Explainable Model of Hybrid Ensemble Learning for Prostate Cancer RNA-Seq Classification via Targeted Feature Selection
Abstract
1. Introduction
- A hybrid ensemble classifier combining KNN, RF and SVM with majority voting and feature selecting to select the most influential genes is proposed for prostate cancer classification.
- A process is proposed that reduces high-dimensional RNA-seq features to the 30 most effective features using systematic feature selection methods MI, MSE, RFE, Lasso and Ridge.
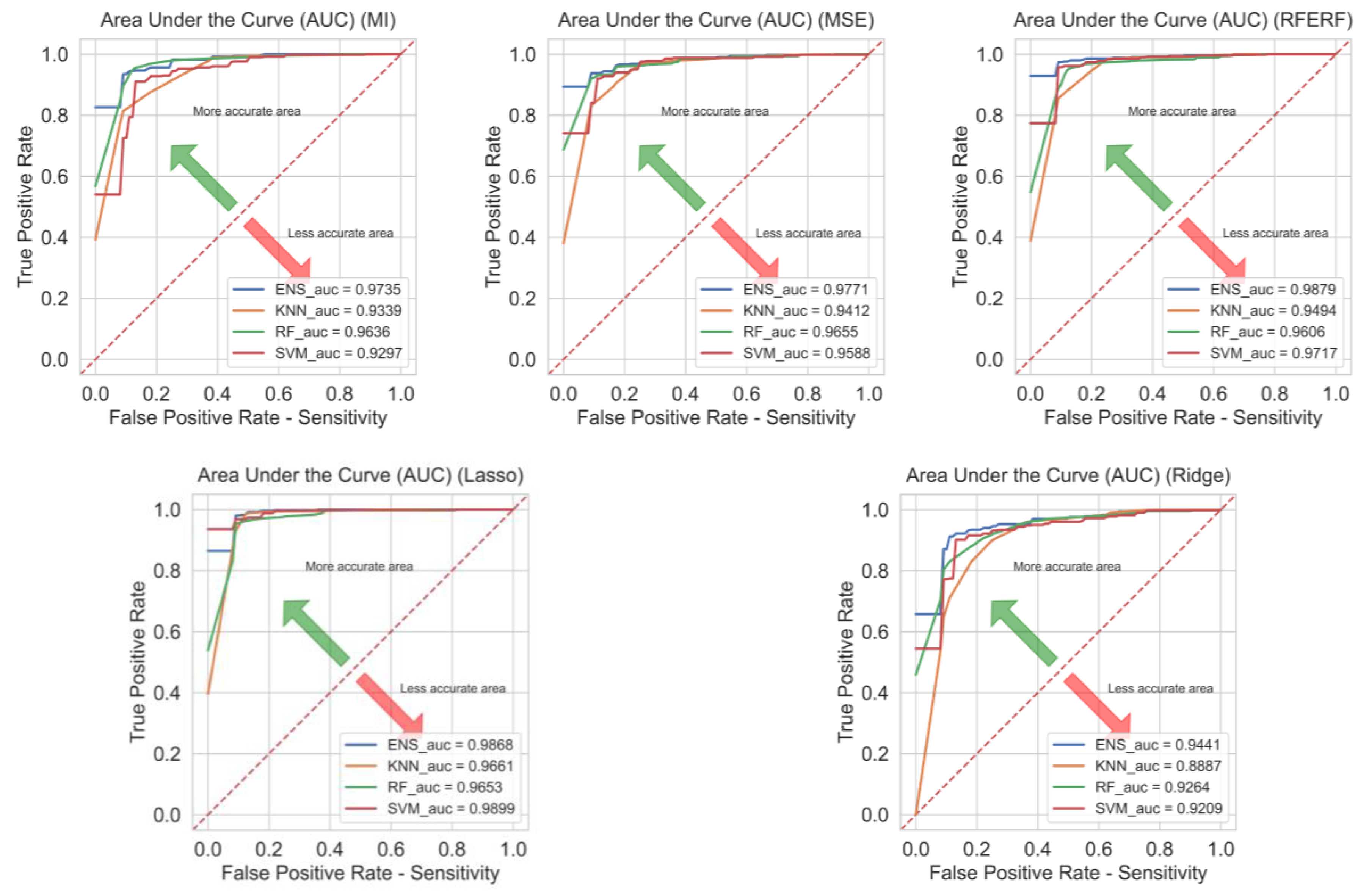
- Experimentally, Lasso is shown to be the most effective selector among classifiers, RFE can be used as an alternative and Ridge is shown to be the weakest selector. This highlights the centrality of feature selection in small-sample, high-dimensional genomics.
- To investigate generalizability beyond the PRAD dataset, the proposed model was tested on cross-validated liver, lung and thyroid cancer datasets and yielded high results.
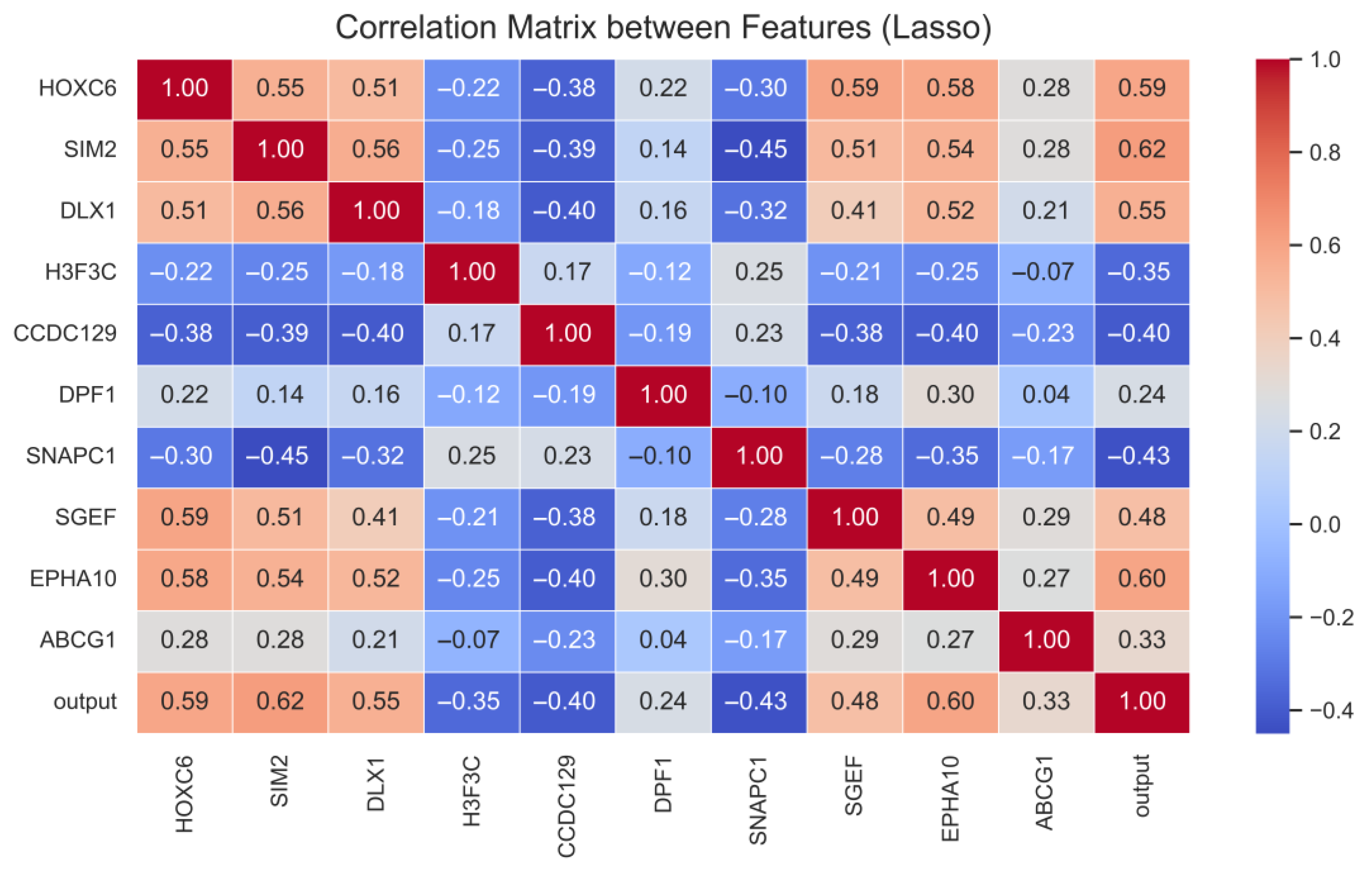
- SHAP and LIME analyses within XAI and correlation analyses revealed that the highly expressed EPHA10, HOXC6 and DLX1 genes were the most effective genes in classifying cancerous samples, while the highly expressed H3F3C and low-expressed ABCG1 and DPF1 genes were the most effective genes in classifying normal samples.
2. Related Works
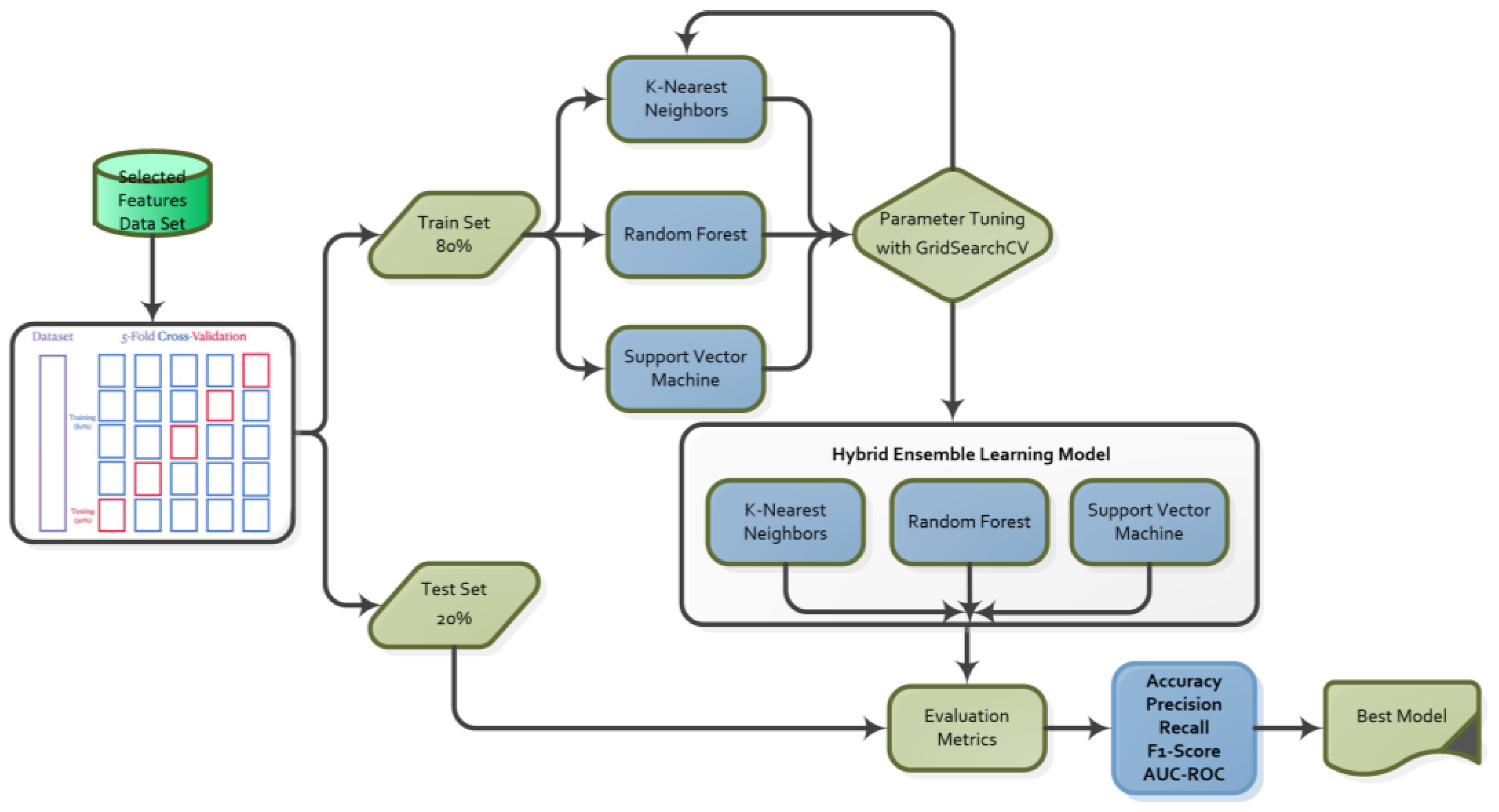
3. Materials and Methods
3.1. Dataset and Pre-Processing
3.2. Feature Selection Methods
3.3. Machine Learning Algorithms
3.3.1. Traditional Machine Learning Algorithms
3.3.2. Ensemble Learning
3.3.3. The Proposed Hybrid Ensemble Learning Model
3.4. Evaluation Metrics
4. Results and Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- James, N.D.; Tannock, I.; N’Dow, J.; Feng, F.; Gillessen, S.; Ali, S.A.; Trujillo, B.; Al-Lazikani, B.; Attard, G.; Bray, F.; et al. The Lancet Commission on Prostate Cancer: Planning for the Surge in Cases. Lancet 2024, 403, 1683–1722. [Google Scholar] [CrossRef]
- Hong, M.; Tao, S.; Zhang, L.; Diao, L.T.; Huang, X.; Huang, S.; Xie, S.J.; Xiao, Z.D.; Zhang, H. RNA Sequencing: New Technologies and Applications in Cancer Research. J. Hematol. Oncol. 2020, 13, 166. [Google Scholar] [CrossRef]
- Giganti, F.; Moreira da Silva, N.; Yeung, M.; Davies, L.; Frary, A.; Ferrer Rodriguez, M.; Sushentsev, N.; Ashley, N.; Andreou, A.; Bradley, A.; et al. AI-Powered Prostate Cancer Detection: A Multi-Centre, Multi-Scanner Validation Study. Eur. Radiol. 2025, 35, 4915–4924. [Google Scholar] [CrossRef]
- Saha, A.; Bosma, J.S.; Twilt, J.J.; van Ginneken, B.; Bjartell, A.; Padhani, A.R.; Bonekamp, D.; Villeirs, G.; Salomon, G.; Giannarini, G.; et al. Artificial Intelligence and Radiologists in Prostate Cancer Detection on MRI (PI-CAI): An International, Paired, Non-Inferiority, Confirmatory Study. Lancet Oncol. 2024, 25, 879–887. [Google Scholar] [CrossRef]
- Teke, M.; Etem, T. Cascading GLCM and T-SNE for Detecting Tumor on Kidney CT Images with Lightweight Machine Learning Design. Eur. Phys. J. Spec. Top. 2025, 1–16. [Google Scholar] [CrossRef]
- Goldenberg, S.L.; Nir, G.; Salcudean, S.E. A New Era: Artificial Intelligence and Machine Learning in Prostate Cancer. Nat. Rev. Urol. 2019, 16, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Jain, A. Feature Selection: Evaluation, Application, and Small Sample Performance. IEEE Trans. Pattern Anal. Mach. Intell. 1997, 19, 153–158. [Google Scholar] [CrossRef]
- Golub, T.R.; Slonim, D.K.; Tamayo, P.; Huard, C.; Gaasenbeek, M.; Mesirov, J.P.; Coller, H.; Loh, M.L.; Downing, J.R.; Caligiuri, M.A.; et al. Molecular Classification of Cancer: Class Discovery and Class Prediction by Gene Expression Monitoring. Science 1999, 286, 531–537. [Google Scholar] [CrossRef]
- Berrar, D.; Bradbury, I.; Dubitzky, W. Avoiding Model Selection Bias in Small-Sample Genomic Datasets. Bioinformatics 2006, 22, 1245–1250. [Google Scholar] [CrossRef] [PubMed]
- Goldman, M.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. The UCSC Xena Platform for Public and Private Cancer Genomics Data Visualization and Interpretation. bioRxiv 2019. bioRxiv:326470. [Google Scholar] [CrossRef]
- Dhumkekar, A.A.; Ghorpade, S.; Awale, R.N. Performance Analysis of Various Cancers Using Genetic Data with Variance Threshold. In Proceedings of the 2022 OITS International Conference on Information Technology (OCIT), Bhubaneswar, India, 14–16 December 2022; IEEE: New York, NY, USA, 2022; pp. 67–72. [Google Scholar] [CrossRef]
- Shamsara, E.; Shamsara, J. Bioinformatics Analysis of the Genes Involved in the Extension of Prostate Cancer to Adjacent Lymph Nodes by Supervised and Unsupervised Machine Learning Methods: The Role of SPAG1 and PLEKHF2. Genomics 2020, 112, 3871–3882. [Google Scholar] [CrossRef]
- Begum, S.; Sarkar, R.; Chakraborty, D.; Sen, S.; Maulik, U. Application of Active Learning in DNA Microarray Data for Cancerous Gene Identification. Expert Syst. Appl. 2021, 177, 114914. [Google Scholar] [CrossRef]
- Gumaei, A.; Sammouda, R.; Al-Rakhami, M.; AlSalman, H.; El-Zaart, A. Feature Selection with Ensemble Learning for Prostate Cancer Diagnosis from Microarray Gene Expression. Health Inform. J. 2021, 27, 1460458221989402. [Google Scholar] [CrossRef]
- Fathi, H.; AlSalman, H.; Gumaei, A.; Manhrawy, I.I.M.; Hussien, A.G.; El-Kafrawy, P. An Efficient Cancer Classification Model Using Microarray and High-Dimensional Data. Comput. Intell. Neurosci. 2021, 2021, 7231126. [Google Scholar] [CrossRef]
- He, B.; Zhang, Y.; Zhou, Z.; Wang, B.; Liang, Y.; Lang, J.; Lin, H.; Bing, P.; Yu, L.; Sun, D.; et al. A Neural Network Framework for Predicting the Tissue-of-Origin of 15 Common Cancer Types Based on RNA-Seq Data. Front. Bioeng. Biotechnol. 2020, 8, 737. [Google Scholar] [CrossRef]
- Santo, G.D.; Frasca, M.; Bertoli, G.; Castiglioni, I.; Cava, C. Identification of Key MiRNAs in Prostate Cancer Progression Based on MiRNA-MRNA Network Construction. Comput. Struct. Biotechnol. J. 2022, 20, 864–873. [Google Scholar] [CrossRef] [PubMed]
- Senbagamalar, L.; Logeswari, S. Genetic Clustering Algorithm-Based Feature Selection and Divergent Random Forest for Multiclass Cancer Classification Using Gene Expression Data. Int. J. Comput. Intell. Syst. 2024, 17, 23. [Google Scholar] [CrossRef]
- Razzaque, A.; Badholia, D.A. PCA Based Feature Extraction and MPSO Based Feature Selection for Gene Expression Microarray Medical Data Classification. Meas. Sens. 2024, 31, 100945. [Google Scholar] [CrossRef]
- Venkataramana, L.; Jacob, S.G.; Saraswathi, S.; Venkata Vara Prasad, D. Identification of Common and Dissimilar Biomarkers for Different Cancer Types from Gene Expressions of RNA-Sequencing Data. Gene Rep. 2020, 19, 100654. [Google Scholar] [CrossRef]
- Yaping, Z.; Changyin, Z. Gene Feature Selection Method Based on ReliefF and Pearson Correlation. In Proceedings of the 2021 3rd International Conference on Applied Machine Learning (ICAML), Changsha, China, 23–25 July 2021; IEEE: New York, NY, USA, 2021; pp. 15–19. [Google Scholar] [CrossRef]
- Ali, N.M.; Hanafi, A.N.; Karis, M.S.; Shamsudin, N.H.; Shair, E.F.; Abdul Aziz, N.H. Hybrid Feature Selection of Microarray Prostate Cancer Diagnostic System. Indones. J. Electr. Eng. Comput. Sci. 2024, 36, 1884. [Google Scholar] [CrossRef]
- Bhonde, S.B.; Wagh, S.K.; Prasad, J.R. Identification of Cancer Types from Gene Expressions Using Learning Techniques. Comput. Methods Biomech. Biomed. Eng. 2023, 26, 1951–1965. [Google Scholar] [CrossRef]
- Mostavi, M.; Chiu, Y.C.; Huang, Y.; Chen, Y. Convolutional Neural Network Models for Cancer Type Prediction Based on Gene Expression. BMC Med. Genomics 2020, 13, 44. [Google Scholar] [CrossRef]
- Petinrin, O.O.; Saeed, F.; Salim, N.; Toseef, M.; Liu, Z.; Muyide, I.O. Dimension Reduction and Classifier-Based Feature Selection for Oversampled Gene Expression Data and Cancer Classification. Processes 2023, 11, 1940. [Google Scholar] [CrossRef]
- Alkhanbouli, R.; Matar Abdulla Almadhaani, H.; Alhosani, F.; Simsekler, M.C.E. The Role of Explainable Artificial Intelligence in Disease Prediction: A Systematic Literature Review and Future Research Directions. BMC Med. Inform. Decis. Mak. 2025, 25, 110. [Google Scholar] [CrossRef]
- Antunes, M.E.; Araújo, T.G.; Till, T.M.; Pantaleão, E.; Mancera, P.F.A.; de Oliveira, M.H. Machine Learning Models for Predicting Prostate Cancer Recurrence and Identifying Potential Molecular Biomarkers. Front. Oncol. 2025, 15, 1535091. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Program (TCGA)—NCI. Available online: https://www.cancer.gov/ccg/research/genome-sequencing/tcga (accessed on 6 September 2025).
- Oğuzlar, A. Veri Ön İşleme. Erciyes Üniv. İktisadi ve İdari Bilim. Fakültesi Derg. 2003, 21, 67–76. Available online: https://dergipark.org.tr/tr/pub/erciyesiibd/issue/5878/77794 (accessed on 8 October 2025).
- Feature Extraction: Foundations and Applications—Google Kitaplar. Available online: https://books.google.com.tr/books?hl=tr&lr=&id=FOTzBwAAQBAJ&oi=fnd&pg=PA1&dq=Feature+Extraction:+Foundations+and+Applications&ots=5Vj9N4aokV&sig=oialtKbQKDqW869D9Ts2LrH7DvA&redir_esc=y#v=onepage&q=Feature%20Extraction%3A%20Foundations%20and%20Applications&f=false (accessed on 7 January 2025).
- Bolón-Canedo, V.; Sánchez-Maroño, N.; Alonso-Betanzos, A.; Benítez, J.M.; Herrera, F. A Review of Microarray Datasets and Applied Feature Selection Methods. Inf. Sci. 2014, 282, 111–135. [Google Scholar] [CrossRef]
- Applying Filter Methods in Python for Feature Selection. Available online: https://stackabuse.com/applying-filter-methods-in-python-for-feature-selection/ (accessed on 7 January 2025).
- Landset, S.; Khoshgoftaar, T.M.; Richter, A.N.; Hasanin, T. A Survey of Open Source Tools for Machine Learning with Big Data in the Hadoop Ecosystem. J. Big Data 2015, 2, 24. [Google Scholar] [CrossRef]
- Novaković, J.; Strbac, P.; Bulatović, D. Toward Optimal Feature Selection Using Ranking Methods and Classification Algorithms. Yugosl. J. Oper. Res. 2011, 21, 119–135. [Google Scholar] [CrossRef]
- Kushmerick, N.; Weld, D.S.; Doorenbos, R.B. Wrapper Induction for Information Extraction. Int. Jt. Conf. Artif. Intell. 1997, 1, 729–735. Available online: https://api.semanticscholar.org/CorpusID:5119155 (accessed on 8 October 2025).
- Makine Öğrenmesinde Değişken Seçimi (Feature Selection) Yazı Serisi: Gömülü (Embedded) Yöntemler ve Python Kodları by Yiğit Şener Medium. Available online: https://yigitsener.medium.com/makine-%C3%B6%C4%9Frenmesinde-de%C4%9Fi%C5%9Fken-se%C3%A7imi-feature-selection-yaz%C4%B1-serisi-g%C3%B6m%C3%BCl%C3%BC-embedded-y%C3%B6ntemler-c23293915b39 (accessed on 7 January 2025).
- Aydın Atasoy, N.; Demiröz, A. Makine Öğrenmesi Algoritmaları Kullanılarak Prostat Kanseri Tümör Oluşumunun İncelenmesi. Avrupa Bilim Ve Teknol. Derg. 2021, 29, 87–92. [Google Scholar] [CrossRef]
- Machine Learning for Dummies: IBM Limited Edition: Free Download, Borrow, and Streaming: Internet Archive. Available online: https://archive.org/details/machine-learning-for-dummies/page/19/mode/2up (accessed on 7 January 2025).
- Understanding Machine Learning: From Theory to Algorithms—Shai Shalev-Shwartz, Shai Ben-David—Google Kitaplar. Available online: https://books.google.com.tr/books?hl=tr&lr=&id=ttJkAwAAQBAJ&oi=fnd&pg=PR15&dq=Understanding+machine+learning:+From+theory+to+algorithms&ots=PIOaYFPaS1&sig=8WXRKn3VjR6_LRxAjCrO0NihxDs&redir_esc=y#v=onepage&q=Understanding%20machine%20learning%3A%20From%20theory%20to%20algorithms&f=false (accessed on 7 January 2025).
- Machine Learning Tutorial: Learn ML for Free. Available online: https://www.tutorialspoint.com/machine_learning/index.htm (accessed on 7 January 2025).
- Introduction to Machine Learning—Wikipedia PDF Statistics Mathematical Analysis. Available online: https://www.scribd.com/document/340091580/Introduction-to-Machine-Learning-Wikipedia (accessed on 7 January 2025).
- Latha, C.B.C.; Jeeva, S.C. Improving the Accuracy of Prediction of Heart Disease Risk Based on Ensemble Classification Techniques. Inform. Med. Unlocked 2019, 16, 100203. [Google Scholar] [CrossRef]
- Mahajan, P.; Uddin, S.; Hajati, F.; Moni, M.A. Ensemble Learning for Disease Prediction: A Review. Healthcare 2023, 11, 1808. [Google Scholar] [CrossRef]
- Huang, F.; Xie, G.; Xiao, R. Research on Ensemble Learning. In Proceedings of the 2009 International Conference on Artificial Intelligence and Computational Intelligence, AICI 2009, Shanghai, China, 7–8 November 2009; pp. 249–252. [Google Scholar] [CrossRef]
- Moldovanu, S.; Munteanu, D.; Sîrbu, C. Impact on Classification Process Generated by Corrupted Features. Big Data Cogn. Comput. 2025, 9, 45. [Google Scholar] [CrossRef]
- Ali, S.; Abuhmed, T.; El-Sappagh, S.; Muhammad, K.; Alonso-Moral, J.M.; Confalonieri, R.; Guidotti, R.; Del Ser, J.; Díaz-Rodríguez, N.; Herrera, F. Explainable Artificial Intelligence (XAI): What We Know and What Is Left to Attain Trustworthy Artificial Intelligence. Inf. Fusion 2023, 99, 101805. [Google Scholar] [CrossRef]
- Etem, T. Interpretable Machine Learning for Battery Health Insights: A LIME and SHAP-Based Study on EIS-Derived Features. Bull. Pol. Acad. Sci. Tech. Sci. 2025, 73, e155033. [Google Scholar] [CrossRef]
- Salih, A.M.; Raisi-Estabragh, Z.; Galazzo, I.B.; Radeva, P.; Petersen, S.E.; Lekadir, K.; Menegaz, G. A Perspective on Explainable Artificial Intelligence Methods: SHAP and LIME. Adv. Intell. Syst. 2025, 7, 2400304. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why Should i Trust You?” Explaining the Predictions of Any Classifier. In KDD ’16: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 2016, San Francisco, CA, USA, 13–17 August 2016; Association for Computing Machinery: New York, NY, USA, 2016; pp. 1135–1144. [Google Scholar] [CrossRef]
- Munteanu, D.; Moldovanu, S.; Miron, M. The Explanation and Sensitivity of AI Algorithms Supplied with Synthetic Medical Data. Electronics 2025, 14, 1270. [Google Scholar] [CrossRef]
- Nagano, K.; Maeda, Y.; Kanasaki, S.I.; Watanabe, T.; Yamashita, T.; Inoue, M.; Higashisaka, K.; Yoshioka, Y.; Abe, Y.; Mukai, Y.; et al. Ephrin Receptor A10 Is a Promising Drug Target Potentially Useful for Breast Cancers Including Triple Negative Breast Cancers. J. Control. Release 2014, 189, 72–79. [Google Scholar] [CrossRef]
- Hughes, C.M.; Rozenblatt-Rosen, O.; Milne, T.A.; Copeland, T.D.; Levine, S.S.; Lee, J.C.; Hayes, D.N.; Shanmugam, K.S.; Bhattacharjee, A.; Biondi, C.A.; et al. Menin Associates with a Trithorax Family Histone Methyltransferase Complex and with the Hoxc8 Locus. Mol. Cell 2004, 13, 587–597. [Google Scholar] [CrossRef] [PubMed]
- Yun, K.; Fischman, S.; Johnson, J.; Hrabe de Angelis, M.; Weinsmaster, G.; Rubenstein, J.L.R. Modulation of the Notch Signalling by Mash1 and Dlx1/2 Regulates Sequential Specification and Differentiation of Progenitor Cell Types in the Subcortical Telencephalon. Development 2002, 129, 5029–5040. [Google Scholar] [CrossRef]
- Schenk, R.; Jenke, A.; Zilbauer, M.; Wirth, S.; Postberg, J. H3.5 Is a Novel Hominid-Specific Histone H3 Variant That Is Specifically Expressed in the Seminiferous Tubules of Human Testes. Chromosoma 2011, 120, 275–285. [Google Scholar] [CrossRef]
- Klucken, J.; Büchler, C.; Orsó, E.; Kaminski, W.E.; Porsch-Özcürümez, M.; Liebisch, G.; Kapinsky, M.; Diederich, W.; Drobnik, W.; Dean, M.; et al. ABCG1 (ABC8), the Human Homolog of the Drosophila White Gene, Is a Regulator of Macrophage Cholesterol and Phospholipid Transport. Proc. Natl. Acad. Sci. USA 2000, 97, 817–822. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.P.; Yan, G.J.; Gu, Z.; Tso, J.K. Localization of EGFP-DPF-1 Expressed and Secreted by HeLa Cells in Oocytes. Shi Yan Sheng Wu Xue Bao 2003, 36, 307–313. [Google Scholar] [PubMed]
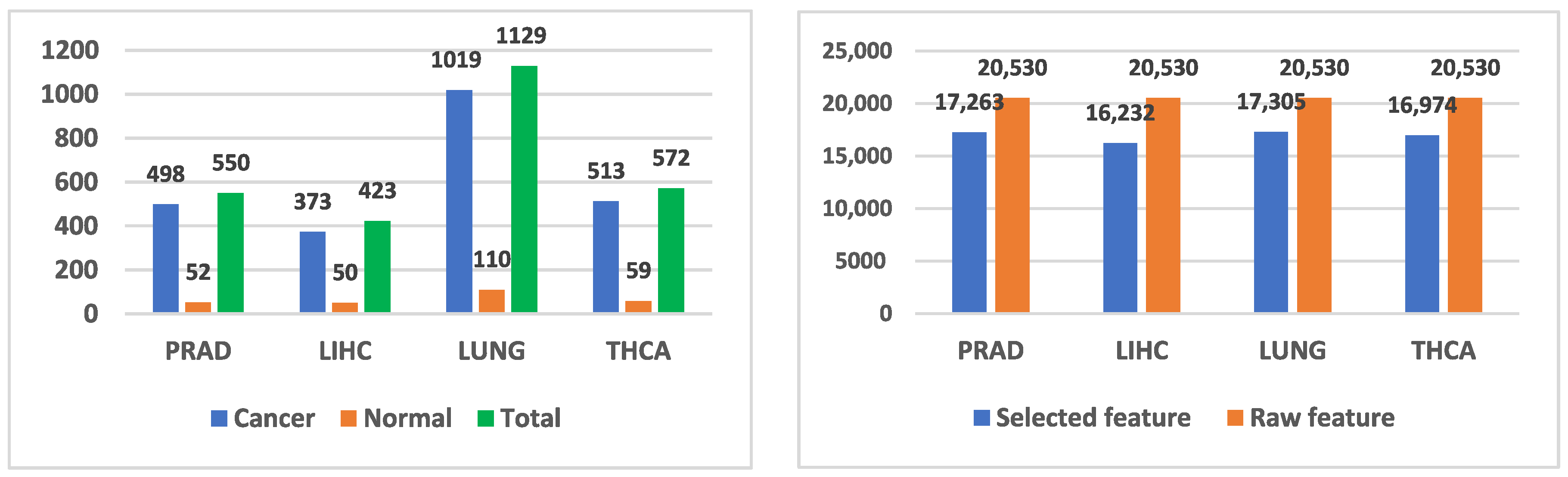
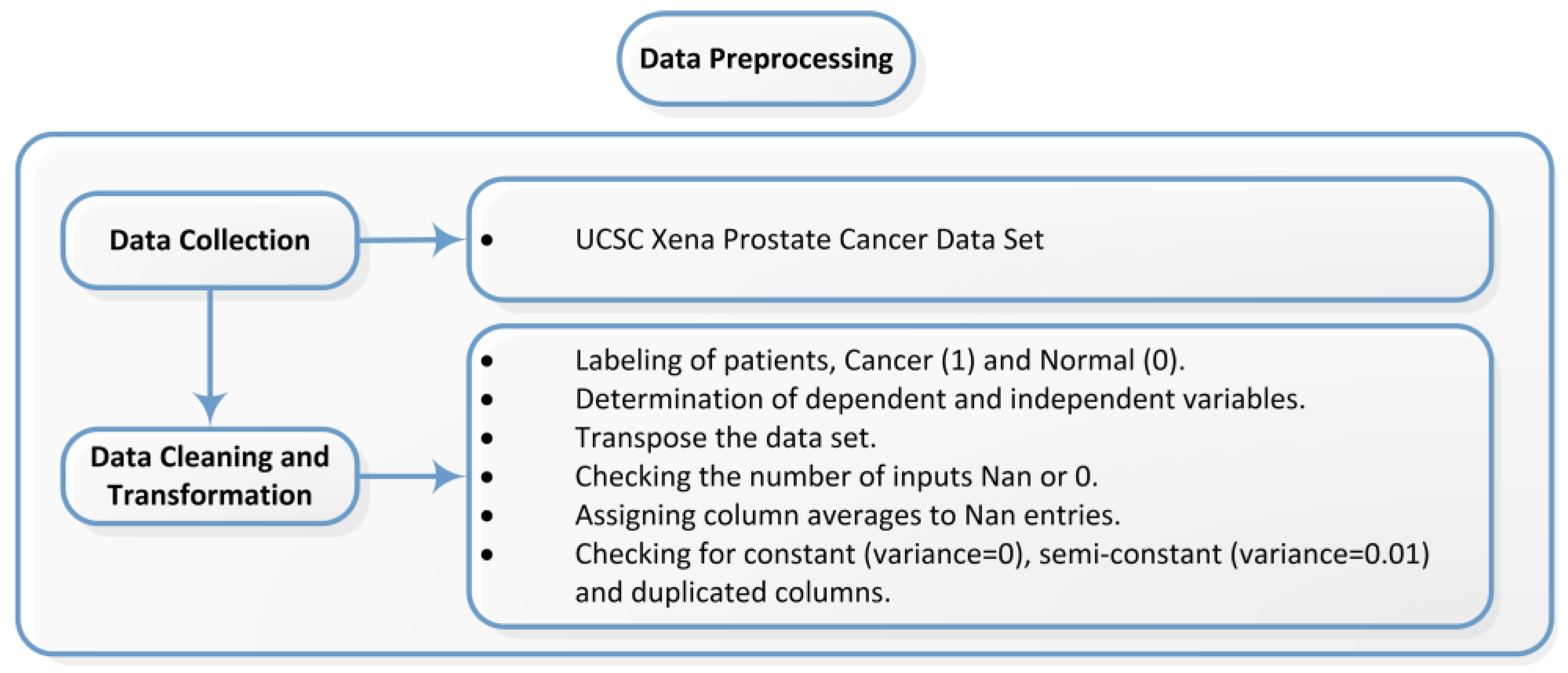
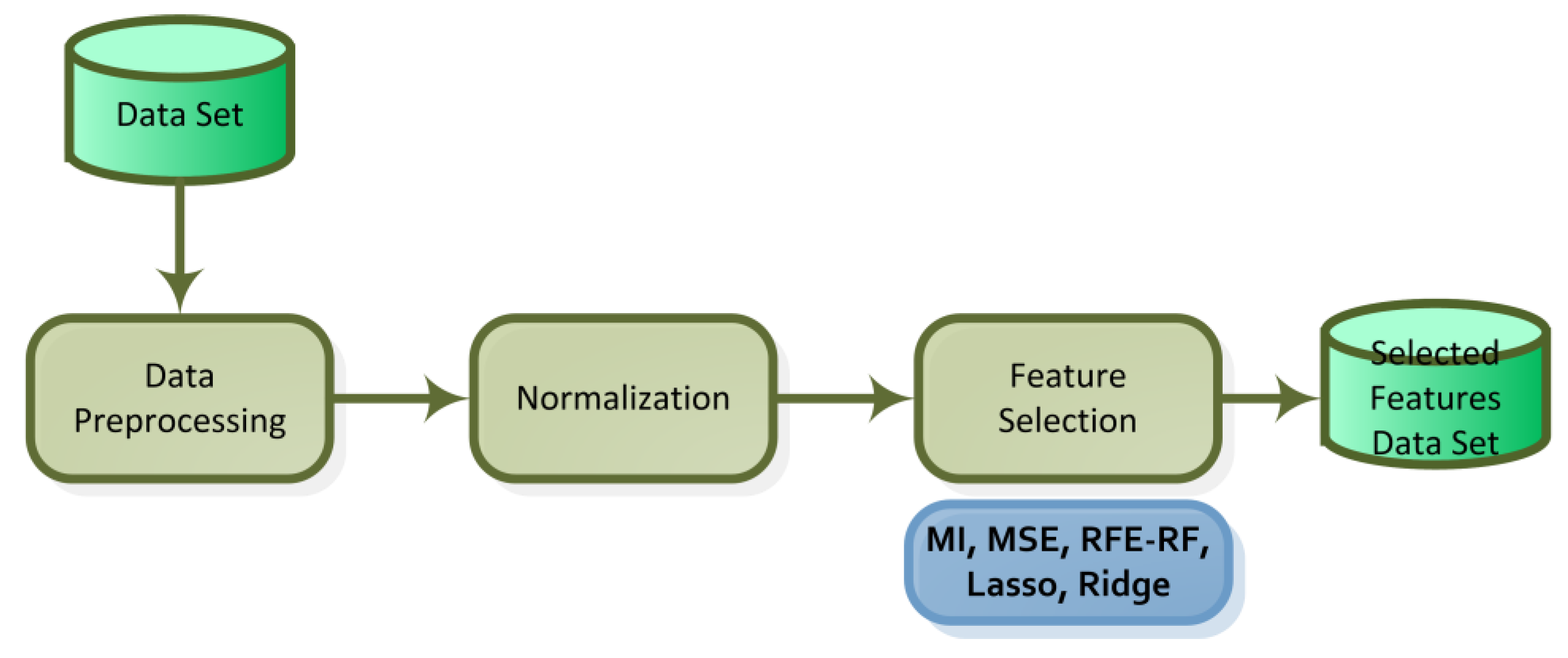
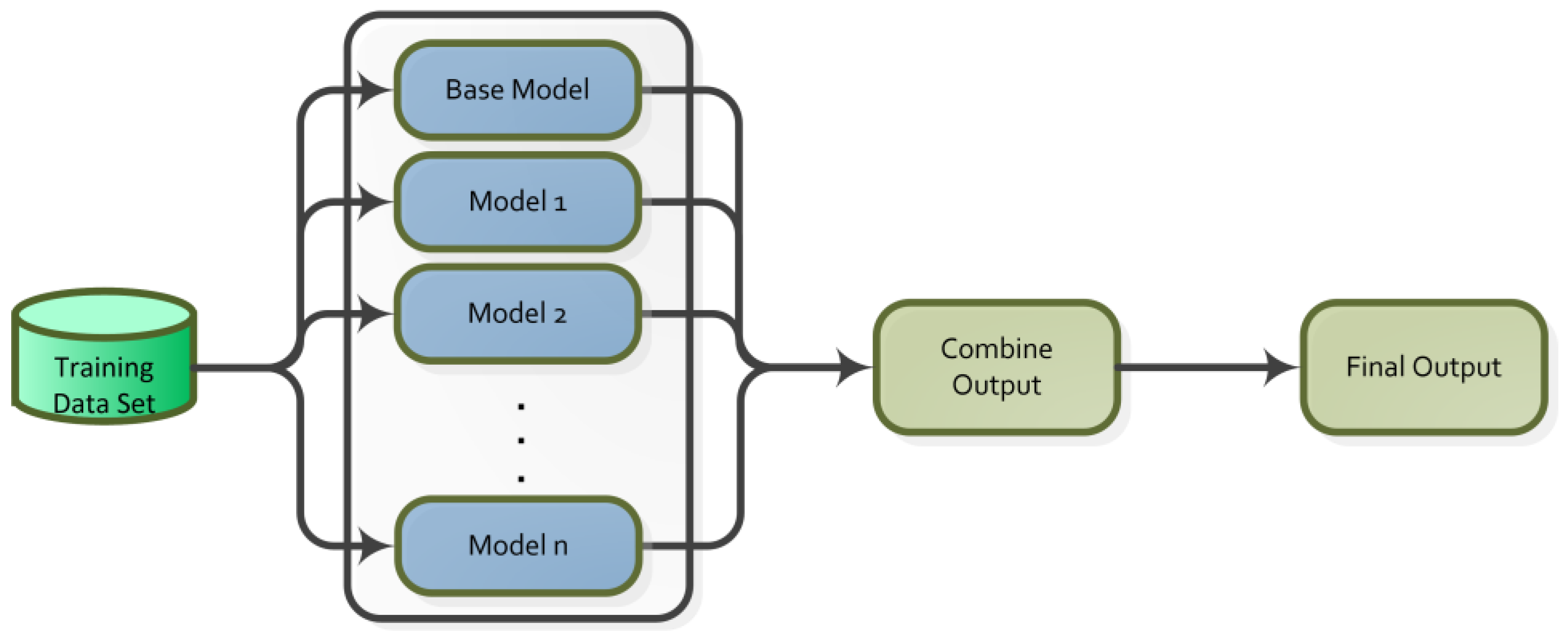

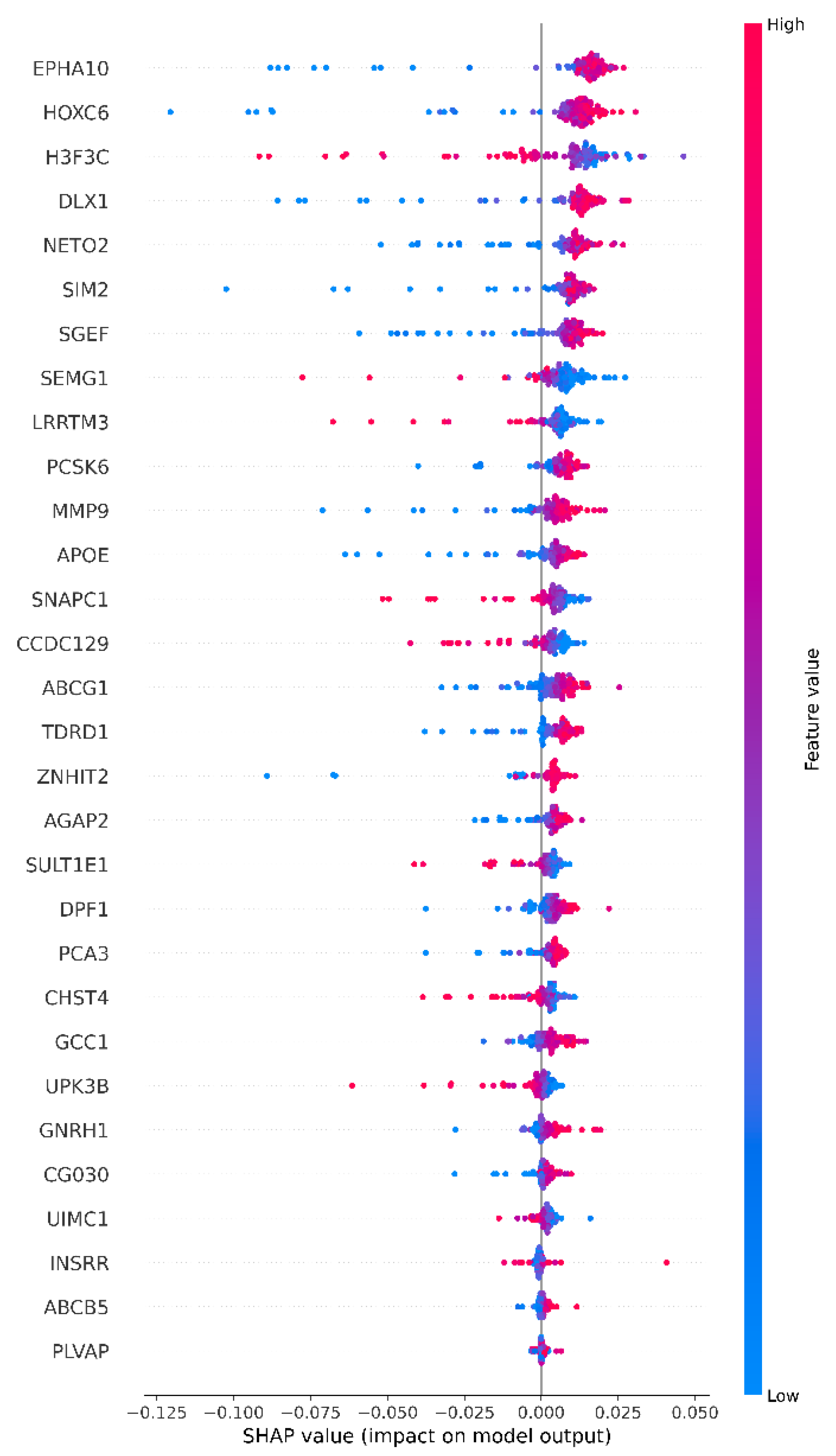
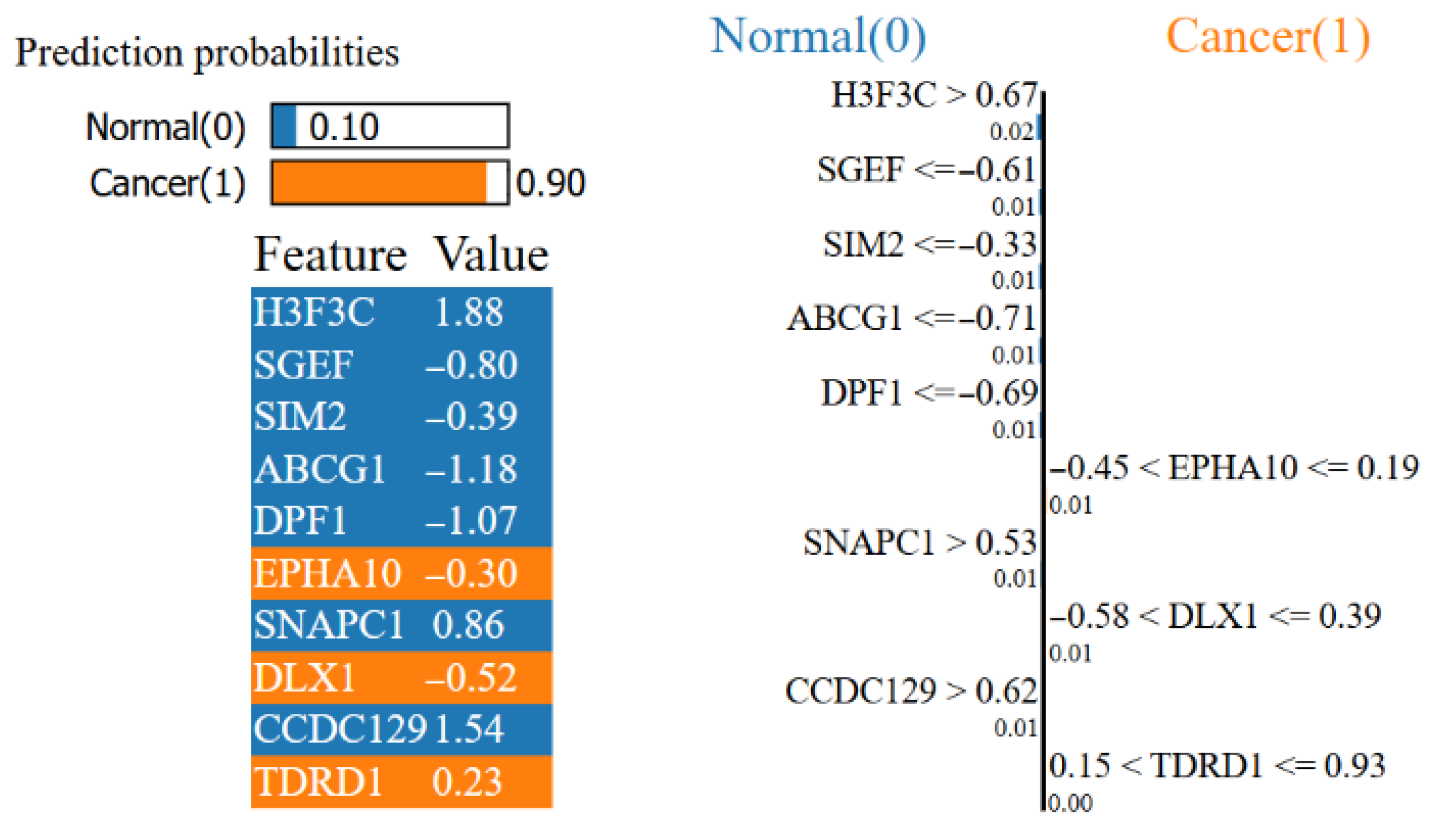
| Study | Dataset | Feature Selection Method | Classification Method | Final Method and Result Score |
|---|---|---|---|---|
| Dhumkekar A.A. et al. [12] | TCGA RNASeq (20,502 features, 8963 samples) | Variance Threshold | DT, NB, SVM, KNN, RF | SVM with Variance Threshold 94% accuracy |
| Santo, G.D. et al. [18] | TCGA (545 samples) | Wilcoxon signed-rank test, Boruta | RF | RF with Wilcoxon signed-rank test average 83.8% accuracy |
| Senbagamala, L. and Logeswari, S. [19] | TCGA RNASeq (16,382 features, 802 samples) | Genetic Clustering Algorithm (GCA) | LR, MLP, RF, YSA, SVM, KNN, DF | DF with GCD 97.74% accuracy |
| Razzaque, A. et al. [20] | Singh (12,600 features, 136 samples) | Modified Particle Swarm Optimization (MPSO) | NB, SVM, KNN | NB with MPSO 93.52% accuracy |
| Ali, N.M. et al. [23] | Singh (12,600 features, 102 samples) | ReliefF-GA, ReliefF-PSO, ReliefF-WOA | SVM | SVM with ReliefF-GA 91.17% accuracy |
| Bhonde, S.B. et al. [24] | TCGA RNASeq (20,501 features, 801 samples) | RF + PSO | RNN-LSTM | RNN-LSTM with RF + PSO 96.89% accuracy |
| Petinrin, O.O. et al. [26] | Singh (12,600 features, 102 samples) | PCA, TSVD, T-tSNE | LR, RF, SVM, GBC, GNB, KNN | LR with PCA and TSVD 92.29% accuracy |
| Antunes, M.E. et al. [28] | TCGA RNASeq (7 features, 489 samples) | Chi-square test | NB, SVM, ANN | SVM with Chi-square test and about 83% |
| Our study (Hybrid Ensemble Learning Model) | TCGA RNASeq (20,530 features, 550 samples) | MI, MSE, RFE-RF, Lasso, Ridge; SHAP and LIME. | KNN, RF, SVM, Ensemble learning with majority voting | Hybrid Ensemble Learning with Lasso and 97.82% accuracy |
| Classifier | Hyperparameter | Value |
|---|---|---|
| KNN | Number of neighbors (k) | 7 |
| Distance metric | minkowski | |
| Weights | Uniform | |
| RF | Number of trees (n_estimators) | 15 |
| Quality measurement (criterion) | entropy | |
| Maximum depth (max_depth) | 30 | |
| Minimum samples split (min_samples_split) | 2 | |
| Minimum samples leaf (min_samples_leaf) | 1 | |
| SVM | Kernel | poly |
| C (regularization parameter) | 1.0 | |
| Gamma | Scale | |
| Probability | True | |
| Tolerance (tol) | 0.001 | |
| Maximum iterations (max_iter) | 1000 |
| Classifier | Feature Selectors | Accuracy | Precision | Recall | F1-Score | AUC-ROC | MCC |
|---|---|---|---|---|---|---|---|
| KNN | MI | 0.9636 | 0.9803 | 0.9799 | 0.9799 | 0.9340 | 0.7993 |
| MSE | 0.9545 | 0.9707 | 0.9799 | 0.9752 | 0.9412 | 0.7773 | |
| RFE by RF | 0.9673 | 0.9768 | 0.9879 | 0.9822 | 0.9494 | 0.7941 | |
| Lasso | 0.9745 | 0.9844 | 0.9879 | 0.9861 | 0.9661 | 0.8130 | |
| Ridge | 0.9436 | 0.9467 | 0.9939 | 0.9697 | 0.8888 | 0.6736 | |
| RF | MI | 0.9618 | 0.9692 | 0.9899 | 0.9793 | 0.9637 | 0.7738 |
| MSE | 0.9545 | 0.9673 | 0.9839 | 0.9753 | 0.9654 | 0.7695 | |
| RFE by RF | 0.9582 | 0.9672 | 0.9879 | 0.9774 | 0.9605 | 0.7899 | |
| Lasso | 0.9673 | 0.9750 | 0.9899 | 0.9822 | 0.9652 | 0.8028 | |
| Ridge | 0.9309 | 0.9389 | 0.9880 | 0.9628 | 0.9261 | 0.6437 | |
| SVM | MI | 0.9436 | 0.9702 | 0.9679 | 0.9690 | 0.9290 | 0.7320 |
| MSE | 0.9545 | 0.9726 | 0.9778 | 0.9750 | 0.9579 | 0.7698 | |
| RFE by RF | 0.9600 | 0.9747 | 0.9818 | 0.9781 | 0.9707 | 0.7699 | |
| Lasso | 0.9727 | 0.9879 | 0.9818 | 0.9848 | 0.9897 | 0.8043 | |
| Ridge | 0.9109 | 0.9612 | 0.9397 | 0.9502 | 0.9206 | 0.6011 | |
| Proposed Hybrid Ensemble Method | MI | 0.9655 | 0.9712 | 0.9920 | 0.9813 | 0.9735 | 0.8217 |
| MSE | 0.9582 | 0.9711 | 0.9839 | 0.9772 | 0.9769 | 0.7775 | |
| RFE by RF | 0.9673 | 0.9732 | 0.9919 | 0.9823 | 0.9878 | 0.8030 | |
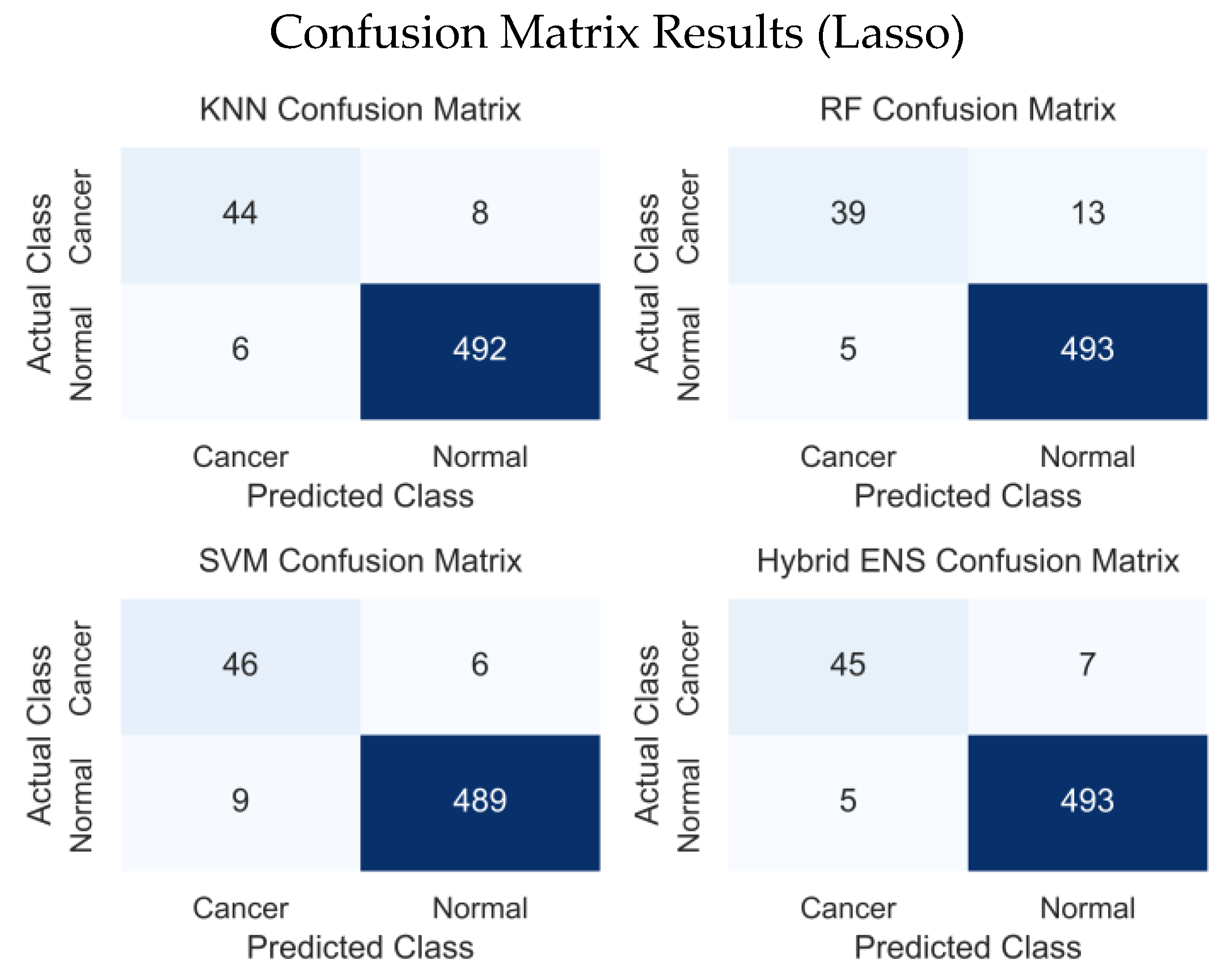
| Lasso | 0.9782 | 0.9862 | 0.9899 | 0.9880 | 0.9868 | 0.8589 | |
| Ridge | 0.9309 | 0.9356 | 0.9919 | 0.9629 | 0.9433 | 0.6983 |
| Classifier | Feature Selection | Dataset | Accuracy | Precision | Recall | F1-Score | AUC-ROC |
|---|---|---|---|---|---|---|---|
| Proposed Hybrid Ensemble Method | Lasso | LIHC | 0.9976 | 1.0000 | 0.9972 | 0.9986 | 1.000 |
| LUNG | 0.9965 | 0.9981 | 0.9981 | 0.9981 | 0.9997 | ||
| THCA | 0.9948 | 0.9980 | 0.9961 | 0.9971 | 0.9990 | ||
| PRAD | 0.9782 | 0.9862 | 0.9899 | 0.9880 | 0.9868 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Demiröz, A.; Aydın Atasoy, N. Explainable Model of Hybrid Ensemble Learning for Prostate Cancer RNA-Seq Classification via Targeted Feature Selection. Electronics 2025, 14, 4050. https://doi.org/10.3390/electronics14204050
Demiröz A, Aydın Atasoy N. Explainable Model of Hybrid Ensemble Learning for Prostate Cancer RNA-Seq Classification via Targeted Feature Selection. Electronics. 2025; 14(20):4050. https://doi.org/10.3390/electronics14204050
Chicago/Turabian StyleDemiröz, Ahmet, and Nesrin Aydın Atasoy. 2025. "Explainable Model of Hybrid Ensemble Learning for Prostate Cancer RNA-Seq Classification via Targeted Feature Selection" Electronics 14, no. 20: 4050. https://doi.org/10.3390/electronics14204050
APA StyleDemiröz, A., & Aydın Atasoy, N. (2025). Explainable Model of Hybrid Ensemble Learning for Prostate Cancer RNA-Seq Classification via Targeted Feature Selection. Electronics, 14(20), 4050. https://doi.org/10.3390/electronics14204050







