1. Introduction
Since colorectal cancer (CRC) is among the top causes of cancer-related deaths globally, improving patient outcomes critically depends on early and accurate diagnosis [
1]. Histopathological image analysis plays a central role in CRC detection, enabling pathologists to examine tissue architecture for malignant features. However, manual assessment is inherently subjective, time-consuming, and subject to inter-observer variability, which can impact diagnostic reliability [
2,
3].
Deep learning approaches, particularly Convolutional Neural Networks (CNNs), have rapidly gained traction for automating and enhancing the classification of colorectal histology images. Architectures like VGG, ResNet, and EfficientNet have shown competitive performance in a range of medical image classification tasks, including CRC diagnosis. Specifically, ResNet50V2 is valued for its identity mapping and residual connections, which strike an effective balance between network depth, classification accuracy, and computational efficiency [
4,
5,
6].
Despite these strengths, traditional CNNs often miss nuanced spatial hierarchies and contextual relationships inherent to histological textures features crucial for distinguishing benign from malignant patterns [
7]. This limitation motivates ongoing research into incorporating attention mechanisms and alternative architectures to capture more complex histopathological features and to further improve classification robustness and interpretability [
8].
Attention mechanisms have become a powerful enhancement to Convolutional Neural Networks (CNNs), particularly in histopathological image analysis. They enable models to selectively suppress less informative regions while focusing on diagnostically significant patterns, thereby advancing feature representation and boosting classification accuracy [
9]. Mechanisms such as channel attention, spatial attention, and composite approaches like the Convolutional Block Attention Module (CBAM) have demonstrated significant efficacy in guiding the network towards the most relevant tissue areas, which is critical for reliable cancer detection and grading. When combined with robust architectures such as ResNet50V2, attention modules can address limitations in conventional CNNs by capturing nuanced spatial hierarchies and contextual relationships within histological images. This results in more accurate identification of malignant vs. benign structures, a cornerstone for clinical applicability.
Complementing these methods, multi-scale visualization approaches including sliding windows, hierarchical patch extraction, and advanced aggregation modules promote resilience against spatial diversity in tissue morphology. By facilitating analysis at multiple magnifications and aggregating local and global context, these strategies support more interpretable outcomes [
10]. For example, they enable the reconstruction of interpretable heatmaps that highlight malignant regions and provide comprehensive tissue-level insights, improving performance and helping pathologists in practical diagnostic scenarios.
The recent development of deep learning in histopathology has been noted by a paradigm shift from convolutional networks (CNNs) towards transformer-based architectures, better at modeling long-range spatial dependencies and global context. While foundational, our initial focus on CNNs and attention mechanisms (CBAM) did not fully acknowledge this broader trend. Seminal work now includes hybrid CNN–Transformer models for multi-class classification [
11], self-supervised Vision Transformers for grading across institutions [
12], and dedicated transformer-based MIL frameworks that set new benchmarks in weakly-supervised classification [
13]. DeepFocusNet is architected within this landscape, drawing inspiration from the global context modeling of transformers but seeking to achieve a more computationally efficient and interpretable solution for clinical-grade whole-slide analysis.
In this work, we introduce a tailored DeepFocusNet framework for colorectal cancer histology classification. Enhanced by attention mechanisms and multi-scale visualization, our model not only achieves excellent accuracy but also produces detailed, interpretable heatmaps that identify malignant foci. This approach meaningfully contributes to clinical workflows, offering both quantitative reliability and deep visual interpretability for pathologists in real-world diagnosis.
Figure 1. provides a visual summary of the study, highlighting the key steps and workflow.
DeepFocusNet introduces three key innovations over existing methods.
First, it integrates attention mechanisms that focus on diagnostically relevant regions, improving both accuracy and interpretability.
Second, it employs a scalable tiling strategy, dividing large whole-slide images into high-resolution windows and reconstructing them into probability heatmaps and multi-class overlays for precise spatial localization.
Third, it uses a progressive training pipeline with augmentation, fine-tuning, and selective layer freezing to reduce overfitting and enhance generalization. Together, these innovations enable superior diagnostic precision, stability, and clinical relevance compared to prior models.
The remainder of this paper is organized as follows.
Section 2 provides an overview of related work in deep learning for medical image analysis, with a focus on histopathology.
Section 3 describes the Dataset Acquisition and Description, Data Preprocessing, and data Augmentation pipeline. In
Section 4 we describe the proposed DeepFocusNet architecture and its underlying design principles.
Section 5 shows the experimental setup, training and convergence analysis, qualitative analysis, quantitative performance and comparative analysis of different models. In
Section 6 we discuss interpretation of the results, clinical implications and limitations. Finally,
Section 7 concludes the paper and outlines potential directions for future research.
2. Related Work
Automated histopathological image classification has gained significant momentum in recent years due to its critical role in cancer detection and the growth of digital pathology repositories. To enhance the precision and reliability of colorectal cancer (CRC) histological classification, researchers have explored both traditional machine learning and advanced deep learning strategies.
Early studies in histopathology image classification relied primarily on hand-crafted features such as texture, color histograms, and morphological descriptors. Classifiers like support vector machines (SVM), random forests (RF), and k-nearest neighbors (k-NN) were standard approaches in these studies. For instance, one of the first publicly available colorectal histology datasets, developed by Kather et al., enabled assessment of various feature-based methods, which demonstrated only moderate accuracy and faced challenges in generalizing across different tissue structures and staining variations. Recent studies confirm that these traditional models, even when using sophisticated geometric features, perform notably below deep learning approaches, with CNN-based models achieving higher detection accuracy and sensitivity [
14,
15]. With the advent of Convolutional Neural Networks (CNNs), deep learning has become the dominant paradigm in histopathology image categorization. Following the influential work by Coudray et al. on predicting genetic mutations from lung cancer slides using deep CNNs, similar techniques were adapted for CRC.
State-of-the-art CNN architectures such as VGG16, ResNet50, and InceptionV3 have been fine-tuned and evaluated on CRC datasets, producing superior results compared to earlier methods. These efforts include models that specialize in nuclei or gland detection, though dense manual annotations and a lack of ability to model large-scale tissue context remain limitations [
16]. Given the scarcity of labeled data in medical imaging, transfer learning is prevalent in histopathology. Pretrained networks such as VGG16, ResNet50, and EfficientNet (originally trained on ImageNet) are commonly adapted by fine-tuning on domain-specific datasets. This approach enables networks to generalize to medical image domains with minimal retraining and has been particularly successful in distinguishing glandular from non-glandular tissue in CRC [
17]. Interpretability remains a major concern in medical AI. Deep learning models are frequently interpreted via Grad-CAM, saliency maps, and probability heatmaps, which highlight areas within histology slides that contributed to a model’s prediction. Such techniques not only improve transparency but also facilitate validation of model decisions by clinicians, as demonstrated by overlaying probability maps on tissue images [
18].
Recent years have seen the integration of attention mechanisms into CNNs, empowering models to focus on salient image regions while suppressing less informative areas. For instance, self-attention models for cancer histology have increased classification accuracy by selectively weighting discriminative regions, and dual-attention strategies combining spatial and channel attention have further improved pattern recognition, especially in challenging tissue contexts [
19,
20,
21].
Table 1. summarizes colon cancer classification methods and results.
Due to the enormous size of whole-slide images (WSIs), patch-based and multi-scale analysis have become best practices. By extracting and aggregating patches at various magnifications, models efficiently capture both micro-level details and broader contextual information, markedly improving classification performance. Multi-scale aggregation and hierarchical fusion approaches are reported to further enhance resilience against variability in tissue morphology and staining, allowing networks to integrate complementary features across resolutions [
29,
30,
31,
32].
Prior art in computational histopathology is largely bifurcated: CNN-based approaches, often enhanced with attention, struggle to take long-range dependencies essential for gigapixel WSIs, while emerging pure transformer models usually incur prohibitive computational costs and offer limited interpretability, a critical barrier for clinical adoption [
33,
34,
35]. DeepFocusNet is designed to navigate this trade-off. It organized the transformers’ global contextual awareness characteristic through its hierarchical attention mechanism. Still, it retains the spatial efficiency of CNNs, thereby addressing key limitations in both offspring of research.
In summary, the evolution from hand-crafted feature extraction to advanced deep learning, attention mechanisms, and multi-scale analysis has significantly improved CRC histopathology classification performance, setting the stage for robust AI-aided diagnostic workflows in clinical practice.
3. Dataset
3.1. Dataset Acquisition and Description
The NCT Biobank, made available to the public via its open-access repository by the Department of Pathology at NCT, is the source of the colorectal histology dataset utilized in this investigation. Hematoxylin and eosin (H&E)-stained digital histological images from colorectal tissue samples are included in the collection to aid in computational pathology research.
A carefully selected subset of the collection consists of 5000 color JPEG photos. These images were extracted from whole-slide images scanned at 20× magnification, with each image patch having a spatial resolution of 150 × 150 pixels and a physical size of approximately 75 × 75 µm (0.5 µm/pixel). The collection assigns eight histological tissue classes, tumor, stroma, complex, lympho, debris, mucosa, adipose, and empty, to each image, as shown in
Figure 2. Supervised machine learning for multi-class classification is made possible by these expert-provided labels. The dataset structure follows the rules of the TensorFlow Datasets (TFDS) framework, which facilitates direct comparison with other studies and ensures our results are easily reproducible. We also included large-scale whole-slide images (WSIs) with a size of 5000 × 5000 pixels to further assess the translational applicability of the suggested pipeline. These images more accurately depict the morphological complexity and variety found in actual clinical procedures. When taken as a whole, this dataset offers a strong basis for the creation and assessment of deep learning-based frameworks for the categorisation of colorectal cancer histology.
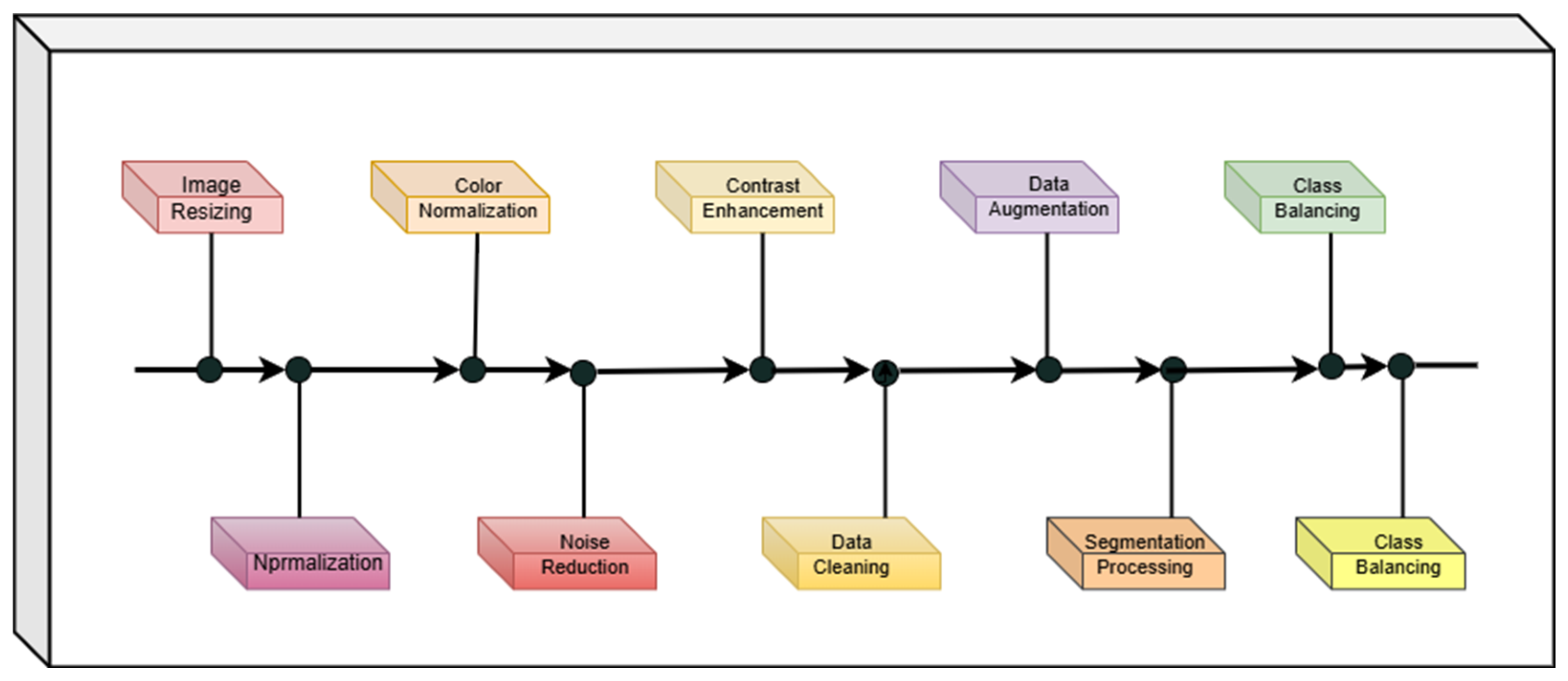
3.2. Data Preprocessing and Augmentation Pipeline
To ensure data quality and improve the robustness of the proposed model, a comprehensive preprocessing and augmentation pipeline was employed. First, all images were resized or resampled to fixed dimensions, with pixel spacing rescaled for consistency across samples.
For histopathological images, stain normalization was performed to minimize inter-sample variability caused by staining protocols. Noise reduction was addressed through median or Gaussian filtering, and in some cases, denoising autoencoders were employed. To further enhance image quality, contrast adjustment was applied using Contrast Limited Adaptive Histogram Equalization (CLAHE). Data cleaning was also performed to ensure the quality of the dataset. Our exclusion criteria involved removing images that were corrupted (e.g., decoding errors), exact duplicates, or of low quality. Low-quality images were defined as those with significant blur, out-of-focus regions covering more than 50% of the patch, or excessive staining artifacts that obscured cellular morphology. This curated a reliable dataset and prevented the model from learning from non-physiological features, which could otherwise bias training. To improve the model’s generalization in the presence of limited labeled data and intrinsic biological variability, a comprehensive data augmentation pipeline was employed, mathematically formalized as a stochastic composite transformation:
where
denotes random horizontal flipping,
represents a rotation operator with θ∼U(−30°, 30°) and
is a zoom scaling with factor s∼U(0.8, 1.2). Pixel intensity normalization was applied to scale inputs within [0, 1], optimizing the network’s convergence properties by standardizing input distributions.
During training, images underwent a series of data augmentations to improve generalization and robustness. Each image had a 50% chance of being randomly flipped horizontally , was rotated by an angle θ uniformly sampled from −30° to 30° (), and was scaled by a zoom factor s sampled from 0.8 to 1.2 () to simulate variations in object size and viewpoint. These transformations increased dataset diversity and reduced overfitting by encouraging the model to learn rotation- and scale-invariant features.
The 5000-image dataset was split at the patient level to prevent information leakage and ensure a robust evaluation of the model’s generalization capability. We used a standard 70-15-15 split, resulting in 3500 images for the training set, 750 images for the validation set, and 750 images for the independent test set. Finally, images were converted into the required format (e.g., DICOM to PNG/JPEG) to standardize input for the deep learning framework.
A schematic overview of the complete preprocessing and augmentation pipeline is presented in
Figure 3, which visually summarizes each step described above.
4. Proposed Methodology
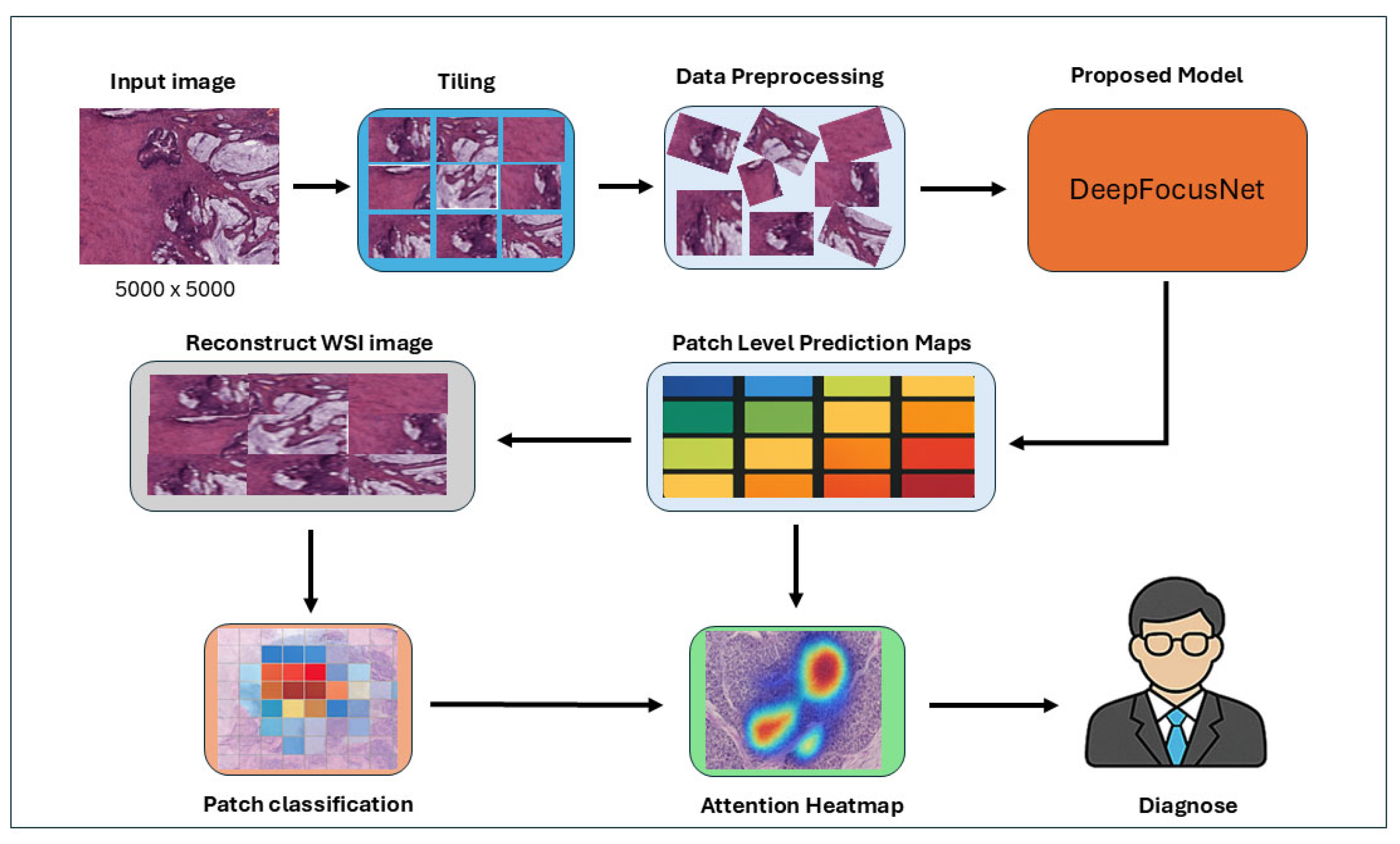
4.1. Overall Pipeline Architecture
The proposed pipeline processes large whole-slide histopathology images by initially dividing them into smaller tiles via a dedicated tiling module. Each slide, a colorectal tissue specimen stained with Hematoxylin and Eosin (H&E), is preprocessed and segmented into regions of interest (ROIs) capturing a broad range of histopathological structures including normal stroma, normal glands, lymphocytic infiltrates, necrotic regions, and multiple grades of adenocarcinoma. Data augmentation techniques are subsequently applied to these tiles, enriching the training dataset and enhancing model generalization. The augmented patches are then fed into a DeepFocusNet architecture for robust feature extraction and classification across eight histological classes, ensuring exposure to both normal and malignant tissue morphologies and supporting the learning of discriminative features throughout the full spectrum.
Patch-level predictions are aggregated to reconstruct a whole-slide prediction map, while an attention heatmap is generated in parallel to highlight diagnostically significant regions within the tissue. This integrated system provides both patch-level classifications and interpretable visual explanations, thereby supporting clinicians in accurate diagnostic decision-making and improving the interpretability of the deep learning process.
Figure 4 provides a schematic overview of the proposed workflow.
4.2. DeepFocusNet Architecture
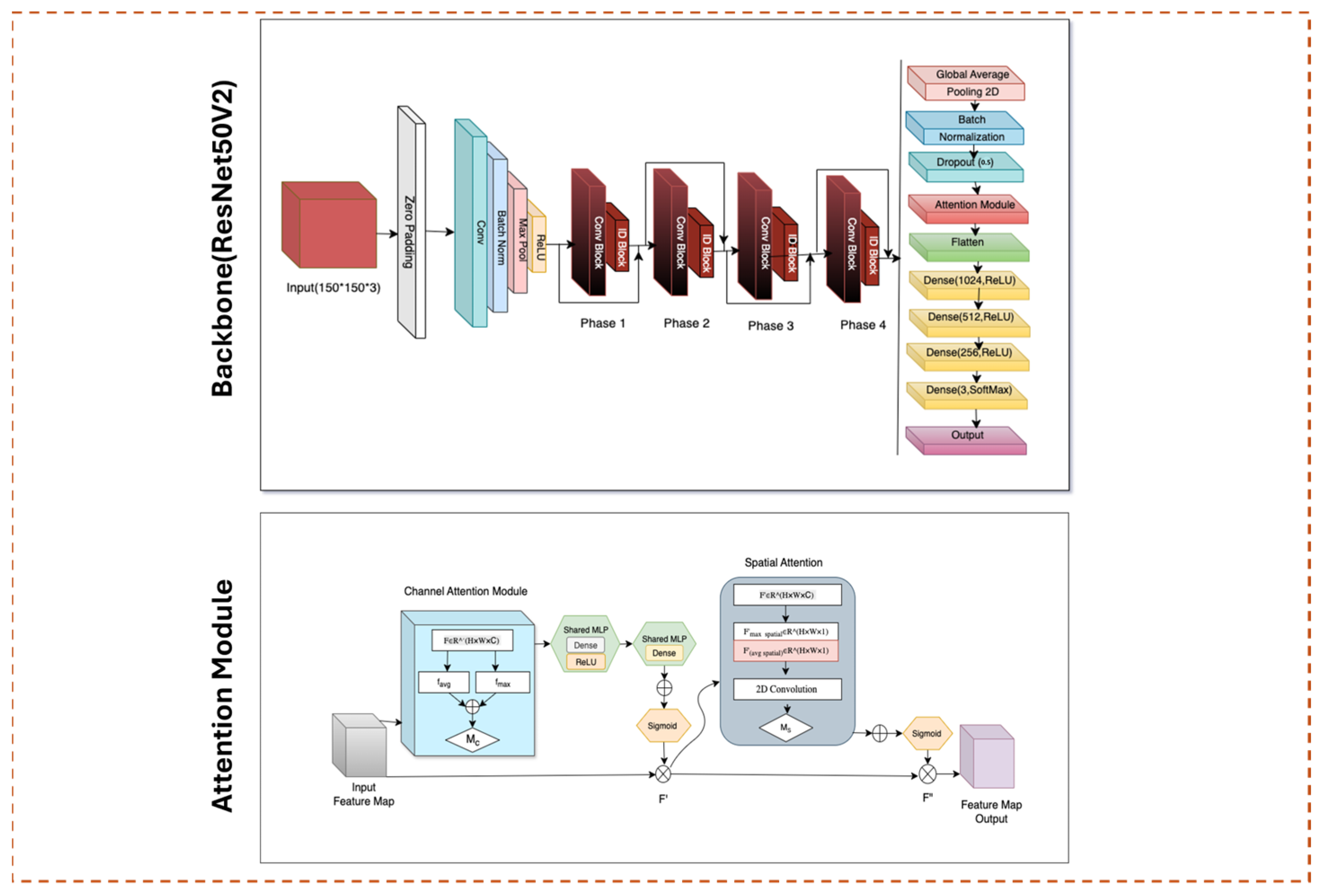
The backbone of our proposed DeepFocusNet framework is the ResNet50V2 architecture, which employs residual learning to address the vanishing gradient problem commonly encountered in deep convolutional neural networks. Residual blocks utilize identity skip connections, allowing the network to learn residual functions relative to the layer inputs. This relationship can be expressed as:
Here, x denotes the input tensor to the residual block, F(⋅) represents the residual mapping learned by convolutional layers with parameters and y is the output of the block. The identity skip connection preserves gradient flow, facilitating the training of deep networks and improving feature propagation.
The pre-activation design of ResNet50V2 further stabilizes gradient flow by applying batch normalization and activation before convolution operations. This structure enhances optimization efficiency and leads to faster and more stable convergence during training.
To enhance the spatial feature extraction capabilities of ResNet50V2, we incorporate the Convolutional Block Attention Module (CBAM), which adaptively recalibrates intermediate feature representations through sequential channel and spatial attention mechanisms as shown in
Figure 5.
The channel attention module computes an importance map
by aggregating spatial features through global average pooling
and global max pooling
These pooled features are passed through a shared multilayer perceptron (MLP) followed by a sigmoid activation function σ\sigma:
where F is the input feature map. This process emphasizes the most informative channels for the classification task.
The spatial attention module subsequently generates a spatial attention map
by concatenating channel-wise average-pooled and max-pooled features along the channel axis, applying a convolutional filter
, and using a sigmoid activation:
where
represents the channel-refined feature map from the previous stage. This spatial attention mechanism enhances the localization of salient regions within the feature maps, enabling the model to focus on diagnostically important morphological structures in histopathological images.
By combining residual connections with CBAM, the model leverages both the preservation of low-level details and enhanced contextual awareness. This synergy creates a robust and discriminative feature hierarchy, thereby improving the accuracy and reliability of colorectal cancer classification.
4.3. Optimization Regime and Convergence Strategies
Model optimization employed the Adam algorithm—an adaptive moment estimation method, with an initial learning rate of η =
, optimizing the categorical cross-entropy loss function:
where
C is the number of classes,
denotes the ground truth, and
the predicted class probability. To mitigate overfitting and ensure convergence stability, early stopping was implemented by monitoring the validation loss with a patience threshold of 10 epochs. A dynamic learning rate scheduler further reduced η by a factor of 0.5 upon plateau detection. Training was conducted for up to 100 epochs with a mini-batch size of 32, balancing computational tractability with the model’s capacity to capture complex histopathological patterns.
5. Experiments and Results
5.1. Experimental Setup
All experiments were implemented in TensorFlow 2.x and executed on a workstation equipped with an NVIDIA GeForce RTX 4060 Ti GPU, 32 GB RAM, and an Intel Core i7 processor, leveraging CUDA and cuDNN libraries for hardware-accelerated computation. The model was trained with a batch size of 32 and an input resolution of 224 × 224 pixels. Real-time data augmentation, including random rotations, shifts, zooms, and horizontal flips, was applied to enhance generalization performance. Model optimization was performed using the Adam optimizer with a learning rate of 1 × 10. Training proceeded until convergence, with early stopping and adaptive learning rate reduction on plateau employed to mitigate overfitting.
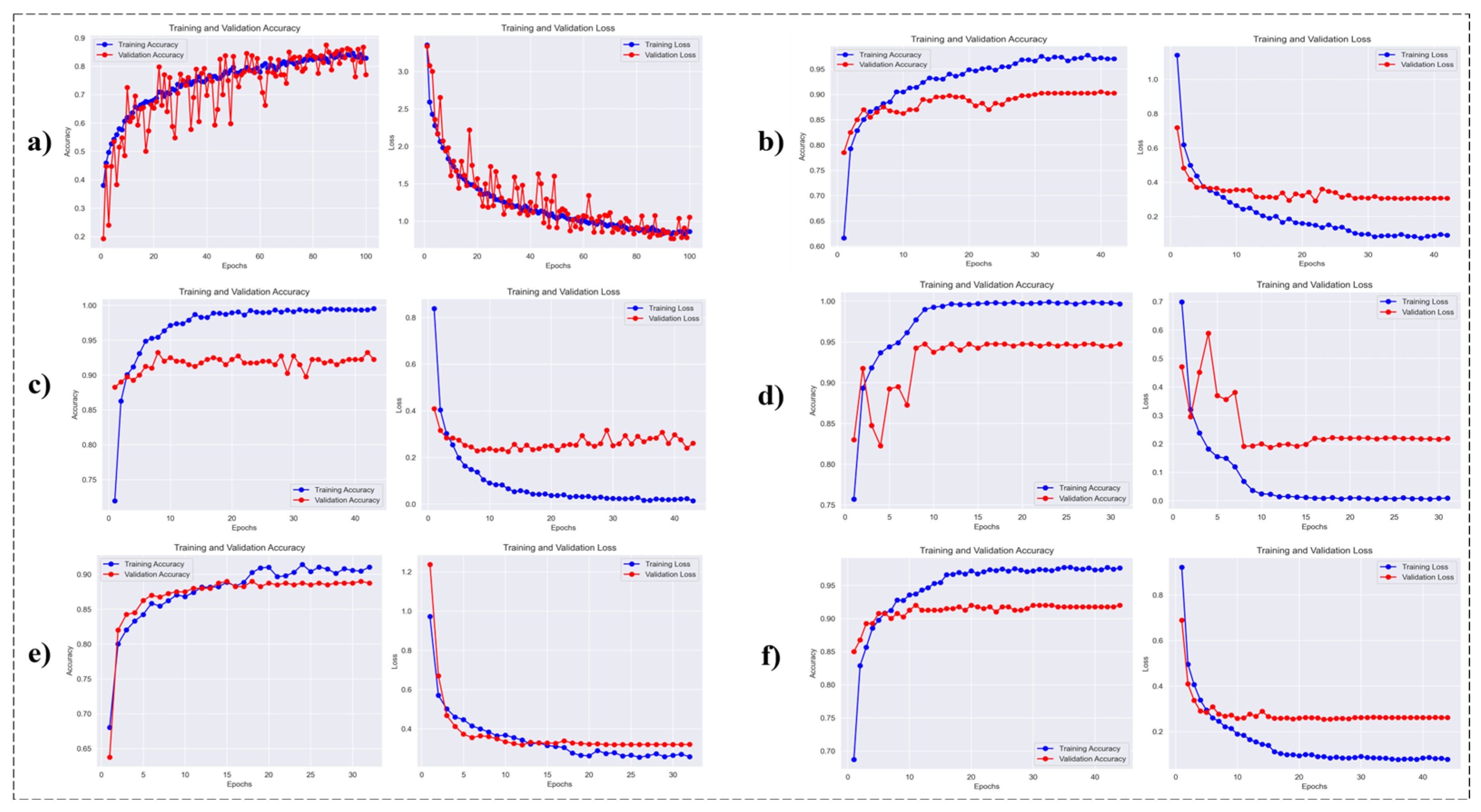
5.2. Training and Convergence Analysis
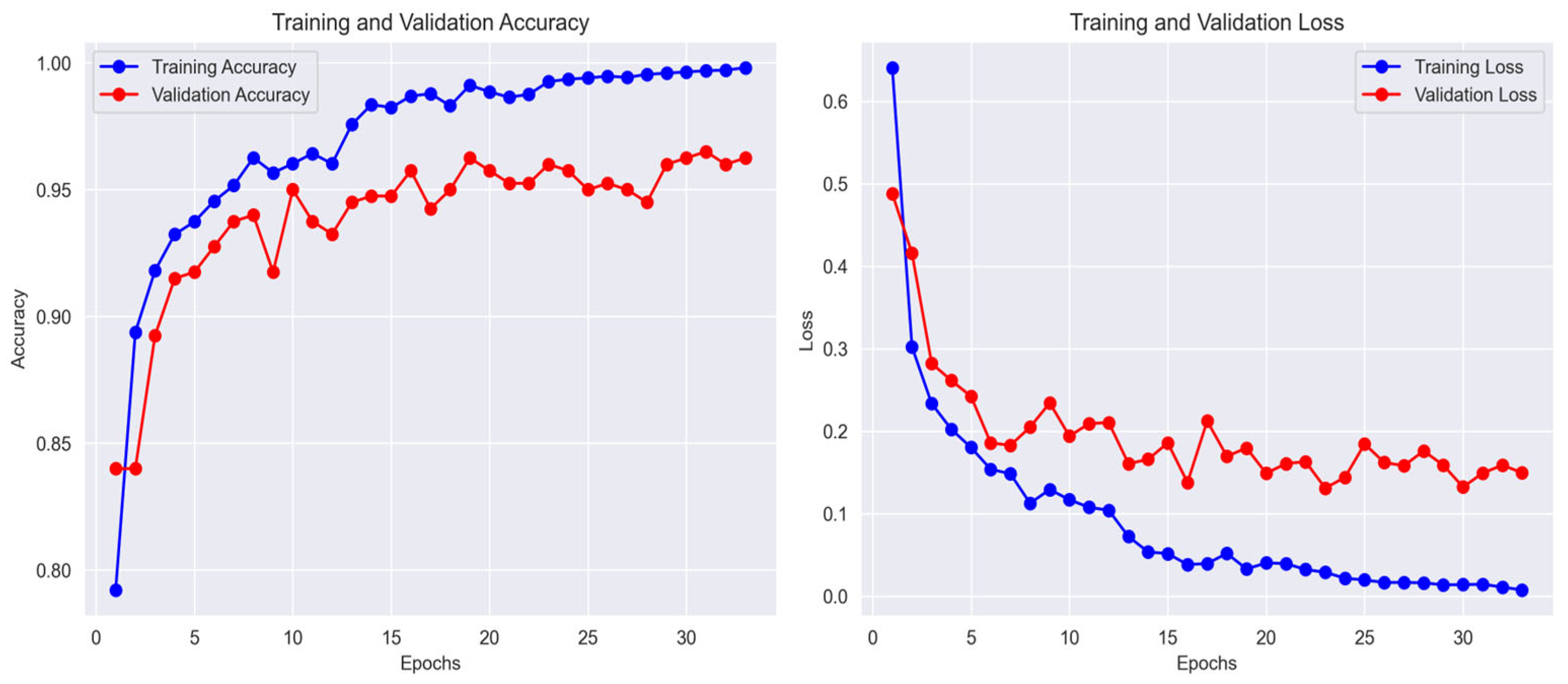
The learning curves of the DeepFocusNet model were analysed alongside five baseline architectures: a simple CNN, Inception, MobileNet, ResNet50V2, and VGG19 to highlight key differences in convergence patterns and generalisation performance. This analysis provides insights into the relative training stability and effectiveness of each network.
As depicted in
Figure 6, the DeepFocusNet model exhibits smooth and stable convergence, with accuracy steadily increasing and loss consistently decreasing during training. Although a modest gap between training and validation accuracy is observed—a common occurrence in deep learning—the proposed model demonstrates strong generalization. Upon completion of training, the model achieved a final training accuracy of 99.8%, a validation accuracy of 98.1%, and a test accuracy of 98.0%. The corresponding final loss values were 0.01 for training, 0.15 for validation, and 0.16 for the test set. This reflects its superior ability to generalize to unseen data compared to the other architectures. The corresponding loss curves also demonstrate a consistent downward trend, confirming the effectiveness of the optimization strategy.
A direct comparison of the final validation accuracies across all models is presented in
Figure 7. These graphs clearly demonstrate the superior performance of the proposed model, which achieves the highest validation accuracy among all evaluated architectures. This comparative analysis including models such as MobileNetV2 and Xception further underscores the advantages of incorporating attention mechanisms for enhancing classification accuracy in colorectal histopathology images.
This figure presents the training and validation accuracy and loss across epochs for the different deep learning architectures evaluated in this study.
5.3. Qualitative Analysis of Model Output
To gain insight into the model’s decision-making process, attention maps were generated and visualized.
Figure 8c presents an attention heatmap superimposed on the original histology image (
Figure 8a). The highlighted regions, shown in bright yellow and white, correspond to areas of high relevance such as dense cellularity, irregular glandular structures, and disorganized tissue hallmarks commonly associated with malignancy.
In addition, the model outputs detailed multi-class segmentation maps, illustrated in
Figure 8b. These maps classify tissue into eight distinct categories (0–7), including normal stroma (orange, Class 1) and malignant regions (purple, green and yellow; Classes 3, 4, and 5). The segmented boundaries are well aligned with morphological features, reflecting the model’s capacity to capture histopathological patterns.
Figure 9 further provides a qualitative grid representation: the first column displays original image patches, the second shows the corresponding multi-class classification maps, and the third depicts probability maps specific to the stroma class. The probability maps indicate the model’s confidence in identifying critical tissue types, thereby underscoring its reliability and practical utility in histopathological analysis.
5.4. Quantitative Performance Metrics
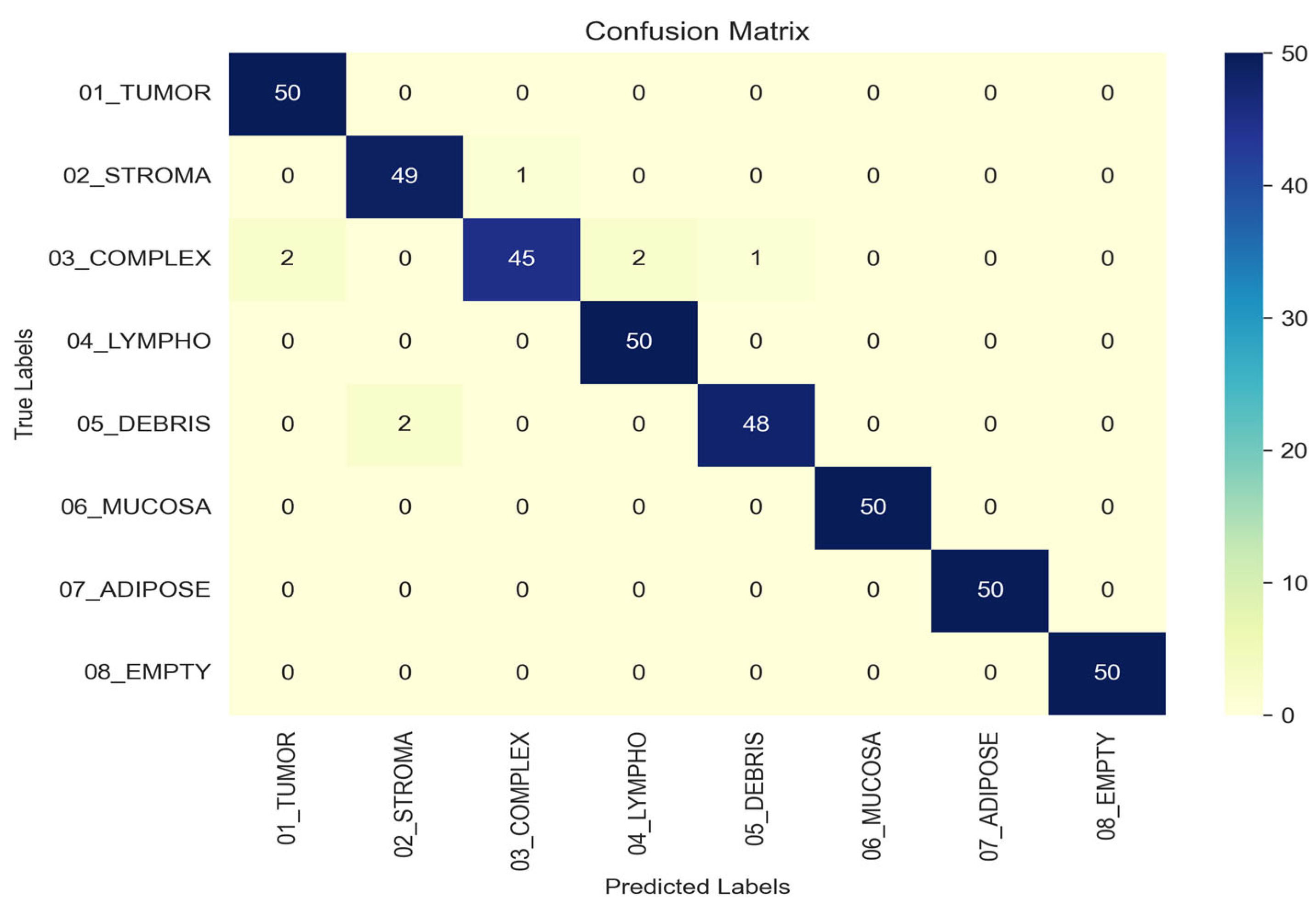
To rigorously assess the performance of the proposed model, we conducted an evaluation using a comprehensive set of quantitative metrics on the independent test set. The confusion matrix, presented in
Figure 10, provides a visual summary of the classification outcomes, where rows correspond to true labels and columns to predicted labels. The strong diagonal dominance observed in the matrix characterized by high values for correct classifications and minimal off-diagonal entries demonstrates the model’s high specificity and its low propensity for misclassifying tissue types. For the TUMOR, LYMPHO, MUCOSA, ADIPOSE, and EMPTY classes, all 50 samples were correctly classified. The STROMA class had 1 misclassification predicted as COMPLEX. The COMPLEX class had 5 misclassifications: 2 predicted as TUMOR, 2 as LYMPHO, and 1 as DEBRIS. The DEBRIS class had 2 misclassifications predicted as STROMA. This detailed breakdown highlights the model’s strengths and specific confusions between classes, enabling a more nuanced interpretation of overall classification performance.
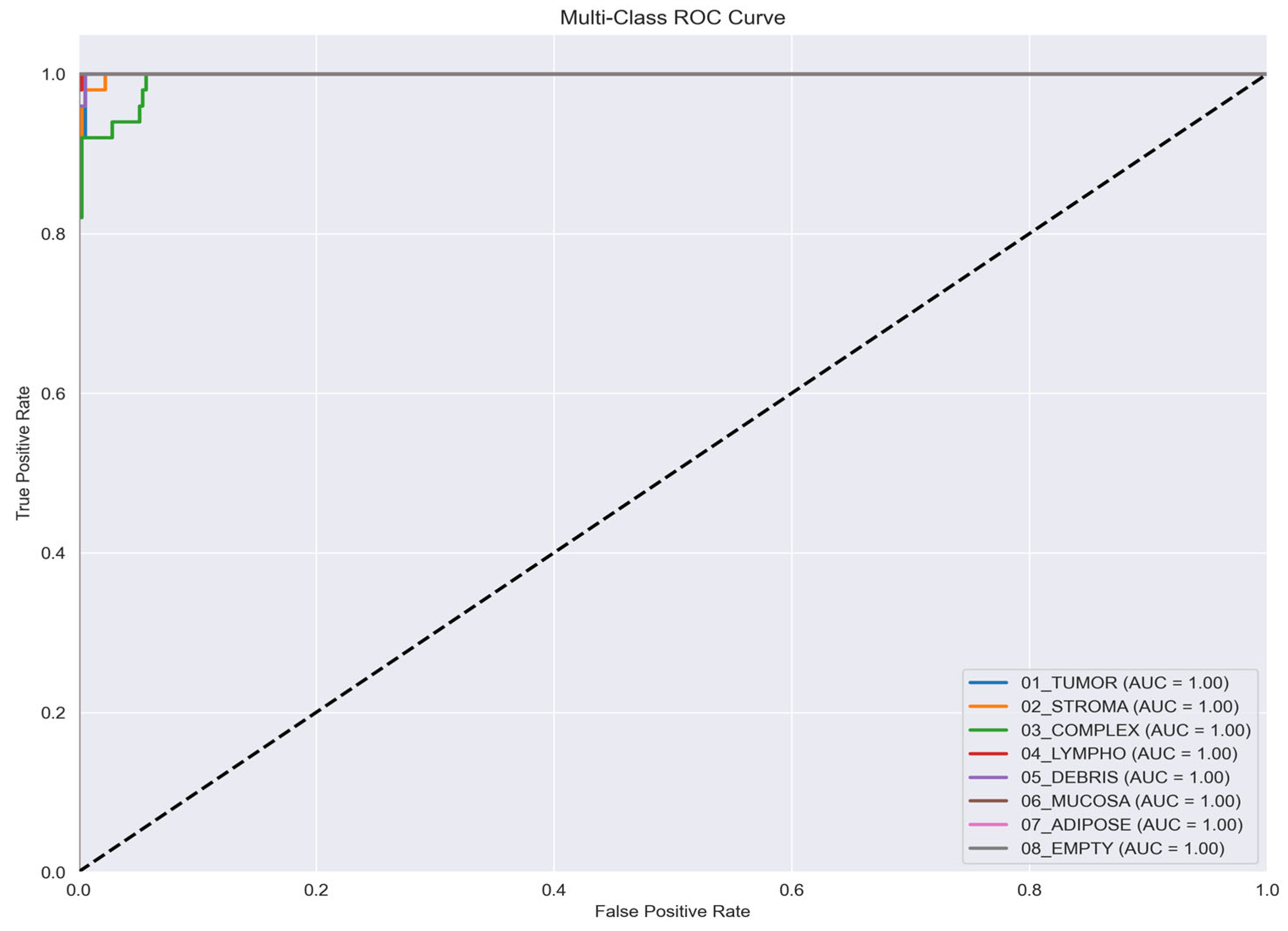
Further evaluation of the model’s discriminatory capacity is presented through the multi-class ROC curves in
Figure 11. The curves exhibit near-perfect Area Under the Curve (AUC) values of 1.00 across all classes, highlighting the model’s exceptional ability to differentiate between distinct tissue pathologies. Such uniformly high AUC scores demonstrate both the robustness of the classification framework and its high level of confidence in discriminating among complex histopathological patterns.
A detailed class-wise evaluation of the model’s performance is presented in
Table 2. The reported metrics Accuracy, Precision, Recall, and F1-Score provide a comprehensive quantitative assessment, reinforcing the robust performance trends illustrated by the confusion matrix and ROC curves. Collectively, these results validate the model’s reliability in achieving consistent and balanced classification across all tissue classes.
5.5. Comparative Analysis of Different Models
To validate the effectiveness of the proposed architecture, we conducted a comparative evaluation against several widely used deep learning models.
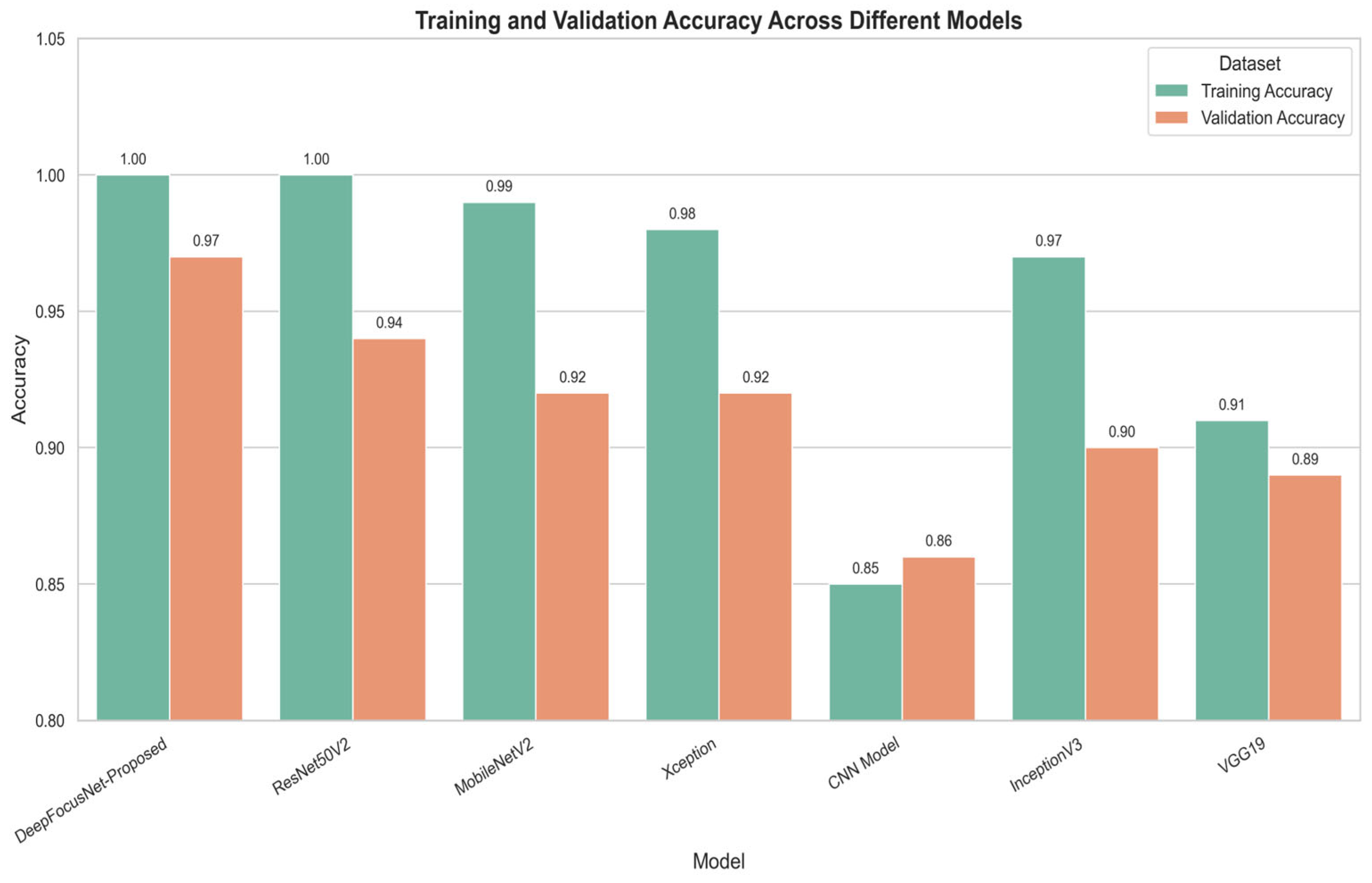
Figure 12 presents the training and validation accuracy of our attention-enhanced DeepFocusNet model in comparison with ResNet50V2, MobileNetV2, VGG19, Xception, a simple CNN, and InceptionV3.
As shown in
Figure 12, the proposed model achieves the highest validation accuracy of 0.97, outperforming all competing architectures. This improvement is particularly notable relative to the baseline ResNet50V2, which attained a validation accuracy of 0.94, thereby demonstrating the clear performance gain achieved through the integration of the CBAM attention mechanism.
The learning patterns further highlight differences in model generalization. While architectures such as MobileNetV2 and Xception achieved high training accuracy, they exhibited larger discrepancies between training and validation performance, suggesting a tendency toward overfitting. In contrast, our proposed model maintains a closer alignment between training and validation accuracy, reflecting superior generalization to unseen data.
Overall, this comparative analysis confirms that incorporating the CBAM attention mechanism substantially enhances classification performance for colorectal histopathology images. Importantly, the improved generalization capability translates to greater reliability in real-world diagnostic scenarios, where minimizing misclassification is critical for ensuring accurate and clinically meaningful outcomes.
6. Discussion
6.1. Interpretation of Results
The proposed DeepFocusNet model achieved consistently high classification accuracy and near-perfect AUC values across all classes, underscoring its strong capability to discriminate among different categories of colorectal tissue images. This superior performance can be attributed to the integration of the Convolutional Block Attention Module (CBAM), which applies channel and spatial attention sequentially to the extracted feature maps.
The channel attention mechanism enables the network to selectively emphasize the most informative feature channels, thereby amplifying critical histopathological signals. In parallel, the spatial attention mechanism directs the model’s focus toward the most diagnostically relevant regions of the tissue image, while suppressing irrelevant background features. This combined attention strategy effectively highlights pathological patterns such as irregular glandular architecture, nuclear atypia, and stromal abnormalities hallmarks essential for colorectal cancer diagnosis.
By jointly enhancing both feature selection and spatial localization, the CBAM substantially improves the model’s ability to capture fine-grained morphological variations. This not only explains the model’s quantitative performance gains but also reinforces its potential clinical utility, where reliable identification of subtle histopathological cues is paramount for accurate and early cancer detection.
6.2. Clinical Implications
The consistently high classification accuracy and near-perfect AUC values indicate that the proposed model has strong potential as a computer-aided diagnostic (CAD) tool for colorectal cancer detection. By delivering rapid and reliable second opinions, the model could assist pathologists in reducing diagnostic workload and minimizing human error, particularly in high-volume screening environments.
A key strength of the framework lies in its ability to generate attention maps that highlight diagnostically relevant regions. This feature not only enhances interpretability but also provides clinicians with greater transparency into the model’s decision-making process. Such explainability is critical for building trust in AI-driven systems and is a prerequisite for their successful adoption in routine clinical practice.
By improving diagnostic efficiency while maintaining interpretability, the model could serve as a valuable adjunct to pathologists, ultimately contributing to earlier detection and more accurate classification of colorectal cancer.
6.3. Limitations
Despite demonstrating strong performance, several important limitations warrant consideration. First, the dataset size, although adequate for experimental validation, may not encompass the full spectrum of variability present in routine clinical practice. Consequently, the model’s performance could be affected by domain shifts when encountering images acquired from different scanners, staining protocols, or diverse patient populations, potentially limiting generalizability. Second, the substantial computational resources required to train and deploy a deep attention-based architecture such as DeepFocusNet may restrict its practical use in low-resource environments. Finally, while attention mechanisms enhance interpretability, they do not fully elucidate the model’s decision-making process, indicating a need for further research to achieve comprehensive clinical explainability.
A key limitation of this study is its dependence on a single-institution dataset from the NCT Biobank. As a result, the model may inadvertently learn institution-specific scanner features and staining patterns rather than purely biologically relevant histopathological features, restricting its generalizability to other clinical centers. Performance may decline on whole-slide images acquired from different scanner manufacturers (Philips, Leica) or using alternative H&E staining protocols. Future work should prioritize external validation across diverse, multi-institutional cohorts to evaluate model portability and mitigate potential sources of bias.
Although the proposed architecture achieves state-of-the-art performance, its perplexity, particularly the attention mechanisms, places substantial computational demands, which could limit acceptance in clinical settings with constrained IT resources. We report the average inference time and GPU memory usage to contextualize these requirements. Future research will investigate model compression strategies, including structured pruning and FP16 quantization, to reduce the computational burden while maintaining diagnostic performance.
7. Conclusions
In this study, we introduced a novel attention-enhanced deep learning framework DeepFocusNet for automated colorectal cancer classification, achieved by integrating the Convolutional Block Attention Module (CBAM) with a ResNet50V2 backbone. The proposed model demonstrated exceptional performance, exhibiting high accuracy, precision, recall, and near-perfect AUC scores across all histological classes. Comparative analyses with standard CNN architectures further highlighted the superior capability of our approach in capturing critical histopathological features.
The principal finding underscores that the synergistic combination of channel and spatial attention markedly enhances the network’s discriminative power, establishing it as a potent and reliable tool for supporting pathologists in the early detection and classification of colorectal cancer.
For future research, we propose validating the model on larger and more heterogeneous datasets, investigating alternative attention mechanisms such as self-attention or transformer-based architectures, and integrating multi-modal data sources including genetic, clinical, and imaging information to further advance diagnostic performance. Furthermore, optimizing the model for computational efficiency would increase its accessibility and facilitate real-time application in clinical environments.