Evaluating the Performance of DenseNet in ECG Report Automation
Abstract
1. Introduction
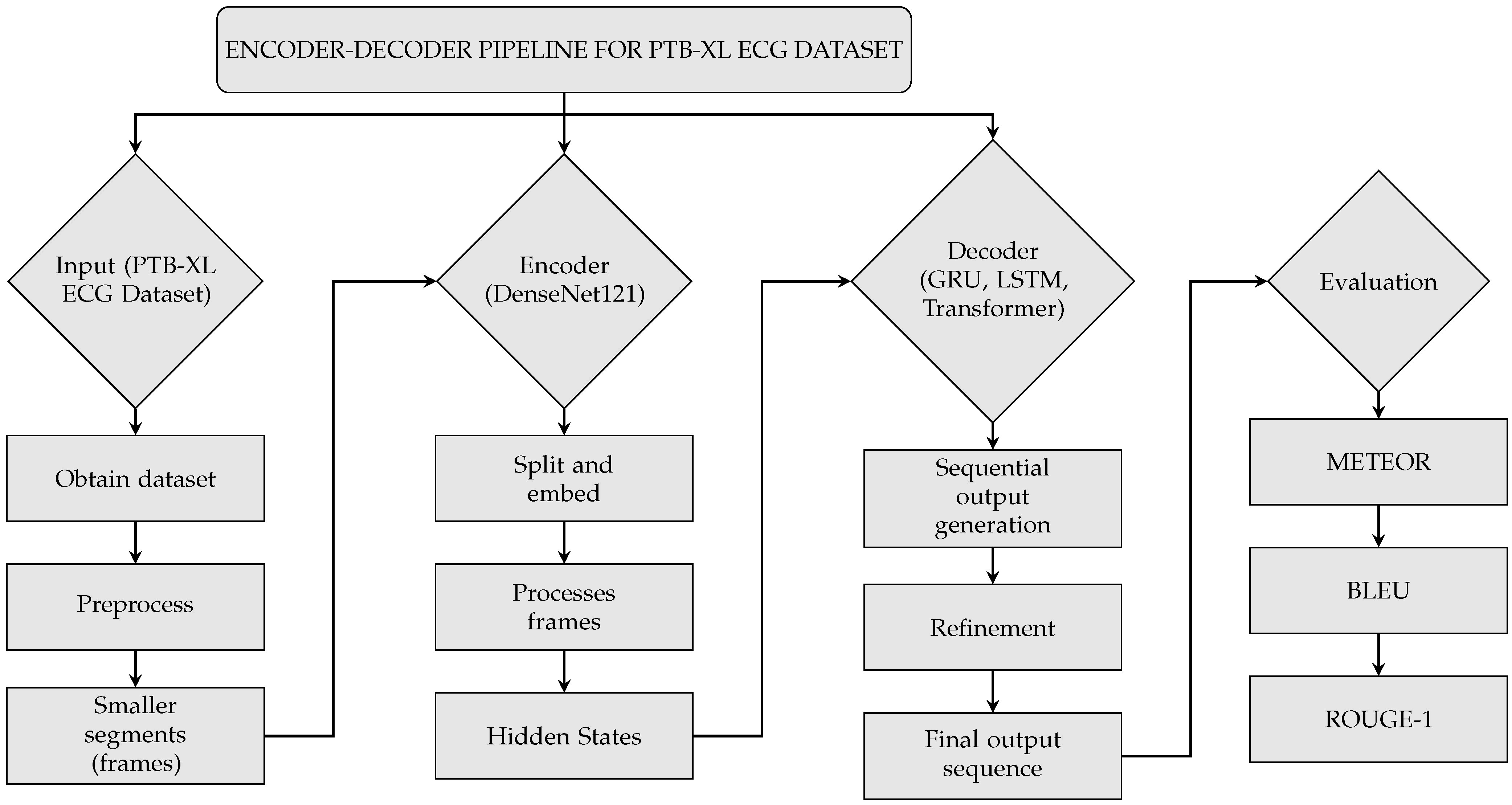
2. Materials and Methods
2.1. Data Preprocessing and Inputting
2.1.1. Dataset
2.1.2. Preprocessing
2.1.3. Noise Reduction
2.1.4. Training, Validation, and Testing
2.2. DenseNet Encoder
2.2.1. Splitting and Embedding
2.2.2. Frames Processing
2.2.3. Hidden States
2.3. Decoders (LSTM, Transformer, GRU)
2.3.1. LSTM Decoder
2.3.2. Transformer Decoder
2.3.3. GRU Decoder
2.4. Evaluation
3. Results
3.1. Cardiovascular Pathologies in PTB-XL Dataset
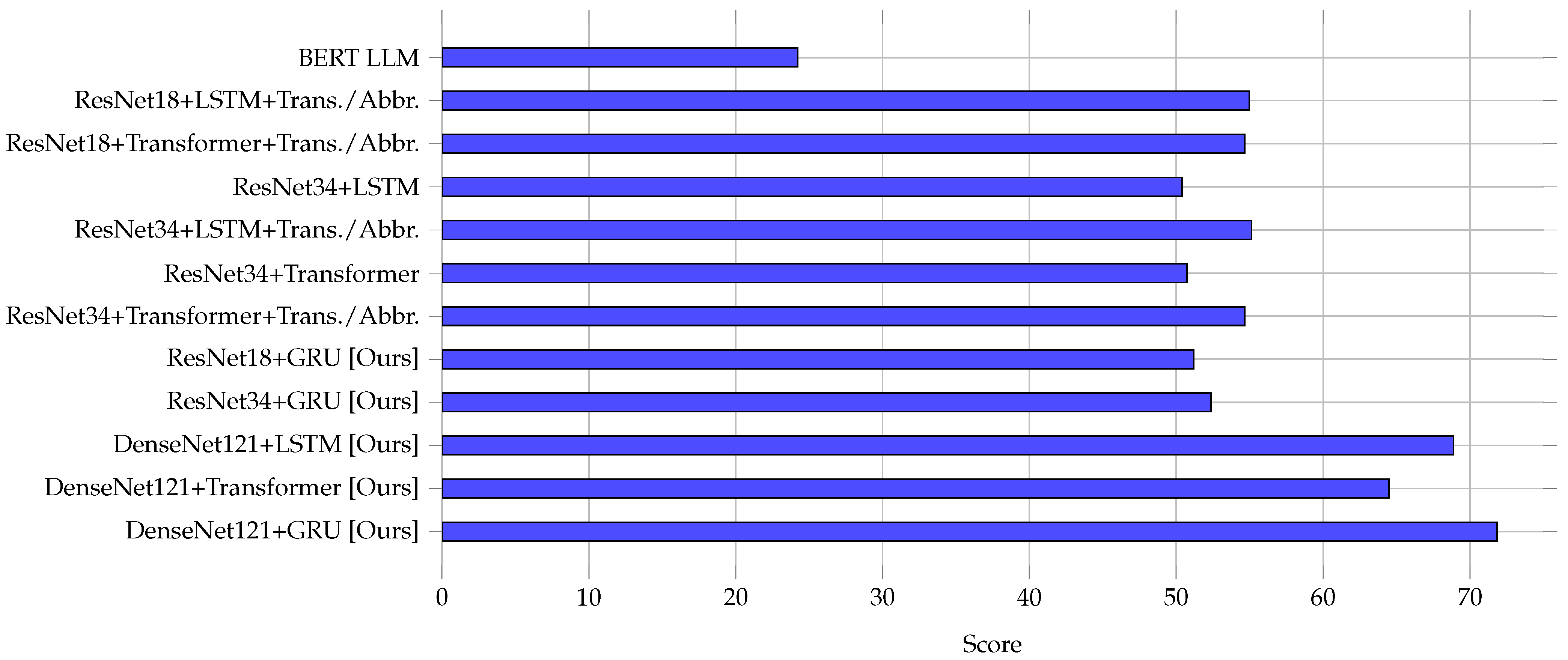
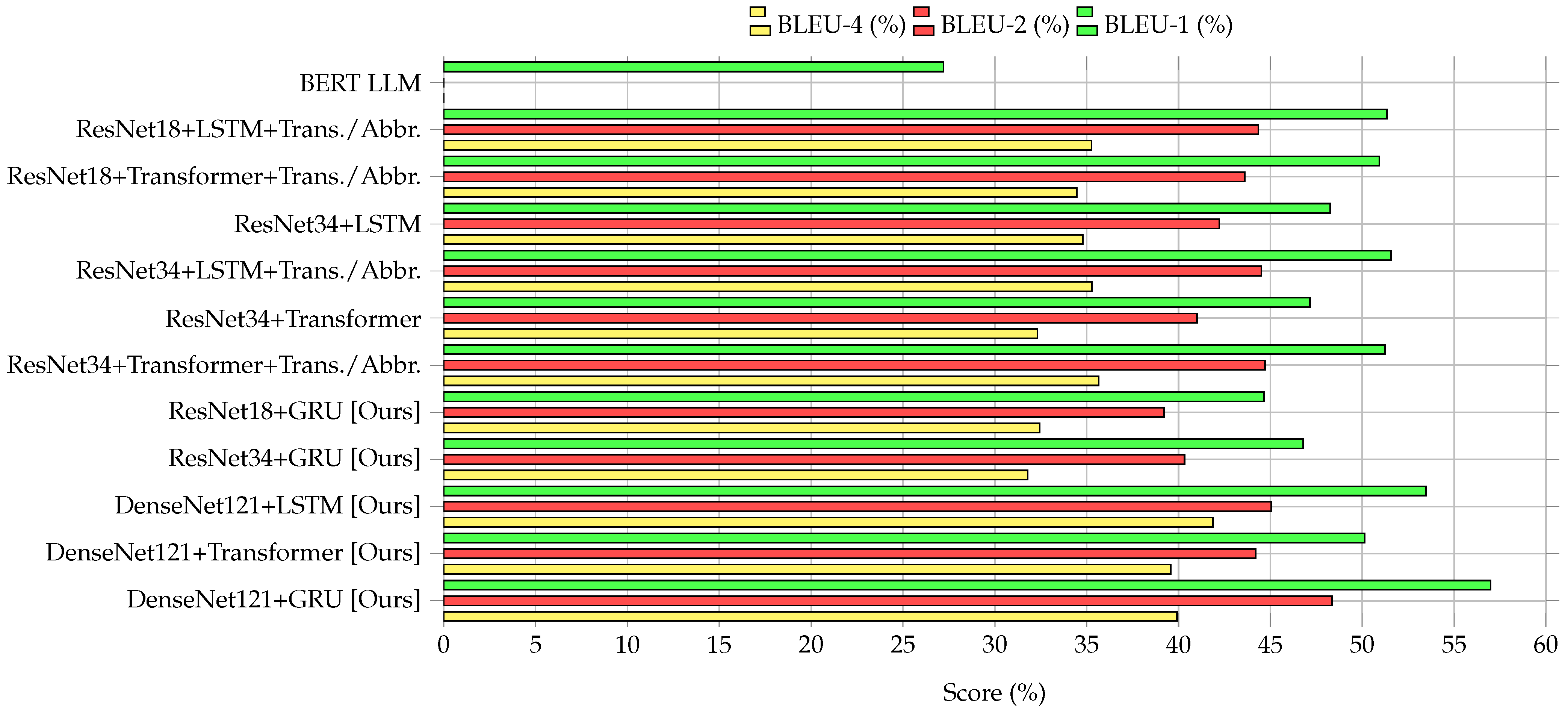
3.2. Performance Comparison of Encoder–Decoder Models
3.3. Visualization of Model Performance
3.4. Comparative Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| AMI | Anterior myocardial infarction |
| AUC | Area under the curve |
| AV | Atrioventricular |
| BERT | Bidirectional Encoder Representations from Transformers |
| BLEU | Bilingual Evaluation Understudy |
| CC BY | Creative Commons Attribution |
| CLBBB | Complete left bundle branch block |
| CNN | Convolutional neural network |
| ECG | Electrocardiogram |
| GRU | Gated Recurrent Unit |
| ILBBB | Incomplete left bundle branch block |
| IMI | Inferior myocardial infarction |
| IRBBB | Incomplete right bundle branch block |
| ISC | Ischemic ST-T-wave changes |
| ISCI | Ischemic changes |
| LAFB | Left anterior fascicular block |
| LMI | Lateral myocardial infarction |
| LLM | Large language model |
| LSTM | Long Short-Term Memory |
| LVH | Left ventricular hypertrophy |
| METEOR | Metric for Evaluation of Translation with Explicit Ordering |
| MI | Myocardial infarction |
| ML | Machine learning |
| NORM | Normal electrocardiogram |
| NST | Nonspecific ST-wave changes |
| PTB-XL | PhysioNet PTB-XL Dataset |
| RAE | Right atrial enlargement |
| RAO | Right atrial overload |
| ResNet | Residual network |
| RNN | Recurrent neural network |
| ROUGE | Recall-Oriented Understudy for Gisting Evaluation |
| STTC | ST-T changes |
| WPW | Wolff–Parkinson–White syndrome |
References
- Khunte, A.; Sangha, V.; Oikonomou, E.K.; Dhingra, L.S.; Aminorroaya, A.; Coppi, A.; Vasisht Shankar, S.; Mortazavi, B.J.; Bhatt, D.L.; Krumholz, H.M.; et al. Automated Diagnostic Reports from Images of Electrocardiograms at the Point-of-Care. medRxiv 2024. [Google Scholar] [CrossRef]
- Bleich, A.; Linnemann, A.; Diem, B.H.; Conrad, T.O. Automated Medical Report Generation for ECG Data: Bridging Medical Text and Signal Processing with Deep Learning. arXiv 2024. [Google Scholar] [CrossRef]
- Nasef, D.; Nasef, D.; Basco, K.J.; Singh, A.; Hartnett, C.; Ruane, M.; Tagliarino, J.; Nizich, M.; Toma, M. Clinical Applicability of Machine Learning Models for Binary and Multi-Class Electrocardiogram Classification. AI 2025, 6, 59. [Google Scholar] [CrossRef]
- Basco, K.J.; Singh, A.; Nasef, D.; Hartnett, C.; Ruane, M.; Tagliarino, J.; Nizich, M.; Toma, M. Electrocardiogram Abnormality Detection Using Machine Learning on Summary Data and Biometric Features. Diagnostics 2025, 15, 903. [Google Scholar] [CrossRef]
- Toma, M.; Husain, G. Algorithm Selection and Data Utilization in Machine Learning for Medical Imaging Classification. In Proceedings of the 2024 IEEE Long Island Systems, Applications and Technology Conference (LISAT), Holtsville, NY, USA, 15 November 2024; IEEE: New York, NY, USA, 2024; pp. 1–6. [Google Scholar] [CrossRef]
- Johnson, L.S.; Zadrozniak, P.; Jasina, G.; Grotek-Cuprjak, A.; Andrade, J.G.; Svennberg, E.; Diederichsen, S.Z.; McIntyre, W.F.; Stavrakis, S.; Benezet-Mazuecos, J.; et al. Artificial intelligence for direct-to-physician reporting of ambulatory electrocardiography. Nat. Med. 2025, 31, 925–931. [Google Scholar] [CrossRef]
- Zhang, C.; Benz, P.; Argaw, D.M.; Lee, S.; Kim, J.; Rameau, F.; Bazin, J.C.; Kweon, I.S. ResNet or DenseNet? Introducing Dense Shortcuts to ResNet. In Proceedings of the 2021 IEEE Winter Conference on Applications of Computer Vision (WACV), Waikoloa, HI, USA, 3–8 January 2021; pp. 3549–3558. [Google Scholar] [CrossRef]
- Hou, Y.; Wu, Z.; Cai, X.; Zhu, T. The application of improved densenet algorithm in accurate image recognition. Sci. Rep. 2024, 14, 8645. [Google Scholar] [CrossRef]
- Qiu, J.; Han, W.; Zhu, J.; Xu, M.; Rosenberg, M.; Liu, E.; Weber, D.; Zhao, D. Transfer Knowledge from Natural Language to Electrocardiography: Can We Detect Cardiovascular Disease Through Language Models? arXiv 2023. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Maaten, L.V.D.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar] [CrossRef]
- Strodthoff, N.; Wagner, P.; Schaeffter, T.; Samek, W. Deep Learning for ECG Analysis: Benchmarks and Insights from PTB-XL. IEEE J. Biomed. Health Inform. 2021, 25, 1519–1528. [Google Scholar] [CrossRef]
- Palczynski, K.; Smigiel, S.; Ledzinski, D.; Bujnowski, S. Study of the Few-Shot Learning for ECG Classification Based on the PTB-XL Dataset. Sensors 2022, 22, 904. [Google Scholar] [CrossRef]
- Laghari, A.A.; Sun, Y.; Alhussein, M.; Aurangzeb, K.; Anwar, M.S.; Rashid, M. Deep residual-dense network based on bidirectional recurrent neural network for atrial fibrillation detection. Sci. Rep. 2023, 13, 15109. [Google Scholar] [CrossRef]
- Zhu, F.; Ye, F.; Fu, Y.; Liu, Q.; Shen, B. Electrocardiogram generation with a bidirectional LSTM-CNN generative adversarial network. Sci. Rep. 2019, 9, 6734. [Google Scholar] [CrossRef] [PubMed]
- Wagner, P.; Strodthoff, N.; Bousseljot, R.D.; Kreiseler, D.; Lunze, F.I.; Samek, W.; Schaeffter, T. PTB-XL, a large publicly available electrocardiography dataset. Sci. Data 2020, 7, 154. [Google Scholar] [CrossRef] [PubMed]
- Zorarpacı, E.; Özel, S.A. Privacy preserving classification over differentially private data. WIREs Data Min. Knowl. Discov. 2020, 11, e1399. [Google Scholar] [CrossRef]
- Yu, H.; Guo, P.; Sano, A. Zero-Shot ECG Diagnosis with Large Language Models and Retrieval-Augmented Generation. In Proceedings of the 3rd Machine Learning for Health Symposium, PMLR, New Orleans, LA, USA, 10 December 2023; Proceedings of Machine Learning Research. Hegselmann, S., Parziale, A., Shanmugam, D., Tang, S., Asiedu, M.N., Chang, S., Hartvigsen, T., Singh, H., Eds.; Volume 225, pp. 650–663. Available online: https://proceedings.mlr.press/v225/yu23b/yu23b.pdf (accessed on 14 April 2025).
- Tian, Y.; Li, Z.; Jin, Y.; Wang, M.; Wei, X.; Zhao, L.; Liu, Y.; Liu, J.; Liu, C. Foundation model of ECG diagnosis: Diagnostics and explanations of any form and rhythm on ECG. Cell Rep. Med. 2024, 5, 101875. [Google Scholar] [CrossRef]
- Irshad, M.S.; Masood, T.; Jaffar, A.; Rashid, M.; Akram, S.; Aljohani, A. Deep Learning-Based ECG Classification for Arterial Fibrillation Detection. Comput. Mater. Contin. 2024, 79, 4805–4824. [Google Scholar] [CrossRef]
- Babu, P.P.S.; Brindha, T. Deep Learning Fusion for Intracranial Hemorrhage Classification in Brain CT Imaging. Int. J. Adv. Comput. Sci. Appl. 2024, 15, 884–894. [Google Scholar] [CrossRef]
- Somani, S.; Russak, A.J.; Richter, F.; Zhao, S.; Vaid, A.; Chaudhry, F.; De Freitas, J.K.; Naik, N.; Miotto, R.; Nadkarni, G.N.; et al. Deep learning and the electrocardiogram: Review of the current state-of-the-art. EP Eur. 2021, 23, 1179–1191. [Google Scholar] [CrossRef]
- Habler, E.; Shabtai, A. Using LSTM encoder-decoder algorithm for detecting anomalous ADS-B messages. Comput. Secur. 2018, 78, 155–173. [Google Scholar] [CrossRef]
- Malhotra, P.; Ramakrishnan, A.; Anand, G.; Vig, L.; Agarwal, P.; Shroff, G. LSTM-based Encoder-Decoder for Multi-sensor Anomaly Detection. arXiv 2016. [Google Scholar] [CrossRef]
- Libovicky, J.; Helcl, J.; Marecek, D. Input Combination Strategies for Multi-Source Transformer Decoder. In Proceedings of the Third Conference on Machine Translation: Research Papers. Association for Computational Linguistics, Brussels, Belgium, 31 October–1 November 2018. [Google Scholar] [CrossRef]
- Li, Y.; Cai, W.; Gao, Y.; Li, C.; Hu, X. More than Encoder: Introducing Transformer Decoder to Upsample. In Proceedings of the 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Las Vegas, NV, USA, 6–8 December 2022; IEEE: New York, NY, USA, 2022; pp. 1597–1602. [Google Scholar] [CrossRef]
- Cho, K.; van Merrienboer, B.; Gulcehre, C.; Bahdanau, D.; Bougares, F.; Schwenk, H.; Bengio, Y. Learning Phrase Representations using RNN Encoder–Decoder for Statistical Machine Translation. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), Doha, Qatar, 25–29 October 2014; Association for Computational Linguistics: Stroudsburg, PA, USA, 2014. [Google Scholar] [CrossRef]
- Lynn, H.M.; Pan, S.B.; Kim, P. A Deep Bidirectional GRU Network Model for Biometric Electrocardiogram Classification Based on Recurrent Neural Networks. IEEE Access 2019, 7, 145395–145405. [Google Scholar] [CrossRef]
- van Halteren, H.; Teufel, S. Examining the consensus between human summaries: Initial experiments with factoid analysis. In Proceedings of the HLT-NAACL 03 Text Summarization Workshop, Stroudsburg, PA, USA, 31 May 2003; pp. 57–64. Available online: https://aclanthology.org/W03-0508/ (accessed on 14 April 2025).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30June 2016; IEEE: New York, NY, USA, 2016. [Google Scholar] [CrossRef]
- Lavie, A.; Denkowski, M.J. The Meteor metric for automatic evaluation of machine translation. Mach. Transl. 2009, 23, 105–115. [Google Scholar] [CrossRef]
- Papineni, K.; Roukos, S.; Ward, T.; Zhu, W.J. BLEU: A method for automatic evaluation of machine translation. In Proceedings of the 40th Annual Meeting on Association for Computational Linguistics—ACL ’02, Philadelphia, PA, USA, 6–12 July 2002; Association for Computational Linguistics: Stroudsburg, PA, USA, 2002. ACL ’02. p. 311. [Google Scholar] [CrossRef]
- Wolk, K.; Marasek, K. Enhanced Bilingual Evaluation Understudy. Lecture Notes on Information Theory 2014, 2, 191–197. [Google Scholar] [CrossRef]
- Liu, F.; Liu, Y. Exploring Correlation Between ROUGE and Human Evaluation on Meeting Summaries. IEEE Trans. Audio, Speech Lang. Process. 2010, 18, 187–196. [Google Scholar] [CrossRef]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Minneapolis, MN, USA, 2–7 June 2019; Association for Computational Linguistics: Stroudsburg, PA, USA, 2019. [Google Scholar] [CrossRef]
- Tandale, S.B.; Stoffel, M. Recurrent and convolutional neural networks in structural dynamics: A modified attention steered encoder–decoder architecture versus LSTM versus GRU versus TCN topologies to predict the response of shock wave-loaded plates. Comput. Mech. 2023, 72, 765–786. [Google Scholar] [CrossRef]
- Mateus, B.C.; Mendes, M.; Farinha, J.T.; Assis, R.; Cardoso, A.M. Comparing LSTM and GRU Models to Predict the Condition of a Pulp Paper Press. Energies 2021, 14, 6958. [Google Scholar] [CrossRef]
- Zeyer, A.; Bahar, P.; Irie, K.; Schluter, R.; Ney, H. A Comparison of Transformer and LSTM Encoder Decoder Models for ASR. In Proceedings of the 2019 IEEE Automatic Speech Recognition and Understanding Workshop (ASRU), Singapore, 14–18 December 2019; IEEE: New York, NY, USA, 2019; pp. 8–15. [Google Scholar] [CrossRef]
- Gao, Y.; Glowacka, D. Deep Gate Recurrent Neural Network. In Proceedings of the 8th Asian Conference on Machine Learning, Hamilton, New Zealand, 16–18 November 2016; Proceedings of Machine Learning Research. Durrant, R.J., Kim, K.E., Eds.; The University of Waikato: Hamilton, New Zealand, 2016; Volume 63, pp. 350–365. Available online: http://proceedings.mlr.press/v63/gao30.pdf (accessed on 14 April 2025).

| Cardiovascular Pathology | Class ID in Dataset | Instances | Description |
|---|---|---|---|
| Normal ECG | NORM | 7185 | Normal (non-pathologic) |
| ST-T changes | STTC | 1713 | T-wave abnormalities (non-diagnostic) |
| Anterior MI | AMI | 1636 | Anterior myocardial infarction |
| Inferior MI | IMI | 1272 | Inferior myocardial infarction |
| Fascicular block | LAFB/LPFB | 881 | Left anterior and posterior fascicular block |
| Incomplete RBBB | IRBBB | 798 | Incomplete right bundle branch block |
| LV hypertrophy | LVH | 733 | Left ventricular hypertrophy |
| Complete LBBB | CLBBB | 527 | Complete left bundle branch block |
| Nonspecific ST | NST_ | 478 | Nonspecific ST-wave changes (non-diagnostic) |
| Ischemic ST-T | ISC | 297 | Ischemic ST-T-wave changes |
| AV block | _AVB | 204 | First-degree, second-degree, and third-degree AV block |
| Ischemic changes | ISCI | 147 | Changes in inferior and inferolateral leads |
| WPW syndrome | WPW | 67 | Wolff–Parkinson–White syndrome |
| Atrial overload | LAO/LAE | 49 | Left atrial overload, left atrial enlargement |
| Incomplete LBBB | ILBBB | 44 | Incomplete left bundle branch block |
| Right atrial overload | RAO/RAE | 33 | Right atrial overload, right atrial enlargement |
| Lateral MI | LMI | 28 | Lateral myocardial infarction |
| Model | METEOR | BLEU-1 | BLEU-2 | BLEU-4 | ROUGE-1 | ||
|---|---|---|---|---|---|---|---|
| P | R | F | |||||
| BERT LLM [9] | 24.51 | 27.21 | – | – | 26.12 | 35.71 | 29.56 |
| ResNet18+LSTM+Trans./Abbr. [2] | 55.3 | 51.4 | 44.39 | 35.37 | 62.4 | 59.57 | 58.33 |
| ResNet18+Transformer+Trans./Abbr. [2] | 55.01 | 50.95 | 43.7 | 34.53 | 63.47 ✗ | 59.39 | 58.82 |
| ResNet34+LSTM [2] | 50.72 | 48.25 | 42.29 | 34.9 | 55.53 | 54.93 | 52.29 |
| ResNet34+LSTM+Trans./Abbr. [2] | 55.53 | 51.63 | 44.54 | 35.29 | 61.47 | 60.65 | 58.33 |
| ResNet34+Transformer [2] | 51.11 | 47.21 | 41.06 | 32.39 | 57.64 | 52.04 | 53.87 |
| ResNet34+Transformer+Trans./Abbr. [2] | 55 | 51.41 | 44.74 | 35.69 | 63.22 | 59.58 ✗ | 58.12 |
| ResNet18+GRU [Ours] | 51.48 | 44.63 | 39.27 | 32.56 | 50.26 | 47.99 | 49.09 |
| ResNet34+GRU [Ours] | 52.68 | 46.91 | 40.42 | 31.86 | 54.3 | 53.57 | 53.93 |
| DenseNet121+LSTM [Ours] | 69.2 ✗ | 53.51 ✗ | 45.09 ✗ | 41.91 * | 61.74 | 58.02 | 59.82 ✗ |
| DenseNet121+Transformer [Ours] | 64.82 | 50.17 | 44.29 | 39.56 | 56.28 | 57.01 | 56.64 |
| DenseNet121+GRU [Ours] | 72.19 * | 57.08 * | 48.33 * | 40.02 ✗ | 66.23 * | 62.54 * | 64.33 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Husain, G.; Siddiqua, A.; Toma, M. Evaluating the Performance of DenseNet in ECG Report Automation. Electronics 2025, 14, 1837. https://doi.org/10.3390/electronics14091837
Husain G, Siddiqua A, Toma M. Evaluating the Performance of DenseNet in ECG Report Automation. Electronics. 2025; 14(9):1837. https://doi.org/10.3390/electronics14091837
Chicago/Turabian StyleHusain, Gazi, Ayesha Siddiqua, and Milan Toma. 2025. "Evaluating the Performance of DenseNet in ECG Report Automation" Electronics 14, no. 9: 1837. https://doi.org/10.3390/electronics14091837
APA StyleHusain, G., Siddiqua, A., & Toma, M. (2025). Evaluating the Performance of DenseNet in ECG Report Automation. Electronics, 14(9), 1837. https://doi.org/10.3390/electronics14091837









