Efficient Compression of Red Blood Cell Image Dataset Using Joint Deep Learning-Based Pattern Classification and Data Compression
Abstract
1. Introduction
2. Materials and Methods
2.1. Autoencoders
2.1.1. Sparse Autoencoders
2.1.2. Convolutional Autoencoders
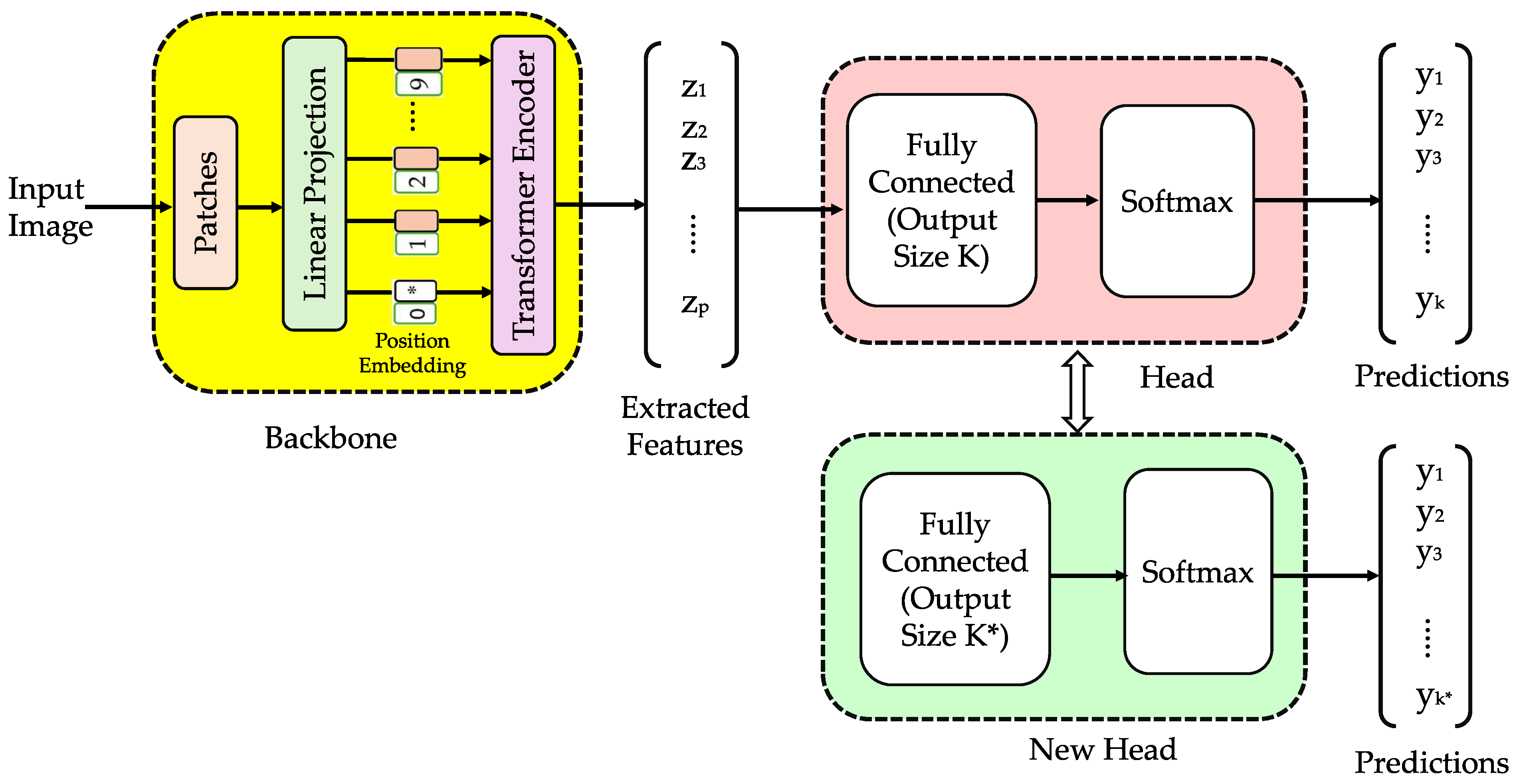
2.2. Vision Transformer
2.3. Development of Deep Learning Models for Image Classification
2.3.1. Stacked Autoencoders for Image Classification
2.3.2. Vision Transformer for Image Classification
2.4. Lossy Image Compression
2.5. Malaria Datasets
2.5.1. UAB_Dataset
2.5.2. NIH_Dataset
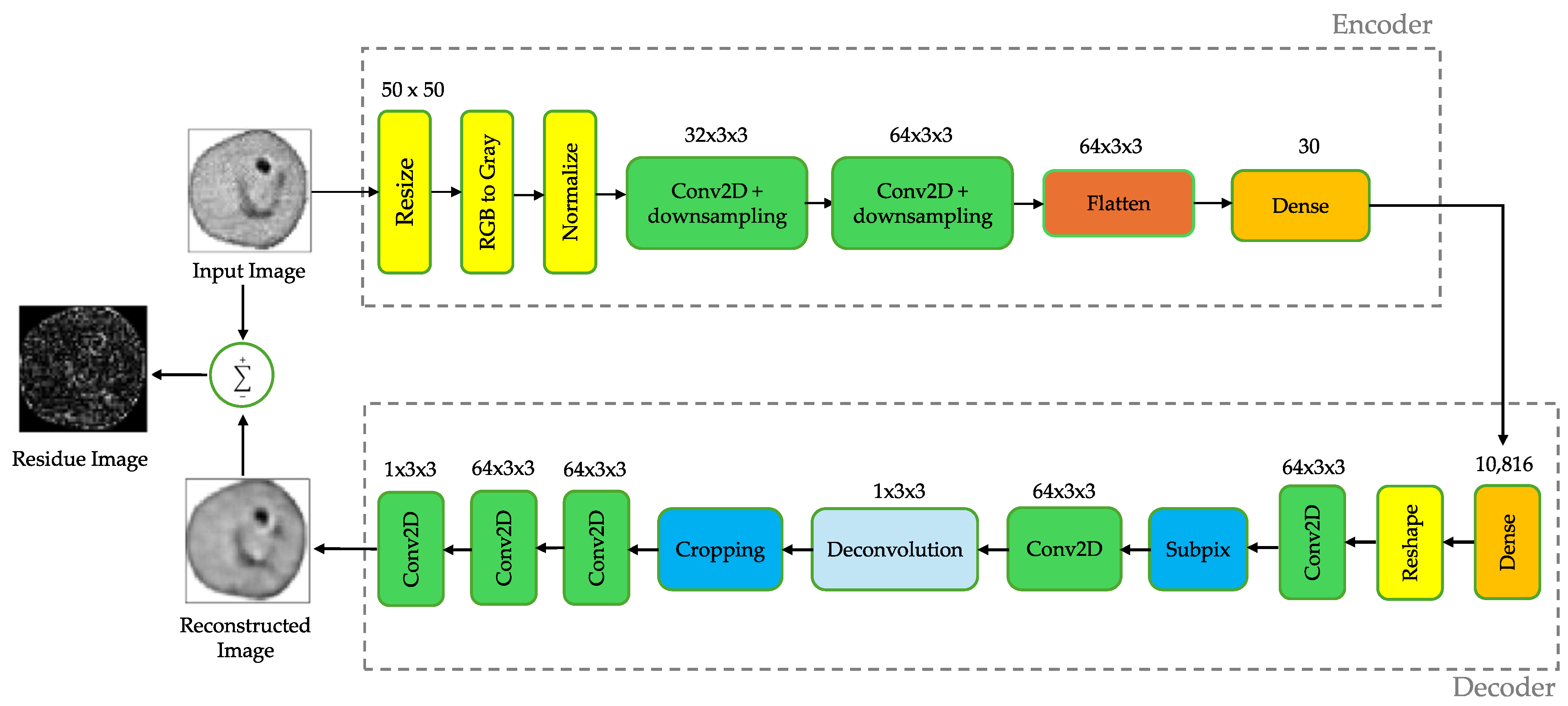
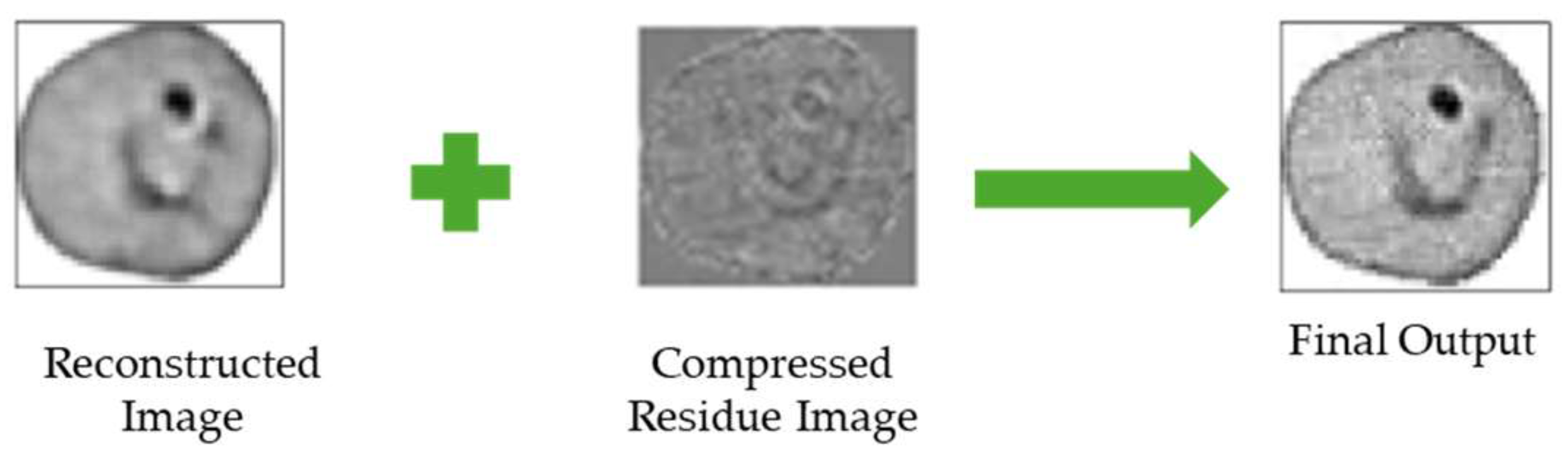
2.6. Proposed Lossy Compression Architecture
2.7. Proposed Joint Classification and Compression Approaches
2.7.1. Hybrid Model (Vision Transformer for Classification and Autoencoder for Compression)
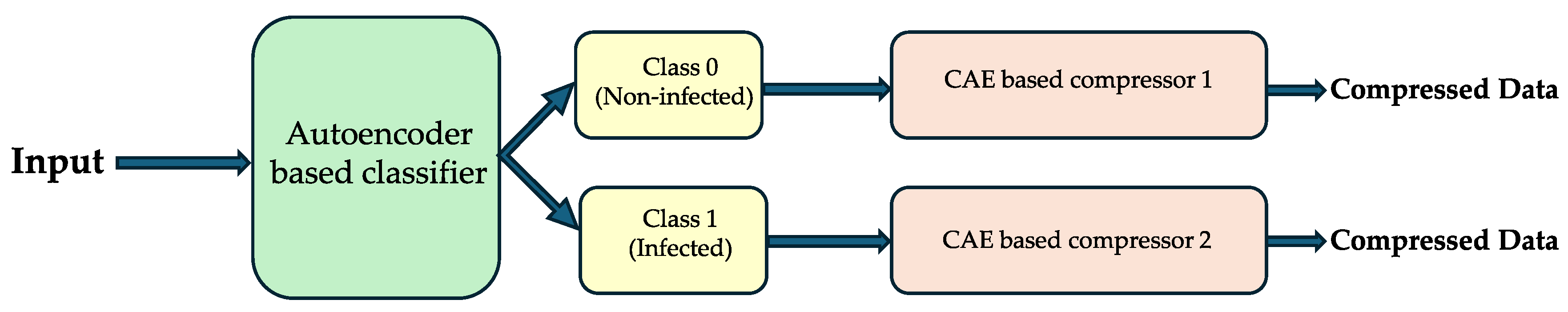
2.7.2. Dual-Purpose Autoencoder Model (Autoencoders for Combined Classification and Compression)
3. Results
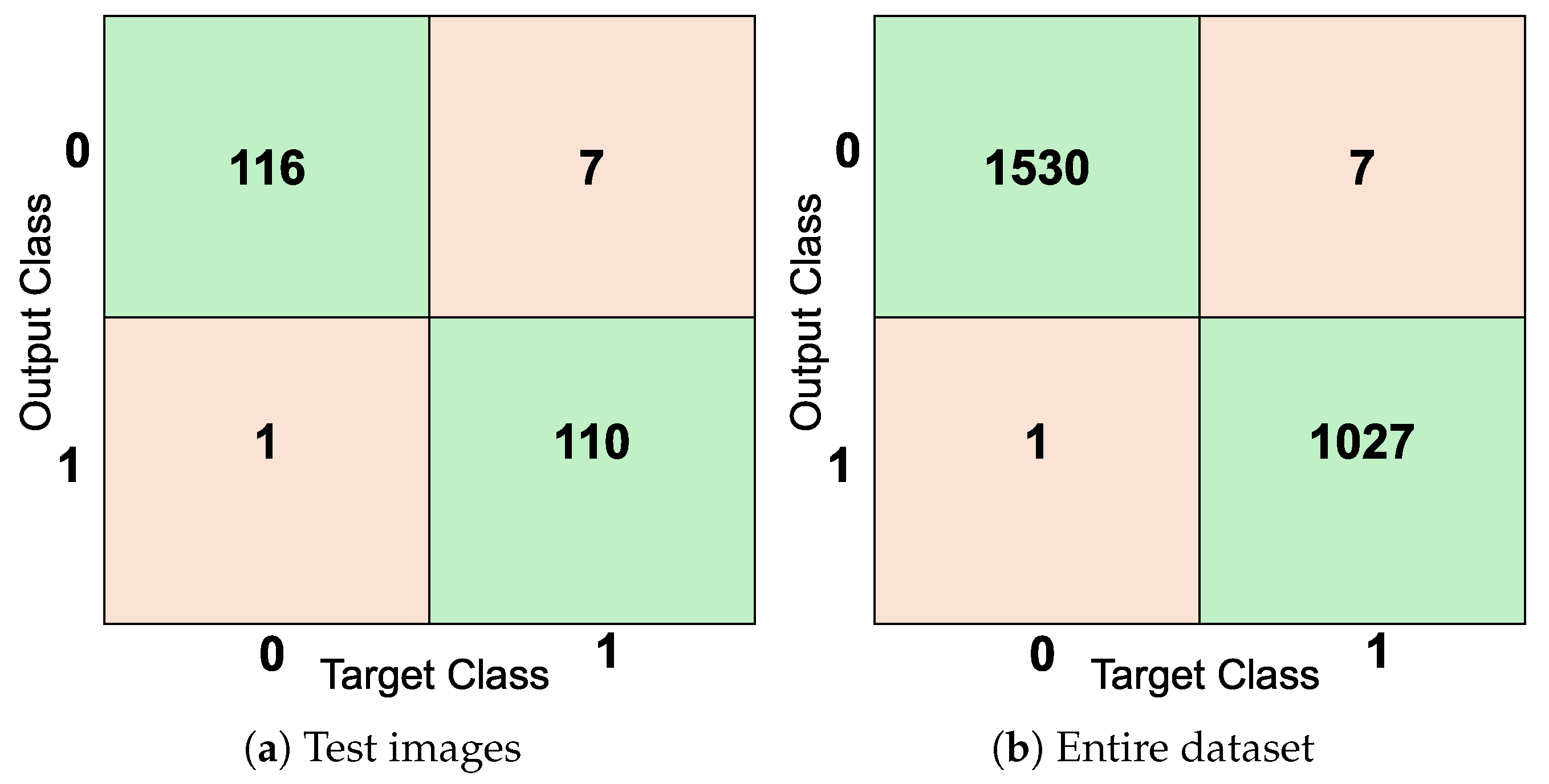
3.1. Evaluation of Classification Performance on UAB_Dataset
3.1.1. Autoencoder Results
3.1.2. Vision Transformer Results
3.2. Evaluation of Classification Performance on NIH_Dataset
3.2.1. Autoencoder Results
3.2.2. Vision Transformer Results
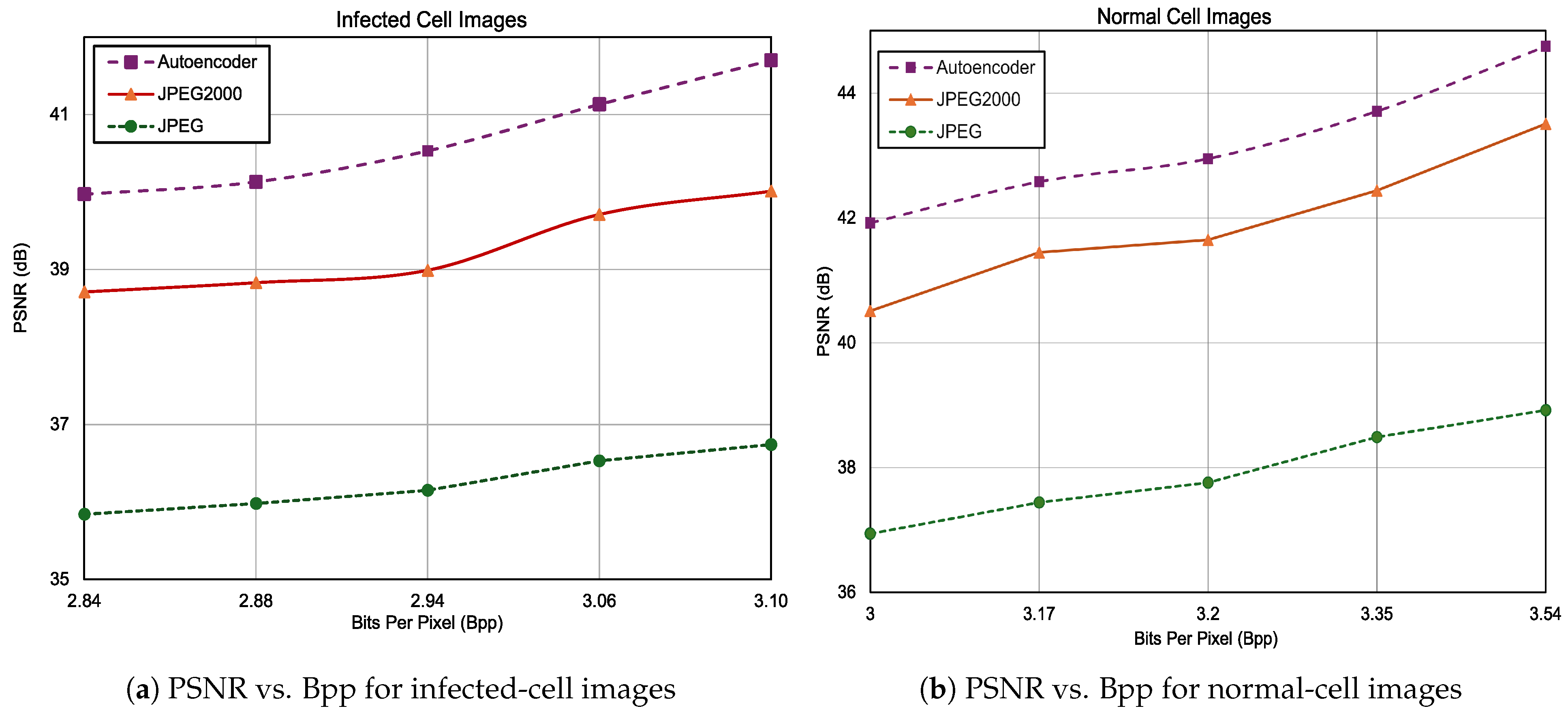
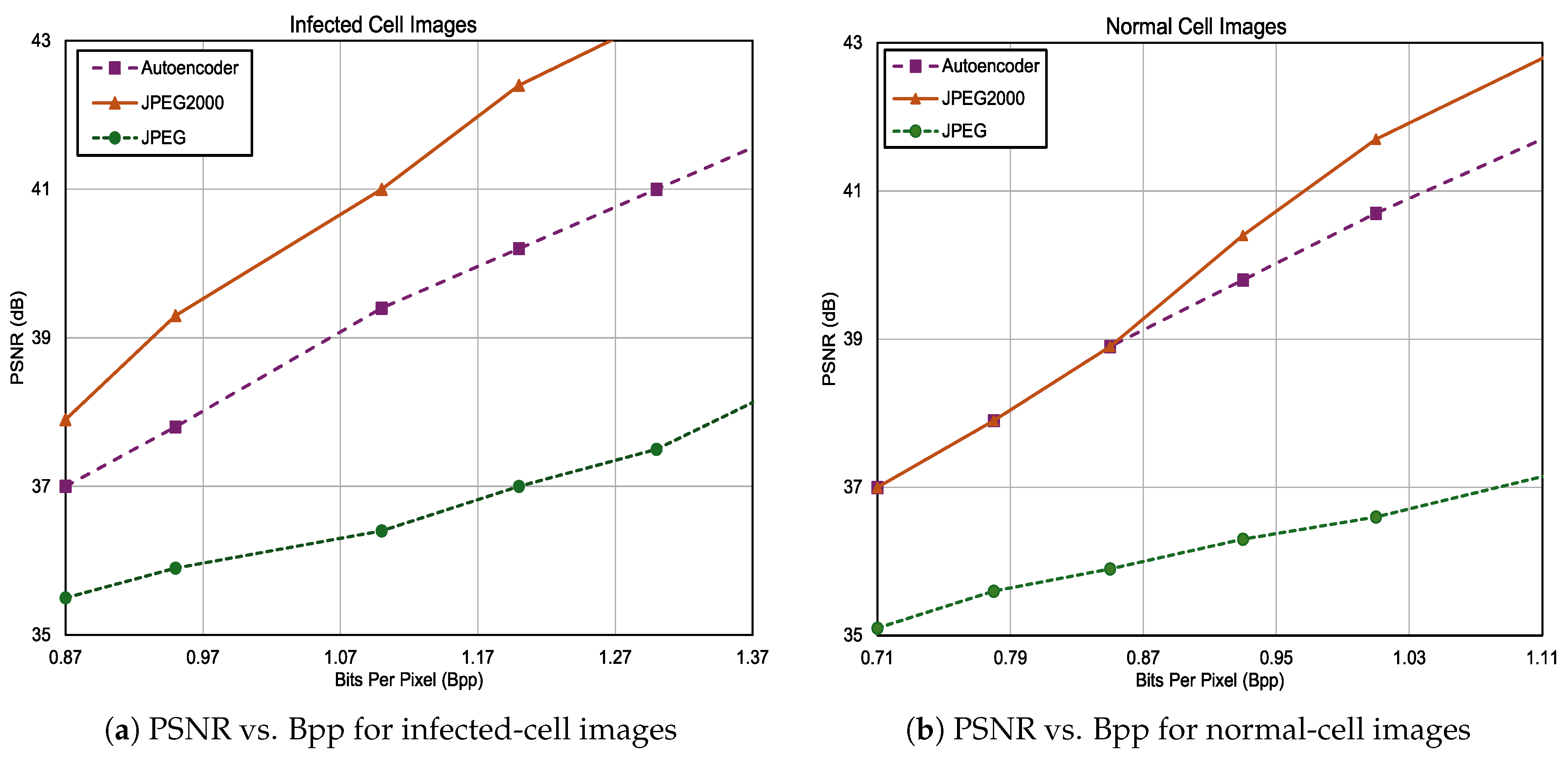
3.3. Compression Results on Precisely Labeled Data
3.3.1. UAB_Dataset
3.3.2. NIH_Dataset
4. Discussion
Quantitative Analysis: How Classification Accuracy Influences Compression
5. Limitations and Future Research Directions
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Centers for Disease Control and Prevention. Malaria’s Impact Worldwide; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2024.
- World Health Organization. World Malaria Report 2023; World Health Organization: Geneva, Switzerland, 2023. [Google Scholar]
- Bronzan, R.N.; McMorrow, M.L.; Patrick Kachur, S. Diagnosis of malaria: Challenges for clinicians in endemic and non-endemic regions. Mol. Diagn. Ther. 2008, 12, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Delahunt, C.B.; Gachuhi, N.; Horning, M.P. Metrics to guide development of machine learning algorithms for malaria diagnosis. Front. Malar. 2024, 2, 1250220. [Google Scholar] [CrossRef]
- Hoyos, K.; Hoyos, W. Supporting Malaria Diagnosis Using Deep Learning and Data Augmentation. Diagnostics 2024, 14, 690. [Google Scholar] [CrossRef]
- Ikerionwu, C.; Ugwuishiwu, C.; Okpala, I.; James, I.; Okoronkwo, M.; Nnadi, C.; Orji, U.; Ebem, D.; Ike, A. Application of machine and deep learning algorithms in optical microscopic detection of Plasmodium: A malaria diagnostic tool for the future. Photodiagn. Photodyn. Ther. 2022, 40, 103198. [Google Scholar] [CrossRef]
- Siłka, W.; Wieczorek, M.; Siłka, J.; Woźniak, M. Malaria detection using advanced deep learning architecture. Sensors 2023, 23, 1501. [Google Scholar] [CrossRef]
- Marques, G.; Ferreras, A.; de la Torre-Diez, I. An ensemble-based approach for automated medical diagnosis of malaria using EfficientNet. Multimed. Tools Appl. 2022, 81, 28061–28078. [Google Scholar] [CrossRef]
- Loh, D.R.; Yong, W.X.; Yapeter, J.; Subburaj, K.; Chandramohanadas, R. A deep learning approach to the screening of malaria infection: Automated and rapid cell counting, object detection and instance segmentation using Mask R-CNN. Comput. Med. Imaging Graph. 2021, 88, 101845. [Google Scholar] [CrossRef] [PubMed]
- Alkhaldi, T.M.; Hashim, A.N. Automatic Detection of Malaria Using Convolutional Neural Network. Math. Stat. Eng. Appl. 2022, 71, 939–947. [Google Scholar]
- Li, S.; Du, Z.; Meng, X.; Zhang, Y. Multi-stage malaria parasite recognition by deep learning. GigaScience 2021, 10, giab040. [Google Scholar] [CrossRef]
- Islam, M.S.B.; Islam, J.; Islam, M.S.; Sumon, M.S.I.; Nahiduzzaman, M.; Murugappan, M.; Hasan, A.; Chowdhury, M.E. Development of Low Cost, Automated Digital Microscopes Allowing Rapid Whole Slide Imaging for Detecting Malaria. In Surveillance, Prevention, and Control of Infectious Diseases: An AI Perspective; Springer: Berlin/Heidelberg, Germany, 2024; pp. 73–96. [Google Scholar]
- Saxena, S.; Sanyal, P.; Bajpai, M.; Prakash, R.; Kumar, S. Trials and tribulations: Developing an artificial intelligence for screening malaria parasite from peripheral blood smears. Med J. Armed Forces India, 2023; in press. [Google Scholar]
- Pan, W.D.; Dong, Y.; Wu, D. Classification of malaria-infected cells using deep convolutional neural networks. In Machine Learning: Advanced Techniques and Emerging Applications; IntechOpen: London, UK, 2018; Volume 159. [Google Scholar]
- Valente, J.; António, J.; Mora, C.; Jardim, S. Developments in image processing using deep learning and reinforcement learning. J. Imaging 2023, 9, 207. [Google Scholar] [CrossRef]
- Mishra, D.; Singh, S.K.; Singh, R.K. Deep architectures for image compression: A critical review. Signal Process. 2022, 191, 108346. [Google Scholar] [CrossRef]
- Yasin, H.M.; Abdulazeez, A.M. Image compression based on deep learning: A review. Asian J. Res. Comput. Sci. 2021, 8, 62–76. [Google Scholar] [CrossRef]
- Bourai, N.E.H.; Merouani, H.F.; Djebbar, A. Deep learning-assisted medical image compression challenges and opportunities: Systematic review. Neural Comput. Appl. 2024, 36, 10067–10108. [Google Scholar] [CrossRef]
- Dimililer, K. DCT-based medical image compression using machine learning. Signal Image Video Process. 2022, 16, 55–62. [Google Scholar] [CrossRef]
- Abd-Alzhra, A.S.; Al-Tamimi, M.S. Image compression using deep learning: Methods and techniques. Iraqi J. Sci. 2022, 63, 1299–1312. [Google Scholar] [CrossRef]
- Yang, E.H.; Amer, H.; Jiang, Y. Compression helps deep learning in image classification. Entropy 2021, 23, 881. [Google Scholar] [CrossRef]
- Silva, J.M.; Almeida, J.R. Enhancing metagenomic classification with compression-based features. Artif. Intell. Med. 2024, 156, 102948. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.I.; Liang, C.C.; Hu, P.F. Iterative Gaussian–Laplacian Pyramid Network for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2024, 62, 5510122. [Google Scholar] [CrossRef]
- Barsi, A.; Nayak, S.C.; Parida, S.; Shukla, R.M. A deep learning-based compression and classification technique for whole slide histopathology images. Int. J. Inf. Technol. 2024, 16, 4517–4526. [Google Scholar] [CrossRef]
- Hamano, G.; Imaizumi, S.; Kiya, H. Effects of JPEG Compression on Vision Transformer Image Classification for Encryption-then-Compression Images. Sensors 2023, 23, 3400. [Google Scholar] [CrossRef]
- Liu, L.; Chen, T.; Liu, H.; Pu, S.; Wang, L.; Shen, Q. 2C-Net: Integrate image compression and classification via deep neural network. Multimed. Syst. 2023, 29, 945–959. [Google Scholar]
- Hurwitz, J.; Nicholas, C.; Raff, E. Neural Normalized Compression Distance and the Disconnect Between Compression and Classification. arXiv 2024, arXiv:2410.15280. [Google Scholar]
- Xie, W.; Fan, X.; Zhang, X.; Li, Y.; Sheng, M.; Fang, L. Co-compression via superior gene for remote sensing scene classification. IEEE Trans. Geosci. Remote Sens. 2023, 61, 5604112. [Google Scholar] [CrossRef]
- Dong, Y.; Pan, W.D.; Wu, D. Impact of misclassification rates on compression efficiency of red blood cell images of malaria infection using deep learning. Entropy 2019, 21, 1062. [Google Scholar] [CrossRef]
- Pintelas, E.; Livieris, I.E.; Pintelas, P.E. A Convolutional Autoencoder Topology for Classification in High-Dimensional Noisy Image Datasets. Sensors 2021, 21, 7731. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Sun, H.; Takeuchi, M.; Katto, J. Deep Convolutional AutoEncoder-based Lossy Image Compression. In Proceedings of the 2018 Picture Coding Symposium (PCS), Francisco, CA, USA, 24–27 June 2018; pp. 253–257. [Google Scholar] [CrossRef]
- Ismail, A.R.; Zulhazmi Rafiqi Azhary, M.; Zaharin Noor Azwan, N.A.; Ismail, A.; Alsaiari, N.A. Performance Evaluation of Medical Image Denoising using Convolutional Autoencoders. In Proceedings of the 2024 3rd International Conference on Creative Communication and Innovative Technology (ICCIT), Tangerang, Indonesia, 7–8 August 2024; pp. 1–6. [Google Scholar] [CrossRef]
- Han, K.; Wang, Y.; Chen, H.; Chen, X.; Guo, J.; Liu, Z.; Tang, Y.; Xiao, A.; Xu, C.; Xu, Y.; et al. A Survey on Vision Transformer. IEEE Trans. Pattern Anal. Mach. Intell. 2023, 45, 87–110. [Google Scholar] [CrossRef]
- Omer, A.A.M. Image Classification Based on Vision Transformer. J. Comput. Commun. 2024, 12, 49–59. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2021, arXiv:2010.11929. [Google Scholar]
- Carion, N.; Massa, F.; Synnaeve, G.; Usunier, N.; Kirillov, A.; Zagoruyko, S. End-to-End Object Detection with Transformers. In Computer Vision—ECCV 2020; Springer: Cham, Switzerland, 2020; pp. 213–229. [Google Scholar] [CrossRef]
- Shivappriya, S.N.; Harikumar, R. Performance Analysis of Deep Neural Network and Stacked Autoencoder for Image Classification. In Computational Intelligence and Sustainable Systems: Intelligence and Sustainable Computing; Anandakumar, H., Arulmurugan, R., Onn, C.C., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 1–16. [Google Scholar] [CrossRef]
- Elngar, A.A.; Arafa, M.; Fathy, A.; Moustafa, B.; Mahmoud, O.; Shaban, M.; Fawzy, N. Image classification based on CNN: A survey. J. Cybersecur. Inf. Manag. 2021, 6, 18–50. [Google Scholar]
- Zhao, Z.Q.; Zheng, P.; Xu, S.T.; Wu, X. Object Detection With Deep Learning: A Review. IEEE Trans. Neural Networks Learn. Syst. 2019, 30, 3212–3232. [Google Scholar] [CrossRef]
- Maurício, J.; Domingues, I.; Bernardino, J. Comparing Vision Transformers and Convolutional Neural Networks for Image Classification: A Literature Review. Appl. Sci. 2023, 13, 5521. [Google Scholar] [CrossRef]
- Fraihat, S.; Al-Betar, M.A. A novel lossy image compression algorithm using multi-models stacked AutoEncoders. Array 2023, 19, 100314. [Google Scholar] [CrossRef]
- Gogoi, M.; Begum, S.A. Image Classification Using Deep Autoencoders. In Proceedings of the 2017 IEEE International Conference on Computational Intelligence and Computing Research (ICCIC), Coimbatore, India, 14–16 December 2017; pp. 1–5. [Google Scholar] [CrossRef]
- Rao, K.R.; Domínguez, H.O. JPEG Series; CRC Press: Boca Raton, FL, USA, 2022. [Google Scholar]
- Sayood, K. Introduction to Data Compression; Morgan Kaufmann: Burlington, MA, USA, 2017. [Google Scholar]
- Jamil, S.; Piran, M.J.; MuhibUrRahman. Learning-Driven Lossy Image Compression; A Comprehensive Survey. arXiv 2022, arXiv:2201.09240. [Google Scholar]
- Valenzise, G.; Purica, A.; Hulusic, V.; Cagnazzo, M. Quality Assessment of Deep-Learning-Based Image Compression. In Proceedings of the 2018 IEEE 20th International Workshop on Multimedia Signal Processing (MMSP), Vancouver, BC, Canada, 29–31 August 2018; pp. 1–6. [Google Scholar] [CrossRef]
- Mishra, D.; Singh, S.K.; Singh, R.K. Lossy medical image compression using residual learning-based dual autoencoder model. In Proceedings of the 2020 IEEE 7th Uttar Pradesh Section International Conference on Electrical, Electronics and Computer Engineering (UPCON), Prayagraj, India, 27–29 November 2020; pp. 1–5. [Google Scholar]
- Dong, Y.; Jiang, Z.; Shen, H.; David Pan, W.; Williams, L.A.; Reddy, V.V.B.; Benjamin, W.H.; Bryan, A.W. Evaluations of deep convolutional neural networks for automatic identification of malaria infected cells. In Proceedings of the 2017 IEEE EMBS International Conference on Biomedical & Health Informatics (BHI), Orland, FL, USA, 16–19 February 2017; pp. 101–104. [Google Scholar] [CrossRef]
- Dataset Used in This Paper. Available online: http://www.ece.uah.edu/~dwpan/malaria_dataset/ (accessed on 3 January 2025).
- National Library of Medicine. Malaria Datasheet—Image Processing Research. 2025. Available online: https://lhncbc.nlm.nih.gov/LHC-research/LHC-projects/image-processing/malaria-datasheet.html (accessed on 2 March 2025).
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. arXiv 2015, arXiv:1502.03167. [Google Scholar]
- Theis, L.; Shi, W.; Cunningham, A.; Huszár, F. Lossy Image Compression with Compressive Autoencoders. arXiv 2017, arXiv:1703.00395. [Google Scholar]
- MathWorks. Train Vision Transformer Network for Image Classification. Available online: https://www.mathworks.com/help/deeplearning/ug/train-vision-transformer-network-for-image-classification.html (accessed on 31 January 2023).
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016; Available online: http://www.deeplearningbook.org (accessed on 3 January 2025).





| Dataset | Method | Accuracy | BPP | PSNR (dB) |
|---|---|---|---|---|
| UAB_Dataset | CAE-based compressor | 100% | 2.90 | 41.35 |
| Dual-purpose autoencoder | 99.6% | 2.91 | 41.20 | |
| Hybrid model | 96.18% | 2.92 | 41.20 | |
| JPEG2000 | – | 3.10 | 41.09 | |
| JPEG | – | 3.97 | 40.73 | |
| NIH_Dataset | CAE-based compressor | 100% | 0.85 | 38.9 |
| Dual-purpose autoencoder | 98.1% | 0.87 | 38.5 | |
| Hybrid model | 94.95% | 0.87 | 38 | |
| JPEG2000 | – | 0.85 | 38.9 | |
| JPEG | – | 1.35 | 38.5 |
| Dataset | Method | Accuracy | BPP | PSNR |
|---|---|---|---|---|
| UAB_Dataset | CAE-based compressor | 100% | 2.90 | 40.58 |
| Dual-purpose autoencoder | 99.6% | 2.92 | 40.56 | |
| Hybrid model | 96.18% | 2.92 | 40.47 | |
| JPEG2000 | – | 3.10 | 40.01 | |
| JPEG | – | 4.00 | 39.82 | |
| NIH_Dataset | CAE-based compressor | 100% | 0.95 | 37.8 |
| Dual-purpose autoencoder | 98.1% | 0.95 | 37 | |
| Hybrid model | 94.95% | 0.96 | 36.87 | |
| JPEG2000 | – | 0.87 | 37.9 | |
| JPEG | – | 1.2 | 37 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nusrat, Z.; Mahmud, M.F.; Pan, W.D. Efficient Compression of Red Blood Cell Image Dataset Using Joint Deep Learning-Based Pattern Classification and Data Compression. Electronics 2025, 14, 1556. https://doi.org/10.3390/electronics14081556
Nusrat Z, Mahmud MF, Pan WD. Efficient Compression of Red Blood Cell Image Dataset Using Joint Deep Learning-Based Pattern Classification and Data Compression. Electronics. 2025; 14(8):1556. https://doi.org/10.3390/electronics14081556
Chicago/Turabian StyleNusrat, Zerin, Md Firoz Mahmud, and W. David Pan. 2025. "Efficient Compression of Red Blood Cell Image Dataset Using Joint Deep Learning-Based Pattern Classification and Data Compression" Electronics 14, no. 8: 1556. https://doi.org/10.3390/electronics14081556
APA StyleNusrat, Z., Mahmud, M. F., & Pan, W. D. (2025). Efficient Compression of Red Blood Cell Image Dataset Using Joint Deep Learning-Based Pattern Classification and Data Compression. Electronics, 14(8), 1556. https://doi.org/10.3390/electronics14081556









