L, Costi M, Lombardi D, Chiarugi F, Varanini M. Automatic Detection of Atrial Fibrillation and Other Arrhythmias in ECG Recordings Acquired by a Smartphone Device. Electronics. 2018; 7(9):199. https://doi.org/10.3390/electronics7090199
Chicago/Turabian StyleBilleci, Lucia, Magda Costi, David Lombardi, Franco Chiarugi, and Maurizio Varanini. 2018. "Automatic Detection of Atrial Fibrillation and Other Arrhythmias in ECG Recordings Acquired by a Smartphone Device" Electronics 7, no. 9: 199. https://doi.org/10.3390/electronics7090199
APA StyleBilleci, L., Costi, M., Lombardi, D., Chiarugi, F., & Varanini, M. (2018). Automatic Detection of Atrial Fibrillation and Other Arrhythmias in ECG Recordings Acquired by a Smartphone Device. Electronics, 7(9), 199. https://doi.org/10.3390/electronics7090199







