1. Introduction
Medical image segmentation is a fundamental task in medical image analysis, aiming to accurately delineate regions of interest, such as lesions or anatomical structures, from medical images [
1]. The quality of segmentation directly influences the reliability of subsequent clinical decisions, including disease diagnosis, prognosis evaluation, and treatment planning. Although manual segmentation is considered the gold standard due to its high accuracy, it is time-consuming, labor-intensive, and highly dependent on expert knowledge, thereby limiting its scalability and consistency in clinical practice [
2]. To address these limitations, automated medical image segmentation has attracted considerable research attention, aiming to improve segmentation efficiency while maintaining or enhancing accuracy [
3].
In recent years, deep learning, particularly convolutional neural networks (CNNs), has achieved remarkable success in medical image segmentation. Notably, UNet [
4] stands out as a representative model, owing to its strong generalization and feature extraction capabilities enabled by multiscale feature propagation and aggregation. Based on UNet, several improved variants have been proposed to further improve the model’s performance in medical image segmentation. For example, UNet++ [
5] introduces nested skip pathways, which reduce the semantic gap between the feature extraction and reconstruction stages and enhance multiscale feature aggregation, thereby improving segmentation performance. Similarly, UNetv2 [
6] introduces semantic information fusion module that leverages skip connections to achieve bidirectional aggregation of semantic and detailed features, significantly enhancing feature representation. Building on these foundations, UDTransNet [
7] improves skip connections with attention-based recalibration, effectively narrowing the semantic gap. Unlike UDTransNet, UTANet [
8] adopts task-adaptive skip connection strategy, flexibly selecting the information flow between the downsampling and upsampling paths to further improve medical image segmentation performance.
More recently, Mamba [
9,
10,
11] and its variants have demonstrated impressive performance in vision tasks by modeling long-range dependencies using selective state space models (SSMs). Inspired by this ability, a series of Mamba-based architectures have been developed to address key challenges in medical image segmentation, including limited data, complex lesion regions or anatomical structures, and the need for efficient global context modeling. For example, UMamba [
12] integrates Mamba modules with convolutional layers in a hybrid framework, using Mamba for global context modeling and CNNs for feature aggregation, achieving competitive segmentation results. In contrast, VMUNet employs Mamba modules for both feature extraction and aggregation, forming a fully SSM-based architecture with higher efficiency. Building on VMUNet, Swin-UMamba [
13] utilizes ImageNet-pretrained weights for transfer learning in medical image segmentation, while H-vmunet [
14] introduces a hierarchical channel-wise interaction mechanism within the Mamba module to suppress redundant information and improve feature representation purity.
These approaches above have demonstrated strong performance across diverse medical image segmentation tasks by learning rich representations from complex inputs. Despite architectural differences, their success fundamentally hinges on two key capabilities: extracting discriminative features that capture local details and/or global context and aggregating these features to produce accurate and coherent segmentation results.
- (a)
Feature Extraction: Feature extraction refers to the process of automatically generating informative representations from raw input data, with the goal of converting the original information into a compact and task-relevant form. In medical image segmentation, CNN-based methods extract multiscale features through stacked convolutions and downsampling, effectively capturing local structures, including edges and textures [
4]. However, due to their limited receptive fields and strong inductive biases, CNNs struggle to model long-range dependencies and global semantics [
3,
14,
15]. In contrast, Mamba-based methods leverage SSMs to capture global information [
10,
16], such as complete object contours, spatial relationships among multiple objects, and scene-level contextual patterns. However, their reliance on flattening 2D images into 1D sequences disrupts spatial continuity and weakens the ability to capture fine-grained local structures [
17]. Consequently, both CNN- and Mamba-based methods face an inherent trade-off: CNNs retain spatial locality but struggle with global context, while Mamba captures global information but compromises local detail.
- (b)
Feature Aggregation: Feature aggregation combines features from multiple levels or scales to produce richer representations, improving model performance [
4]. CNN- and Mamba-based models typically use skip connections to fuse high-resolution shallow features with deep semantic features, mitigating fine-grained information loss caused by downsampling [
8]. However, most aggregation strategies rely on simple operations, such as addition or concatenation with fixed weights, which do not adapt to varying inter-layer content [
18]. As a result, these methods inadequately capture commonalities and subtle differences across layers, compromising feature stability and representation quality. The problem is further amplified by semantic gaps and redundant noise in features from different layers, particularly under challenging conditions such as blurred boundaries, complex structures, or small lesions [
8]. Therefore, inflexible aggregation limits the model’s ability to capture fine structural details, highlighting the need for adaptive methods that explicitly account for inter-layer similarities and fine-grained variations to enhance both stability and boundary sensitivity.
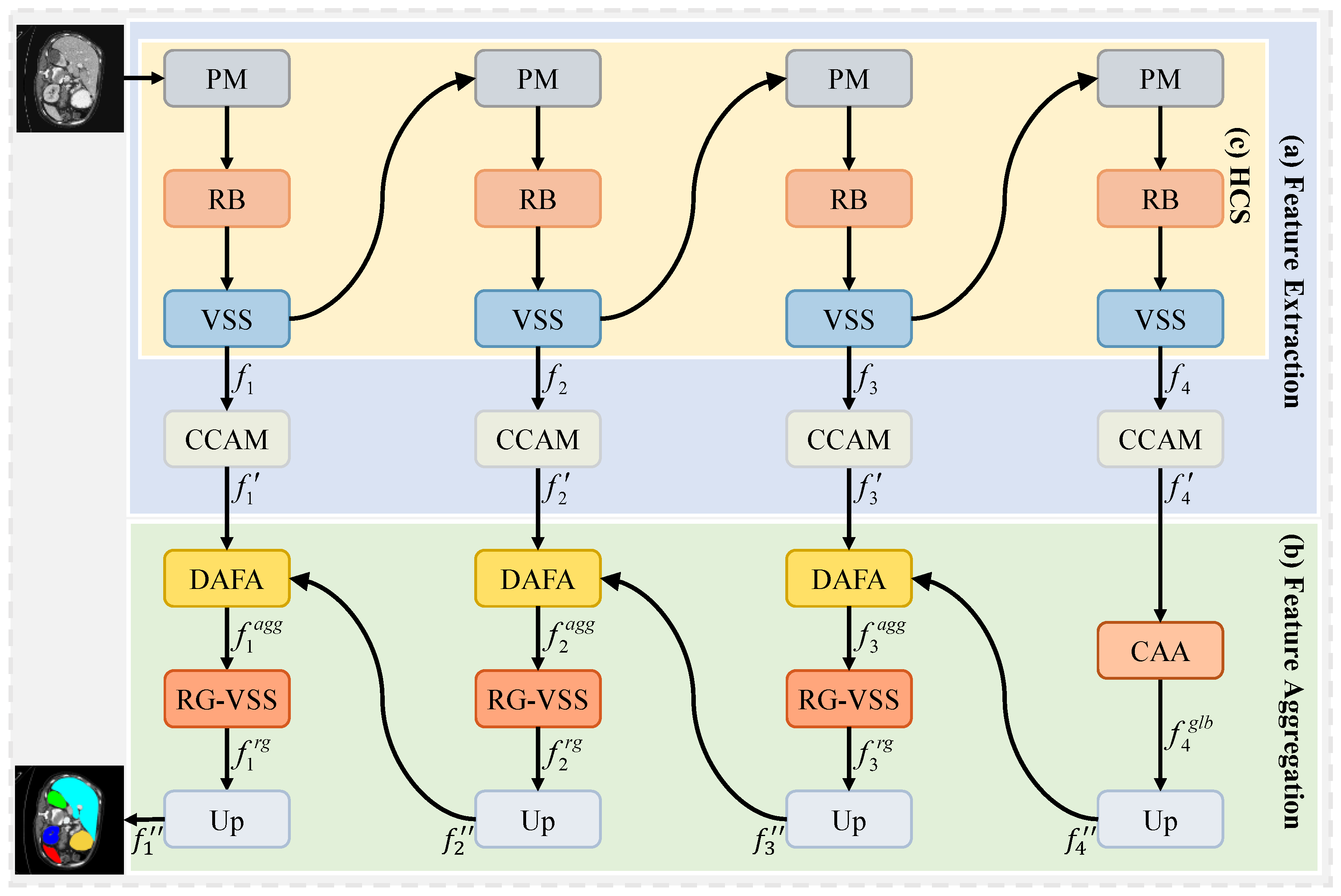
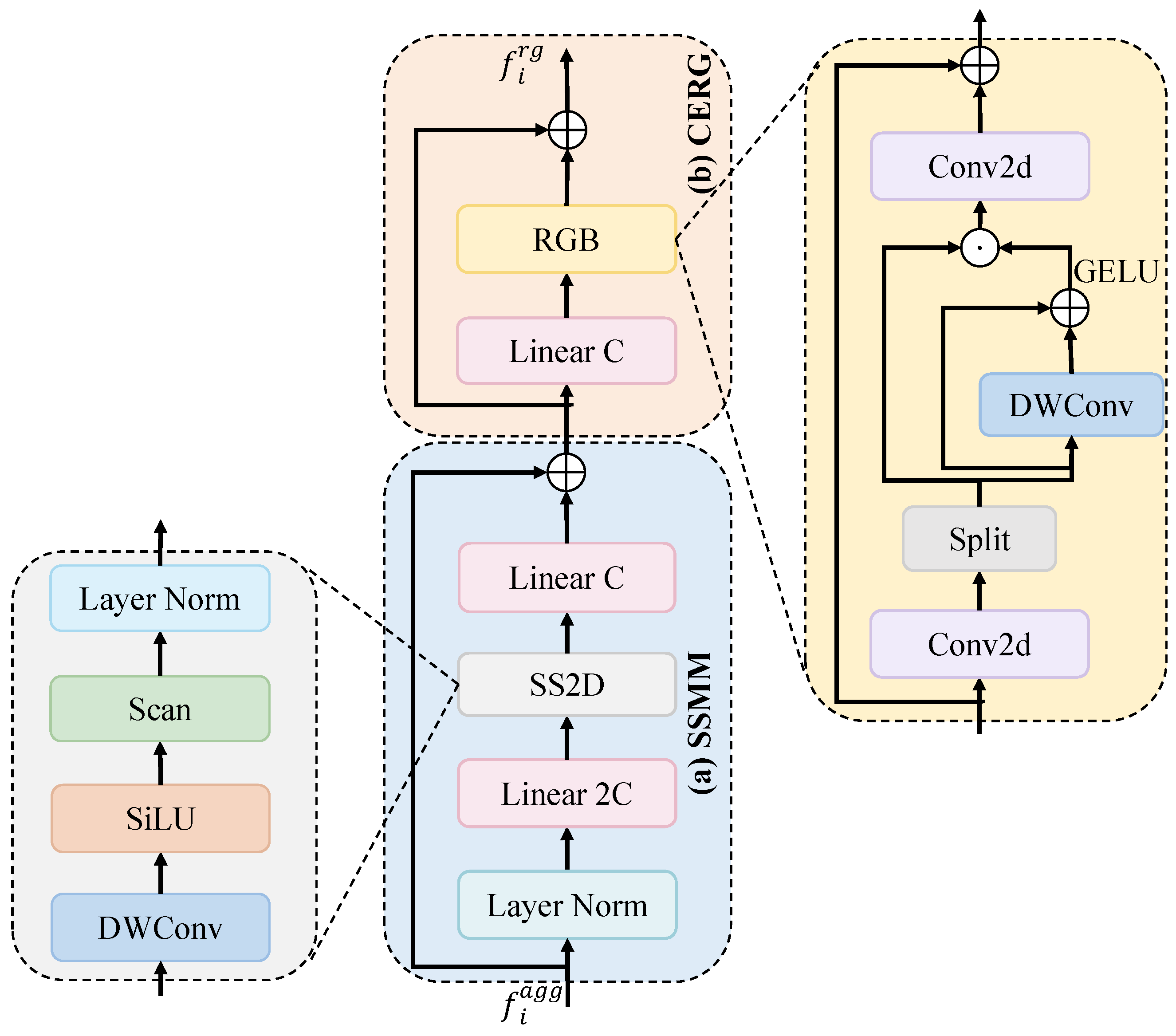
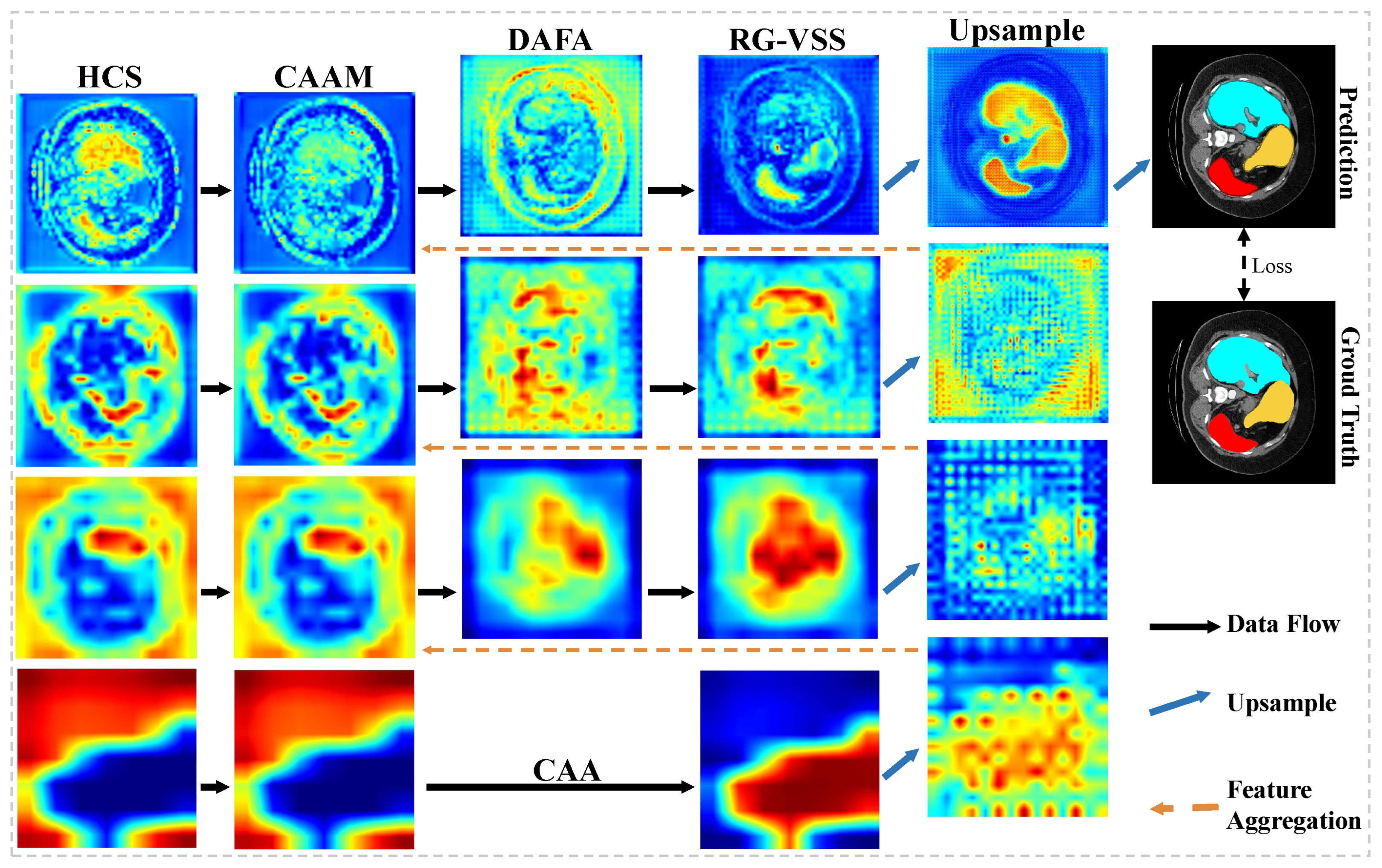
To address issues existing in feature extraction and aggregation, as discussed above, we propose a hybrid cascade and dual-path adaptive aggregation network (HCDAA-Net) for medical image segmentation. For feature extraction, we design a hybrid cascade structure (HCS) that alternately stacks ResNet [
19] and Mamba modules, achieving a spatial balance between short-range detail extraction and long-range semantic modeling. To complement this spatial modeling, we introduce a channel-crossing attention mechanism (CCAM) [
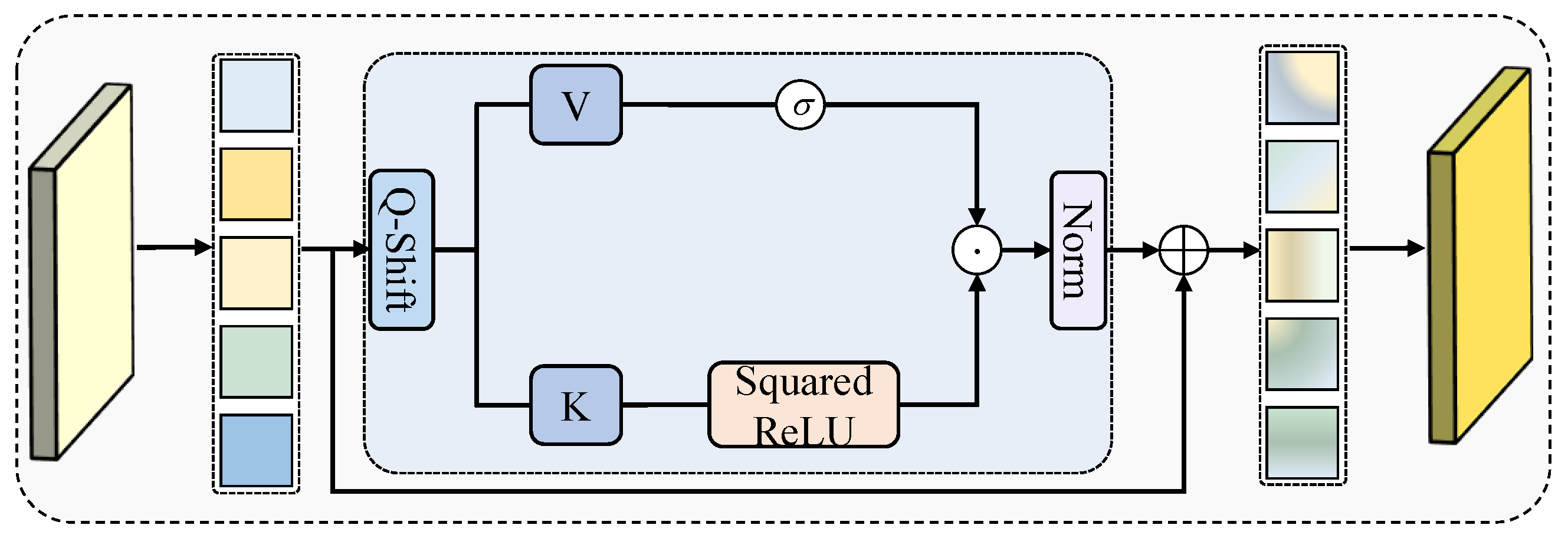
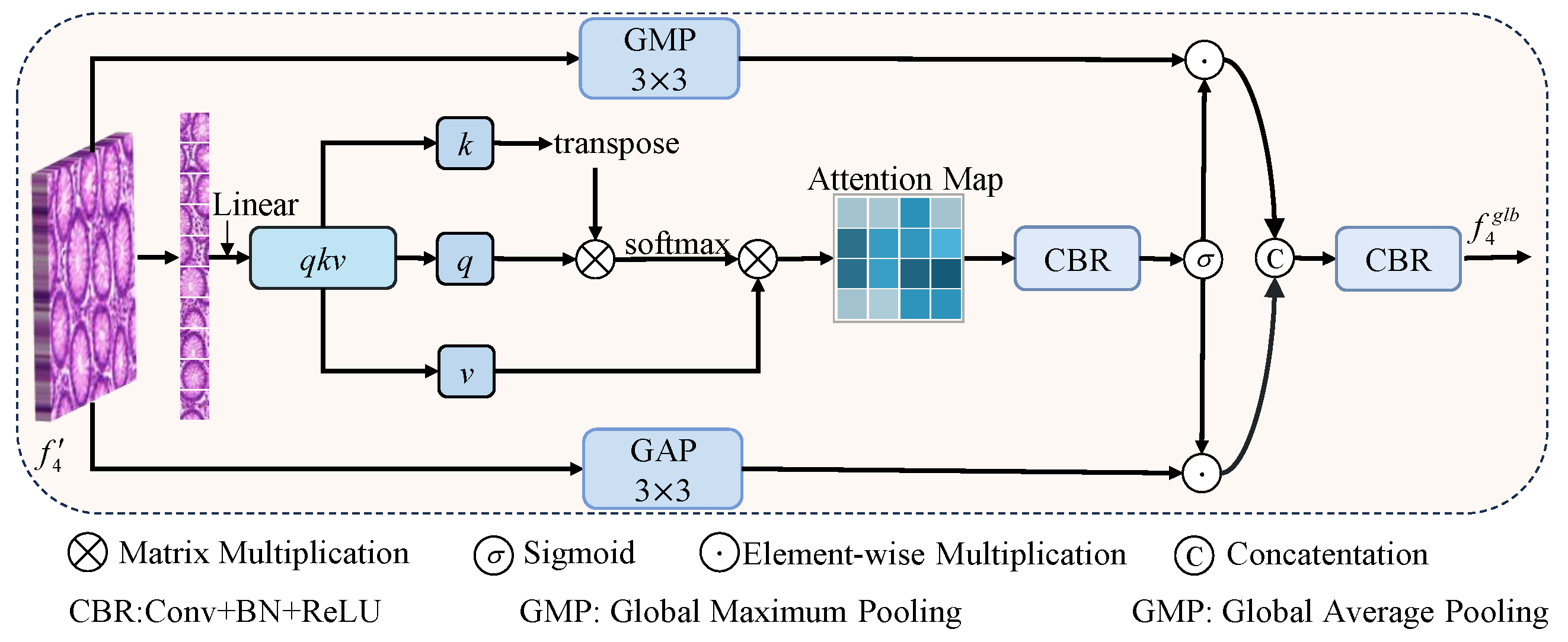
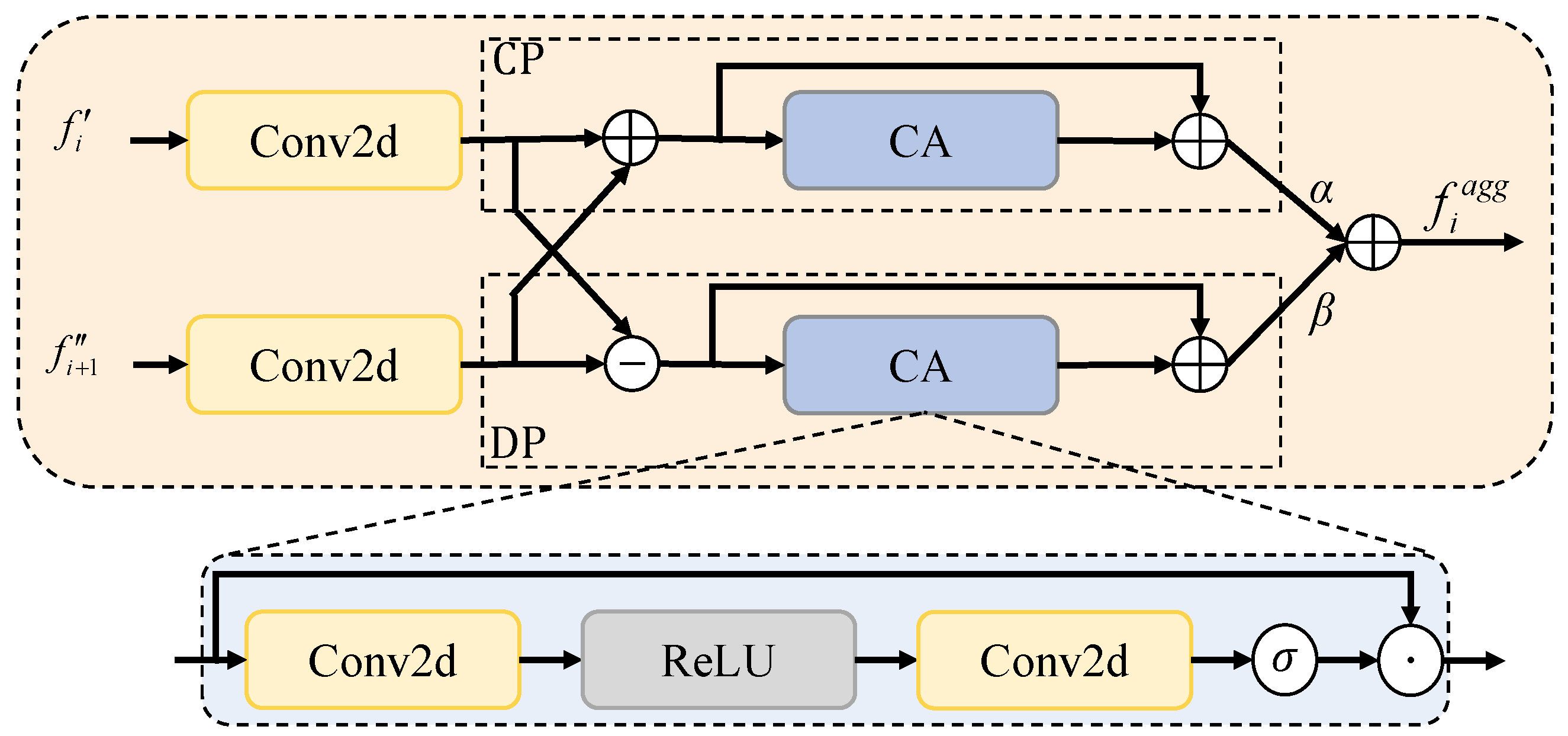
20], which strengthens feature representations and accelerates convergence through inter-channel interactions. For feature aggregation, we first design a correlation-aware aggregation (CAA) module, which captures the global correlations among features of the same class to address the feature representation inconsistency caused by variations in size and morphology, improving the segmentation accuracy of critical structures. We then propose the dual-path adaptive feature aggregation (DAFA) module with two branches: the common path (CP) and delta path (DP). CP employs a sum operation and channel attention to learn common semantics across scales and suppress redundancy, whereas DP adopts a difference operation to highlight fine-grained variations and enhance local lesion extraction. By integrating both paths, DAFA jointly achieves robust representation of common features and precise perception of discrepancies, thereby strengthening multiscale feature fusion. Finally, we propose a novel residual-gated visual state space (RG-VSS) module to further refine the fused features by dynamically modulating information flow through a convolution-enhanced residual gating mechanism.
The main contributions of this paper are as follows:
We design a novel HCDAA-Net for medical image segmentation. We employ an HCS that alternately integrates ResNet and Mamba modules to balance short-range detail extraction and long-range semantic modeling, followed by CCAM for channel-wise feature enhancement.
We propose a CAA to capture global correlations among same-class features to reduce feature inconsistencies from shape and scale variations, enhancing critical structure segmentation.
We introduce a DAFA to integrate a common path for shared semantics with delta path for highlighting fine-grained differences, enabling robust and precise multiscale feature fusion.
We develop a RG-VSS that dynamically modulates information flow through a convolution-enhanced residual gating mechanism to refine feature representations.
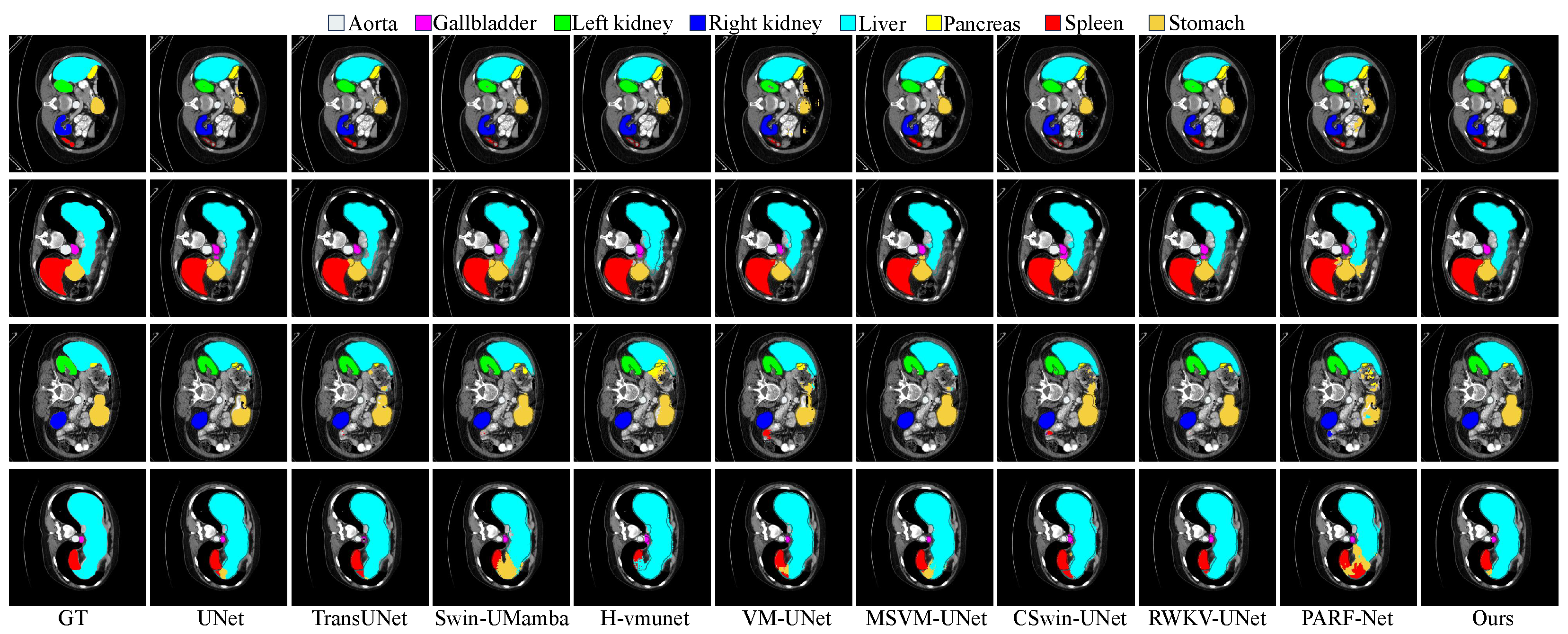
Experiments on four public datasets, covering various anatomical and pathological segmentation tasks, demonstrate that HCDAA-Net achieves SOTA performance.
The structure of this paper is as follows:
Section 2 reviews related research progress.
Section 3 details the proposed HCDAA-Net architecture.
Section 4 presents experimental results.
Section 5 concludes with the main contributions.
5. Conclusions
In this work, we present HCDAA-Net, a hybrid cascade and dual-path adaptive aggregation network, to improve medical image segmentation through enhanced feature extraction and feature aggregation. For feature extraction, we design an HCS to alternately integrate ResNet and Mamba modules, preserving fine edges while capturing global semantics. We further employ CCAM to strengthen feature representation and accelerate convergence. For feature aggregation, we introduce CAA to capture intra-structure correlations and DAFA to combine stable cross-layer semantics with subtle differences, improving sensitivity to fine details. Finally, we introduce RG-VSS to refine fused features through dynamic modulation of information flow. Experimental results demonstrate that HCDAA-Net achieves superior segmentation performance, effectively capturing both fine-grained structures and global context, highlighting its potential for robust and accurate medical image analysis.
Although HCDAA-Net demonstrates strong overall performance, its segmentation accuracy for certain specific organs, such as the liver, remains limited. We attribute this suboptimal performance in segmenting large targets to two main factors: (1) RG-VSS excessively modulates feature flow, overemphasizing fine-grained details and impairing the modeling of overall organ structures and (2) LKPE emphasizes local details via convolution and pixel-shuffle upsampling but lacks global structural modeling, leading to blurred boundaries in large targets. To address these issues, future work will focus on enhancing global structural modeling for large targets by (1) improving RG-VSS to enhance local details while preserving overall structural integrity and avoiding overemphasis on fine-grained features and (2) optimizing the upsampling strategy of the LKPE by incorporating larger receptive fields to reduce boundary blurring and improve contour reconstruction for large targets. Additionally, our HCDAA-Net relies entirely on fully supervised segmentation and does not consider reducing annotation costs or leveraging large Vision-Language Models (VLMs). Given the high cost of medical labels, future work also will explore semi-supervised learning, weakly-supervised segmentation, or VLMs to improve practicality and scalability.