1. Introduction
Medical image segmentation is a critical component of medical image analysis, playing a vital role in clinical diagnosis, treatment planning, and disease research. Its applications range from tumor detection to organ segmentation, making it indispensable in modern medicine. However, obtaining high-quality annotated medical images faces significant challenges. These challenges are particularly pronounced in COVID-19 lesion segmentation, where the visual identification of lesions faces inherent difficulties due to low contrast boundaries between lesions like ground-glass opacities and surrounding lung tissue, heterogeneous manifestations appearing as diverse patterns including nodular, patchy, and diffuse across patients, and ambiguous margins with ill-defined edges that challenge precise delineation even for experts [
1,
2]. Accompanying textual annotations provide critical complementary information by specifying anatomical context, characterizing lesion attributes, and highlighting clinically relevant features that may be visually obscure in the images. On the one hand, annotation requires substantial time and effort from medical professionals, which results in high labor costs. On the other hand, the complexity and specificity of medical images make annotation more difficult compared with conventional images, while consistency across annotations is also hard to ensure. These factors severely limit the performance improvement of medical image segmentation models.
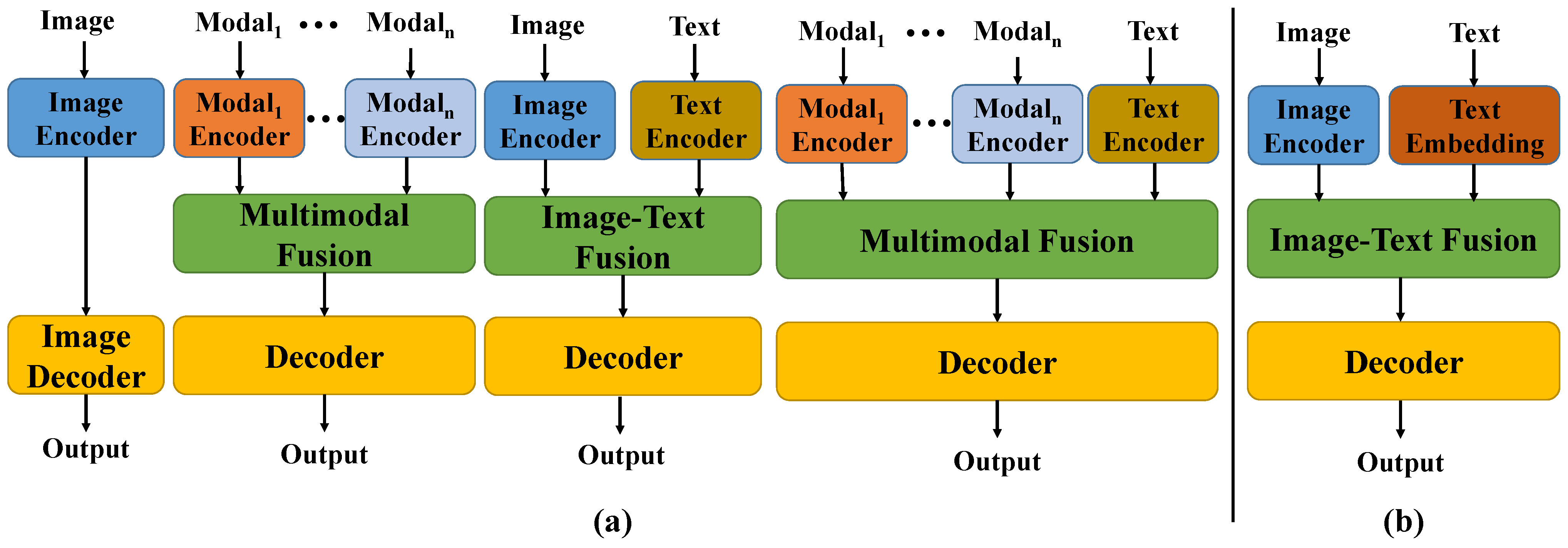
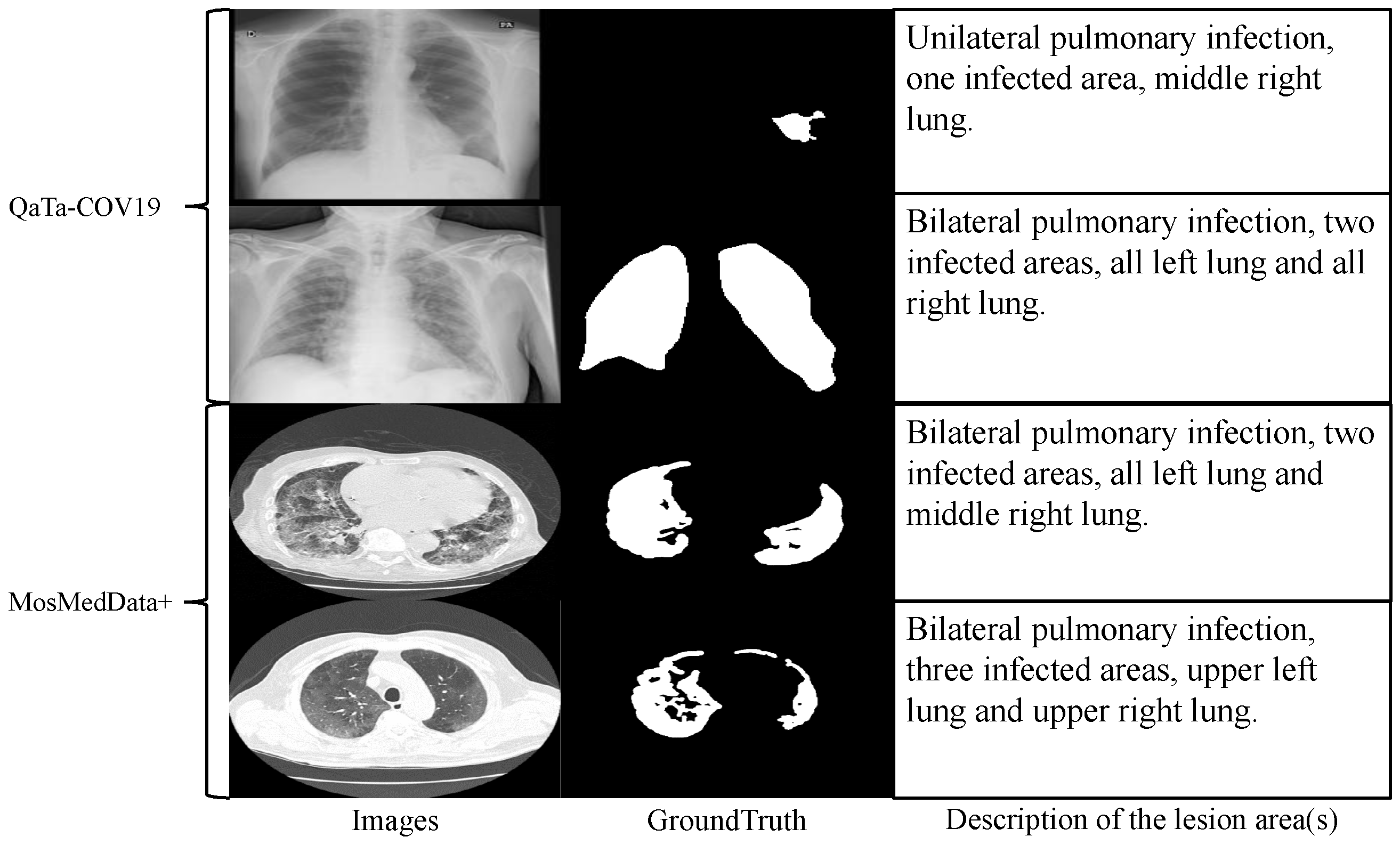
In recent years, although deep learning technologies have achieved remarkable results in this field, most existing models rely on large-scale annotated data for supervised learning and struggle to overcome the data bottleneck. In practical applications, as shown in
Figure 1a, medical images are often accompanied by textual annotations that contain rich semantic information, such as the location, shape, and number of lesions [
3]. Effectively integrating such textual information with visual data through a text encoder could bring new opportunities to segmentation tasks. However, the substantial parameter footprint of text encoders imposes a significant computational burden when used jointly with visual models. Therefore, adopting a lightweight text embedding scheme as an alternative to text encoders can enable high-accuracy text-assisted medical image segmentation with minimal parameter overhead. Meanwhile, medical images often exhibit blurred boundaries between different regions and low grayscale contrast, making accurate segmentation highly challenging [
4]. Therefore, more efficient feature fusion mechanisms and attention strategies are urgently needed to address this issue [
5].
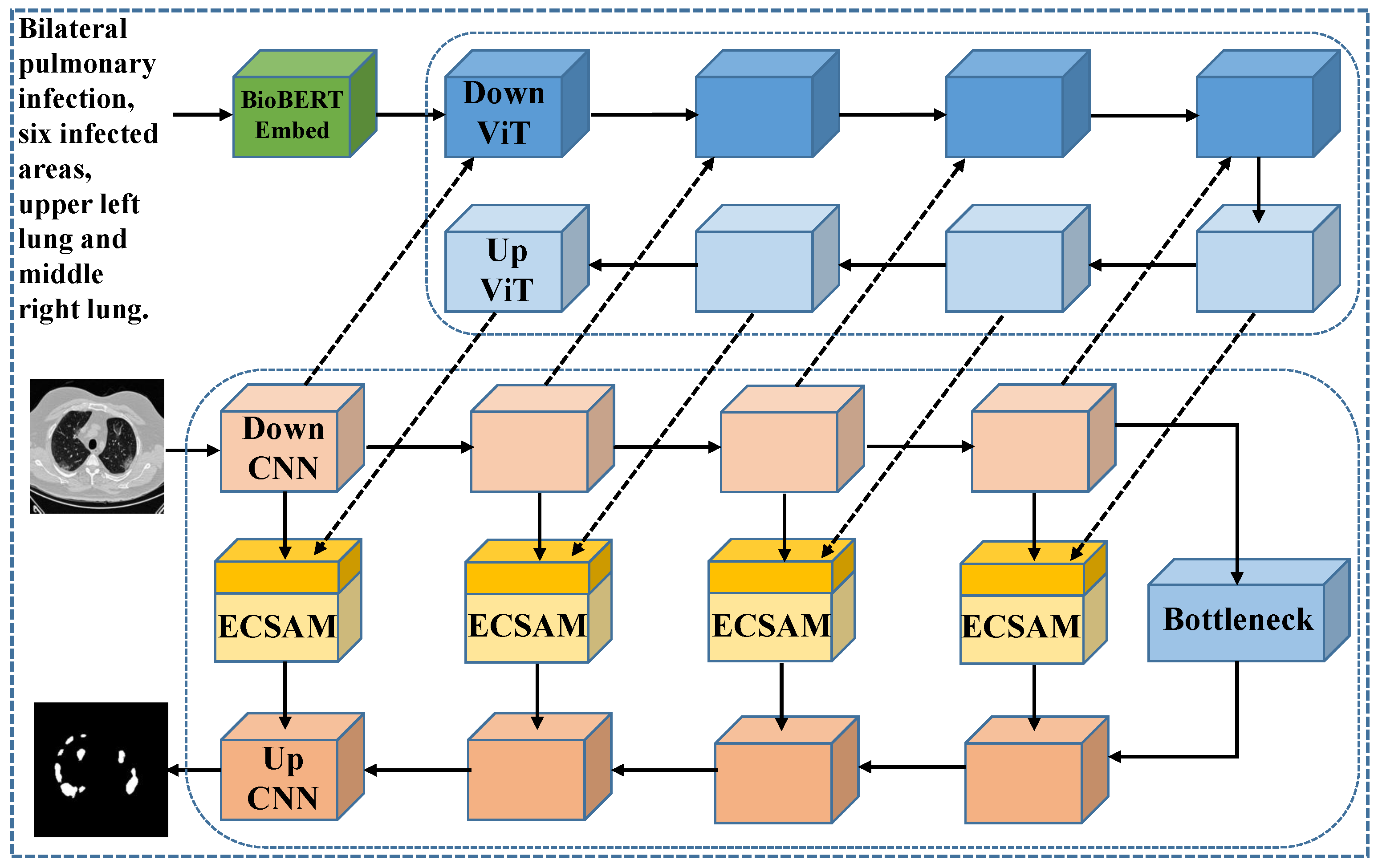
To address these challenges, we propose an innovative medical image segmentation approach that replaces the traditional text encoder with a novel, parameter-efficient text embedding method. Specifically, we introduce BioBERT [
6], a model pre-trained on large-scale medical literature and specifically designed for the biomedical domain. With its profound understanding of medical terminology and semantics, BioBERT outperforms general BERT [
7] in generating more clinically relevant text representations, thus providing a robust foundation for subsequent tasks.
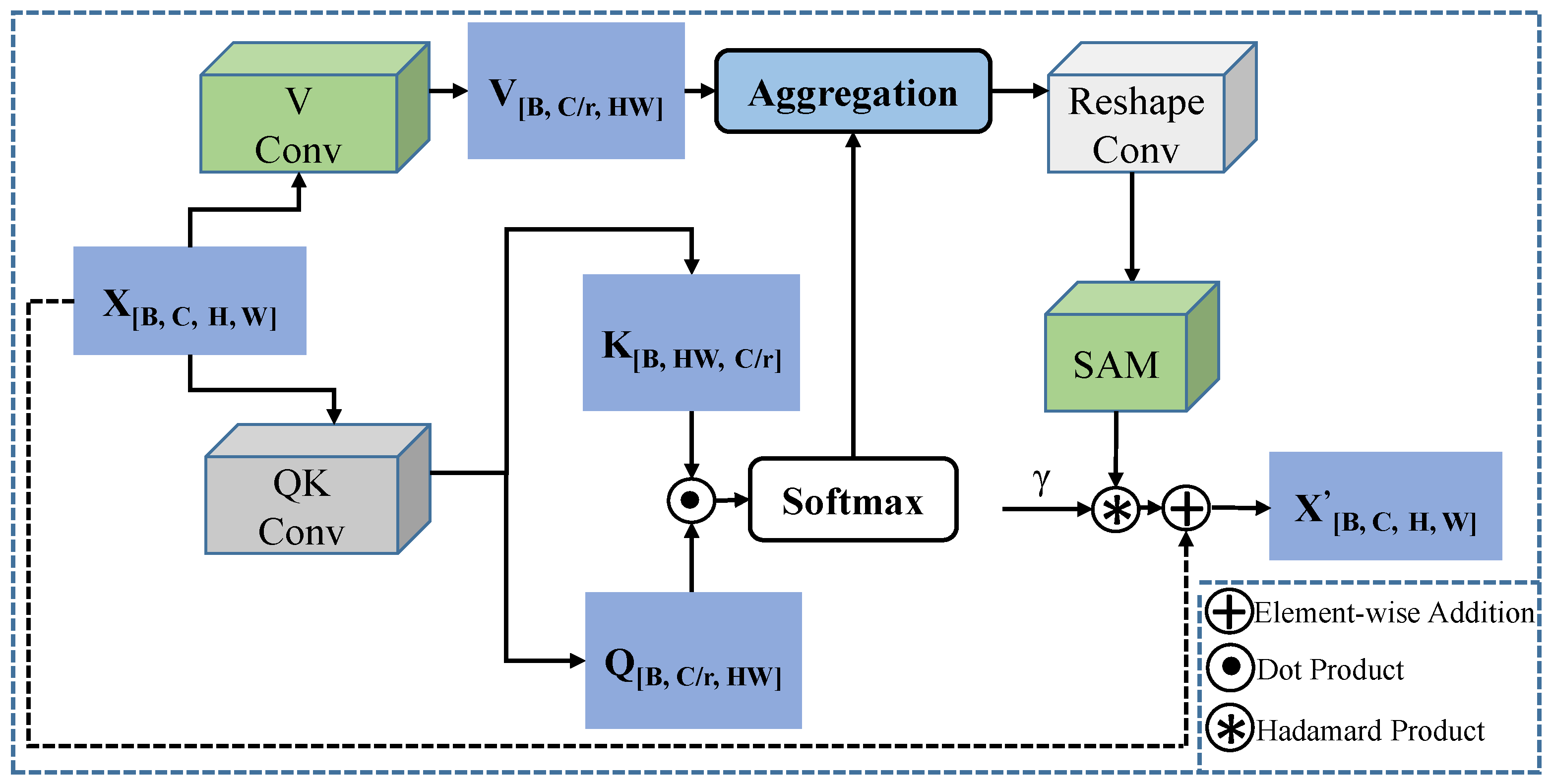
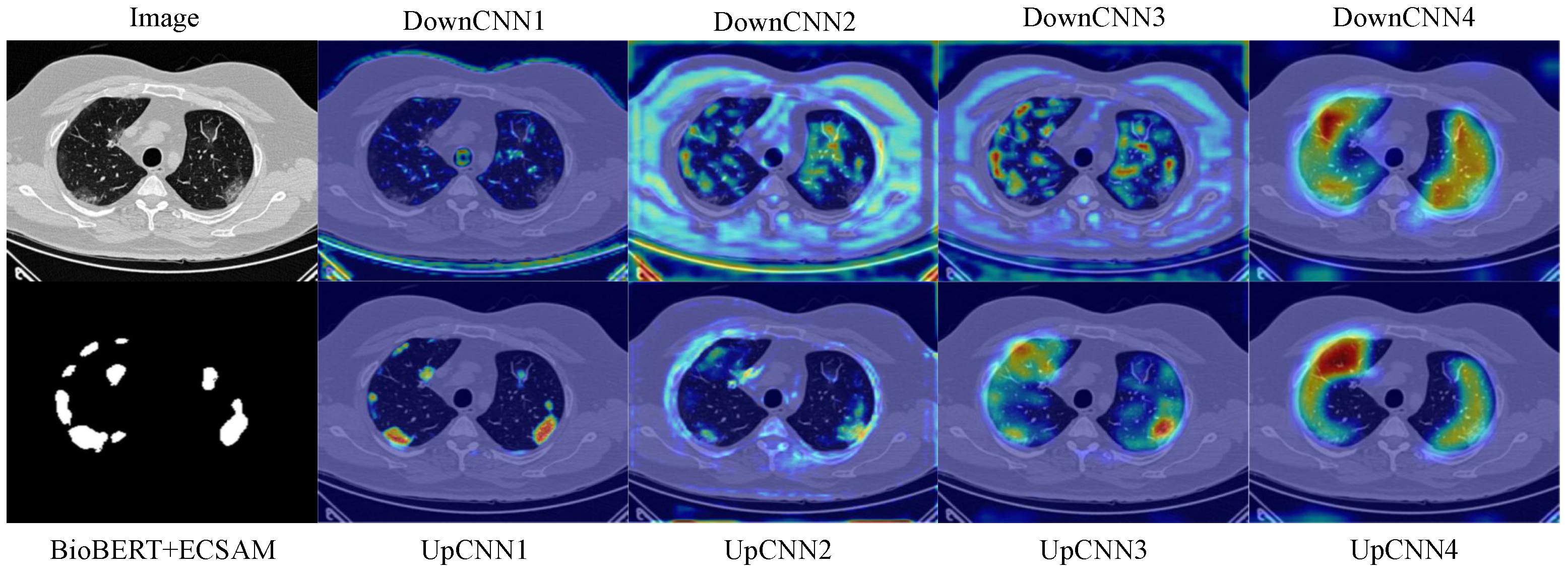
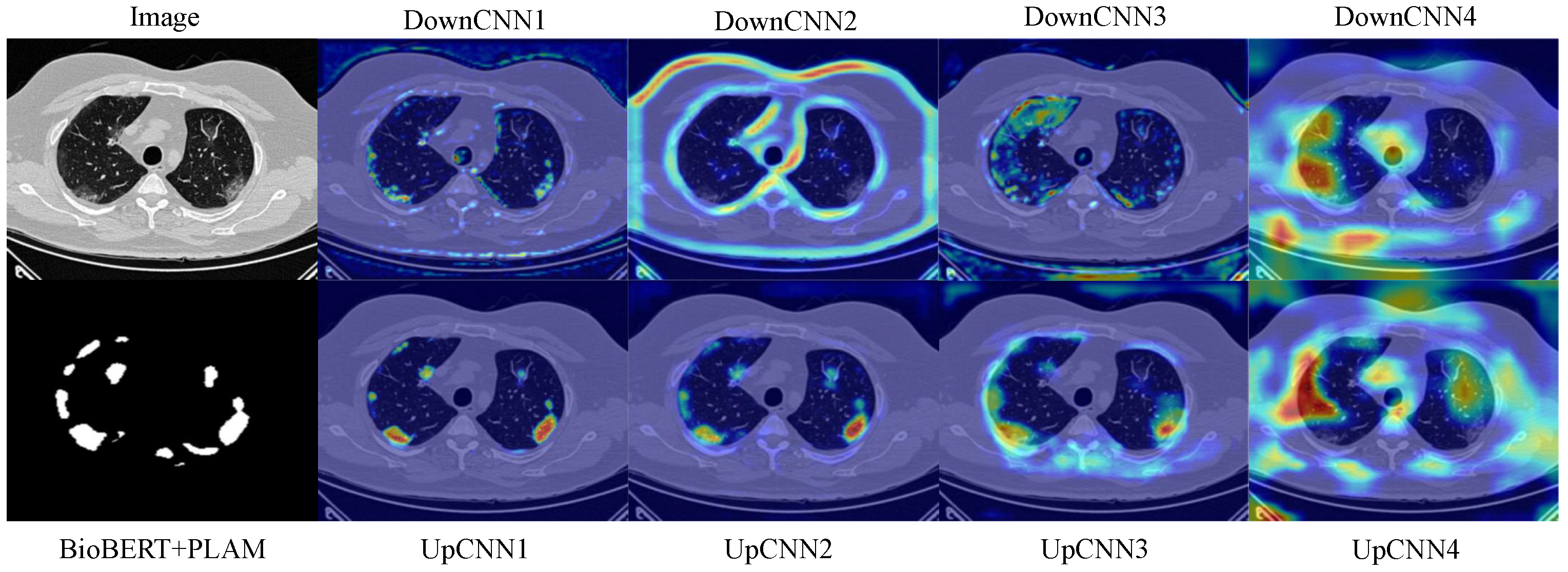
Additionally, we design a lightweight feature fusion module named the Enhanced Channel-Spatial Attention Module (ECSAM), which incorporates critical enhancements for multimodal feature fusion. Through its attention mechanism, ECSAM effectively captures cross-modal associations between images and text while enhancing MedLangViT’s focus on critical areas (e.g., lesion regions) and suppressing irrelevant information. This design significantly improves the discriminative power of feature representations.
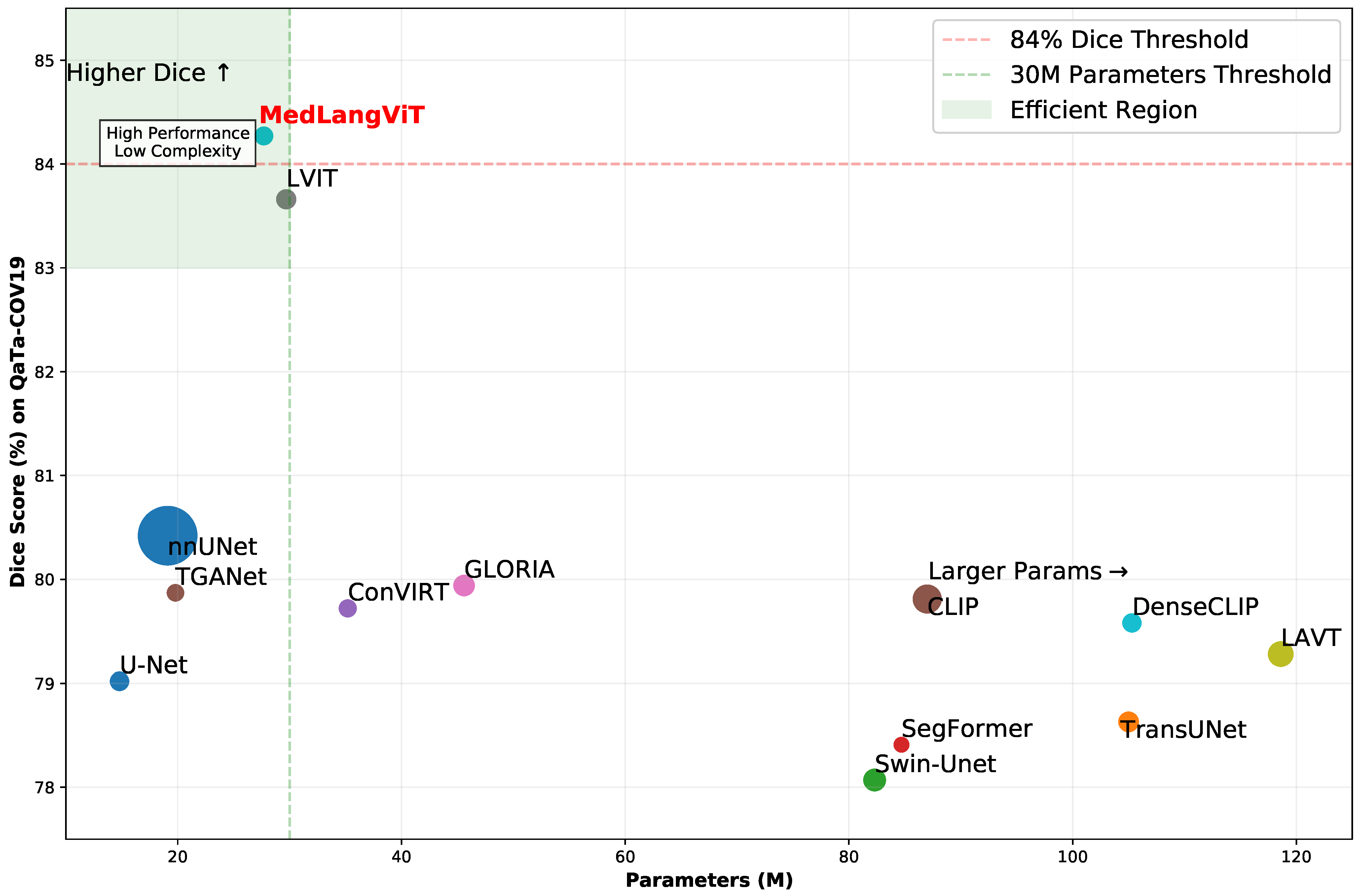
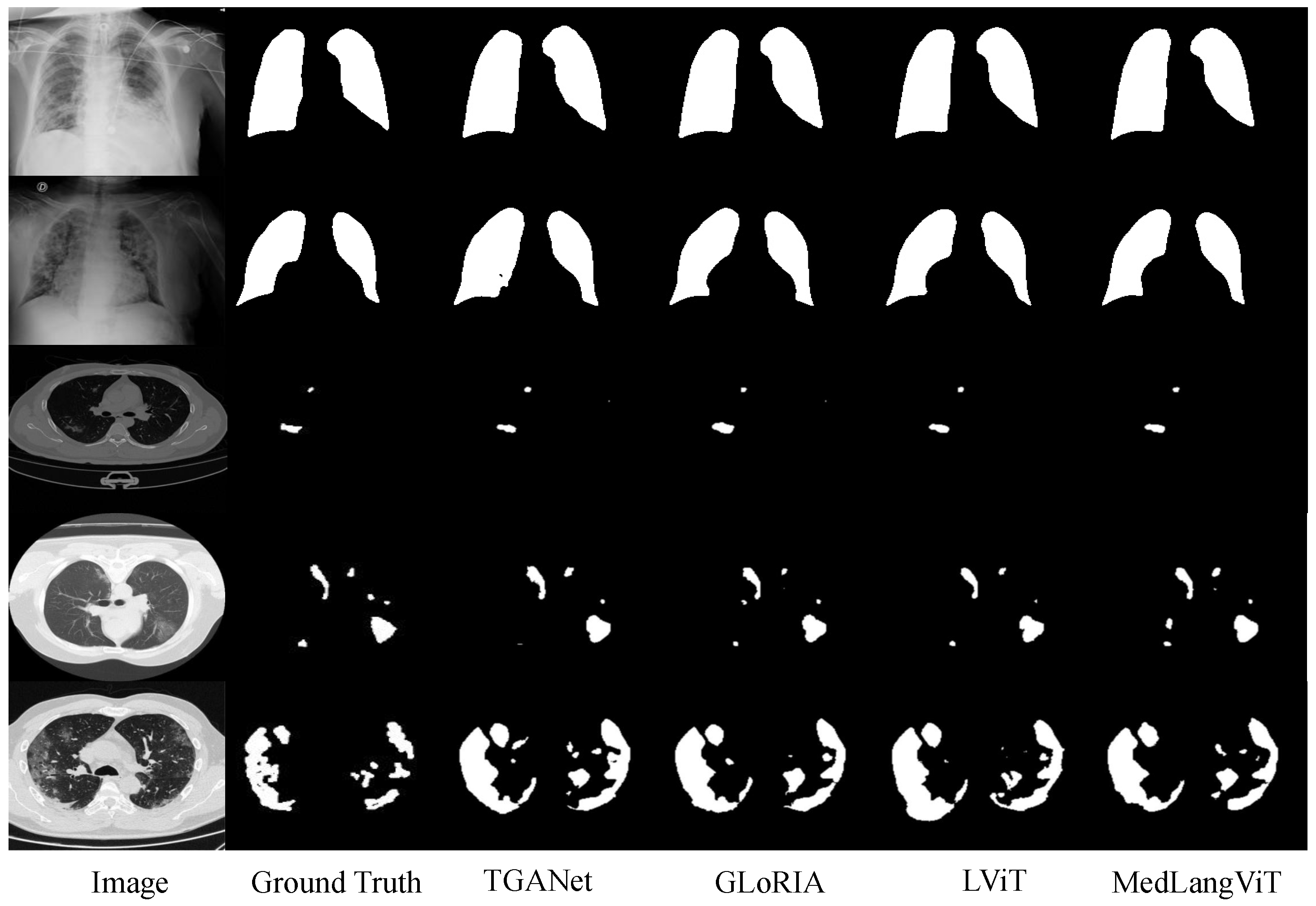
In order to verify the effectiveness of the proposed method, we conducted comprehensive experiments on the MosMedData+ [
8] and QaTa-COV19 [
9,
10] datasets. The MosMedData+ dataset contains extensive lung CT images with diverse infection types and lesion characteristics, while the QaTa-COV19 dataset comprises COVID-19 chest X-ray images with varied lesion patterns and detailed annotations. The experimental results are presented in
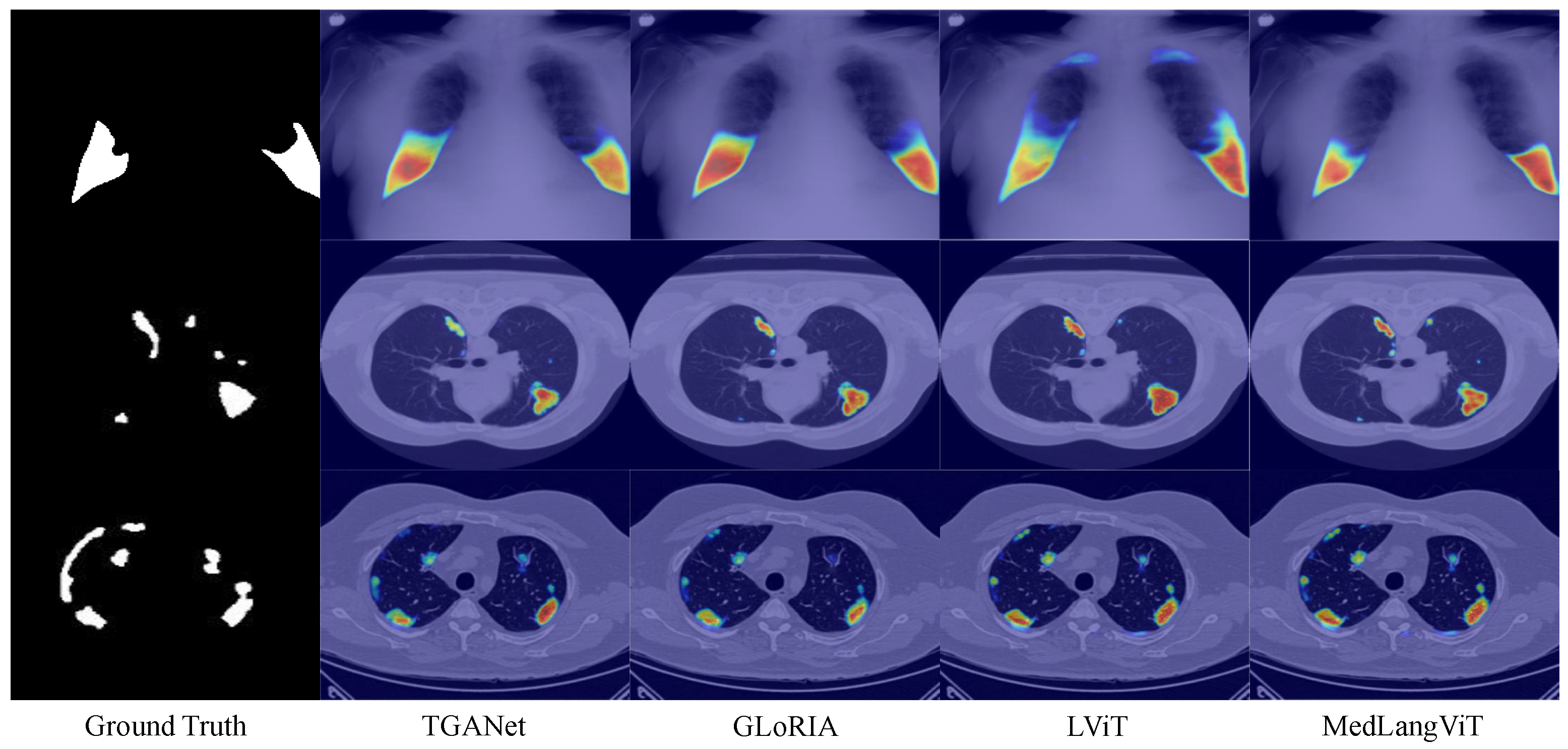
Figure 2, demonstrating that MedLangViT achieves significant improvements in segmentation performance despite using fewer parameters. Key metrics, including the Dice coefficient and mean Intersection over Union (mIoU), surpass those of existing methods while maintaining lower computational complexity. These findings validate the method’s strong clinical applicability and suggest promising new research directions for medical image segmentation. Our key contributions are summarized as follows:
Novel Network Architecture. Leveraging BioBERT as the text embedder, we propose MedLangViT, an innovative image–text framework specifically designed for medical image segmentation tasks enriched with textual annotations.
Innovative Attention Module. We propose the Enhanced Channel-Spatial Attention Module, a lightweight feature-fusion mechanism that effectively captures cross-modal correlations between visual and textual modalities.
Superior Performance. MedLangViT achieves state-of-the-art results on the QaTa-COV19 and MosMedData+ datasets, demonstrating substantial improvements in medical image segmentation accuracy.
The remainder of this paper is organized as follows.
Section 2 reviews related work.
Section 3 presents the proposed MedLangViT method in detail.
Section 4 describes the experimental setup and reports results on multiple datasets, including comprehensive ablation studies.
Section 5 discusses the limitations of our approach and outlines directions for future improvement, and
Section 6 concludes the paper.
5. Discussion
While MedLangViT has achieved substantial progress by integrating clinical text, its upper limit is ultimately constrained by annotation quality. Firstly, inter-clinician variations in descriptive precision, exemplified by ambiguous phrases such as “mildly opaque” or “hazy area”, introduce semantic ambiguity. This challenges BioBERT’s word sense disambiguation capabilities. Additionally, terminology differences across institutions—including British versus American spellings and abbreviation conventions—further amplify these inconsistencies. Secondly, mismatches between textual descriptions and actual visual features (e.g., mentions of invisible lesions or extremely subtle pathologies) can misguide ECSAM’s spatial attention and cause over-activation of attention heatmaps in erroneous regions. Thirdly, MedLangViT inherently assumes clean and complete text–image pairs. However, common retrospective data issues such as spelling errors, missing fields, and copy–paste artifacts (including repeated or conflicting descriptions) directly undermine segmentation robustness, particularly in low-resource settings. Finally, model confidence may sharply decline when encountering real-world low-quality or partially missing text annotations. Future work will quantify these impacts and develop ambiguity-resilient fusion mechanisms.