3.1. Dataset Preprocessing
SciERC was chosen as the dataset for our experiments since it has more completed annotations than its predecessors. For the convenience of data processing, parentheses and brackets in SciERC that were presented as “-LRB-”, “-RRB-”, “-LSB-”, and “-RSB-” were all replaced by their corresponding marks “(“, “)”, “[”, and “]”. After replacing the specified strings, there were some challenges to consider while using SciERC. As seen in
Table 2, a direct challenge was that the distribution of annotated relation types was not even, especially for SciERC and SemEval18, where the number of “Used-of” was larger than other types. The imbalance of data could be solved by adding samples from other datasets. For example, it was observed that “Result” in SemEval18 and “Evaluate-For” in SciERC were fundamentally the same in meaning but the arguments of these two relations were in reverse order [
21]. However, the imbalance could reflect the level of semantic invariance learned by a relation classification model [
34]. Hence, only the original SciERC was adopted without adding other data samples.
Another not obvious yet critical challenge was revealed in a deeper analysis of samples from the training and test set of SciERC [
35]. The analysis denoted an annotated relation triple (head, predicate, tail) in the test set as “Exact Match” if the same triple of it was in the training set, as “Partial Match” if one argument of the triple was in the same position of a relation triple for training with the same type, and otherwise, as “New”. Based on this scheme, it was revealed that more than half of the relation samples in the test set were “New”, which occupied 69% of all test samples. Only 30% of test samples were “Partial Match” and less than 1% of samples were “Exact Match”. Although having a large number of “New” samples was beneficial for reflecting the ability of trained models to generalize, this also created a challenge in this direction. Due to the imbalance of data and the large percentage of “New” samples in the test set of SciERC, it was necessary to make full use of information within the existing data. It was argued that entity type information could provide critical information for relation classification [
36,
37]. However, in specified experiments, the annotation inconsistency of entity span in the SCIERC test set was identified with 26.7% annotation mistakes, in which the boundaries of many annotated entity spans and the type of them were considered as mistakes [
38].
3.2. Task Definition
Previous relation extraction followed the annotation of SCIERC by treating the non-annotated span pairs as a separated relation class. However, they did not present their results on simple relation classification, so they created difficulty in investigating the basic classification ability of models especially given the flaws in the annotation of spans. Given the problems in the SciERC annotation of entity spans, the relation annotation in SciERC was treated from the view of open-world assumption [
22] that potential relation samples not annotated within the sample sentences of SciERC were not treated as “Not a relation”. This modification of relation extraction standards reduced the target to the relation classification in the original extraction standard, to provide a solid basis for effective comparison and discussions in future GNN-based work using dependency graphs. Therefore, the relation extraction task of this work was defined as: given the text of an input sentence
S and the word index representation (i.e., tuple (
,
)) of two continuous spans of words in
S, denoted as
and
, classifying the relation between the two spans as one of {Used-for, Evaluate-for, Feature-of, Hyponym-of, Part-of, Conjunction, Compare}, as in Equation (
1). Normally, a deep learning model is expected to generate logits for each class and then predict by choosing the class index with the largest logit, correspondingly.
3.3. Dependency Path Extraction and Clustering
Dependency parsing adopted in this work was based on Python package Spacy 3.2 [
39] and Spacy model en_core_web_trf 3.2.0. Before extracting the dependency paths that connect the entity pair, the corresponding text span of the entities were shortened in order to simplify the subsequent classification of the relation pair. After shortening the spans, only the root words in the spans were kept, which had no parent words within the original spans. In SciERC, it was observed that the head and tail spans did not always contain one word. Since all the annotated spans were nominal, multi-word spans always contained words providing extra information to a “root” noun, which could be identified by utilizing dependency parser. If a whole span was fed into the classification model, less relevant information and even noises might be introduced. For shortening the spans, the current implementation parsed the head and tail spans before parsing the whole sentence and then kept only the root words of the spans if the spans had multiple words. It was observed that when the span was shortened after parsing the whole sentence, some words within an annotated span might be assigned the wrong dependency labels and part-of-speech (PoS) labels, due to the large sentence length. Focusing on a shorter span could reduce this type of error since:
The spans were always nominal and with a fixed dependency pattern: “pre-modification words + a root word + post-modification words”.
Processing spans instead of a whole sentence avoided introducing noisy context into the spans, which might mislead the dependency parser.
The dependency path between the subject and object spans was extracted using Python package NetworkX 3.1 [
40], after obtaining the dependency graph by parsing an input sentence. Treating the dependency graph as undirected, the simple path between the root words of the spans was extracted, which was a path with no repeated nodes. The obtained dependency graph was treated as undirected since there could be no direct path between the two spans when considering the original directed graph. In this case, both of the two spans can be reached starting from a root word of the input sentence, which is their LCA.
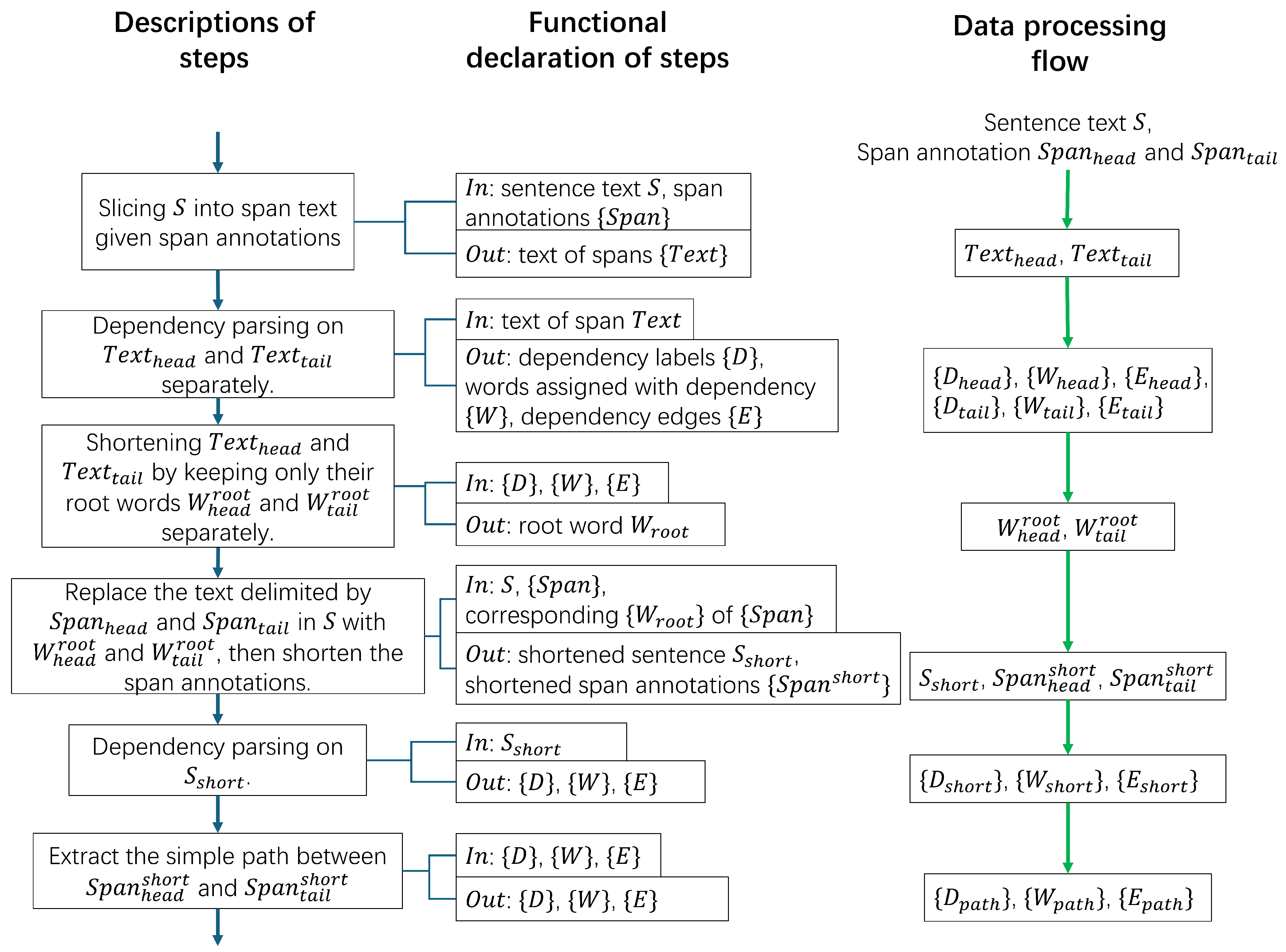
The pipeline from span shortening to path extraction is depicted in
Figure 1. Given the sentence text
S and the word index representations
and
, the text of head and tail entity (
and
) are obtained through text slicing. Then, text spans
and
are shortened to two root words
and
, correspondingly, based on dependency parsing. Next, the original text in
S of the entity pair are replaced with the two root words, correspondingly, which generates the shortened text
and new word index representation
and
. Finally,
,
, and
are parsed again for extracting the simple path between the entity pair, generating words
, dependency labels
, and dependency edges
. Represented by index pairs, edges in
are listed starting from the head entity and ending with the tail entity.
For revealing the contributions of collocation-level patterns, a path clustering scheme was designed to group the words along the path together into collocations, which incorporated the idea of span-based methods and was rarely discussed in the past efforts based on dependency graphs. The design scheme was inspired by the categorization of open class words and close class words in universal dependency scheme (
https://universaldependencies.org/u/pos/index.html [accessed on 30 May 2025]), in which “open class words” referred to words that readily accepted new member words and otherwise “close class words”. Dependency labels in the designed clustering scheme are divided into three main groups, namely, the open group, the end group, and the special group only for dependency root, as in
Table 3. Within an input dependency path, a word
w with a dependency label in the open group is the start of new word collocation, as well as the end of the previous word collocation exclusively (i.e., not including
w itself). Words with dependency labels the close group only marked the end of the current word collocation inclusively (i.e., including the word itself). A word collocation can only contain
w if the previous word collocation does not exist or there are no words after a word with open dependency. The completed table of dependency labels for the path clustering scheme is provided in
Appendix A (
Table A1).
Given the dependency groups, each edge in is traversed to determine the starts and ends of collocations, represented as index pair . When corresponds to a local root (whose parent word is not within the path), only should be clustered to an existing word collocation if is the right/left end of the path. When belongs to the open group, should be clustered as the start of a new word collocation. Only should be clustered to an existing word collocation if has already been processed. If no special cases, and should be both clustered to an existing word collocation. The final output of path clustering can be represented as , where C means collocation.
3.4. GAT for Relation Classification
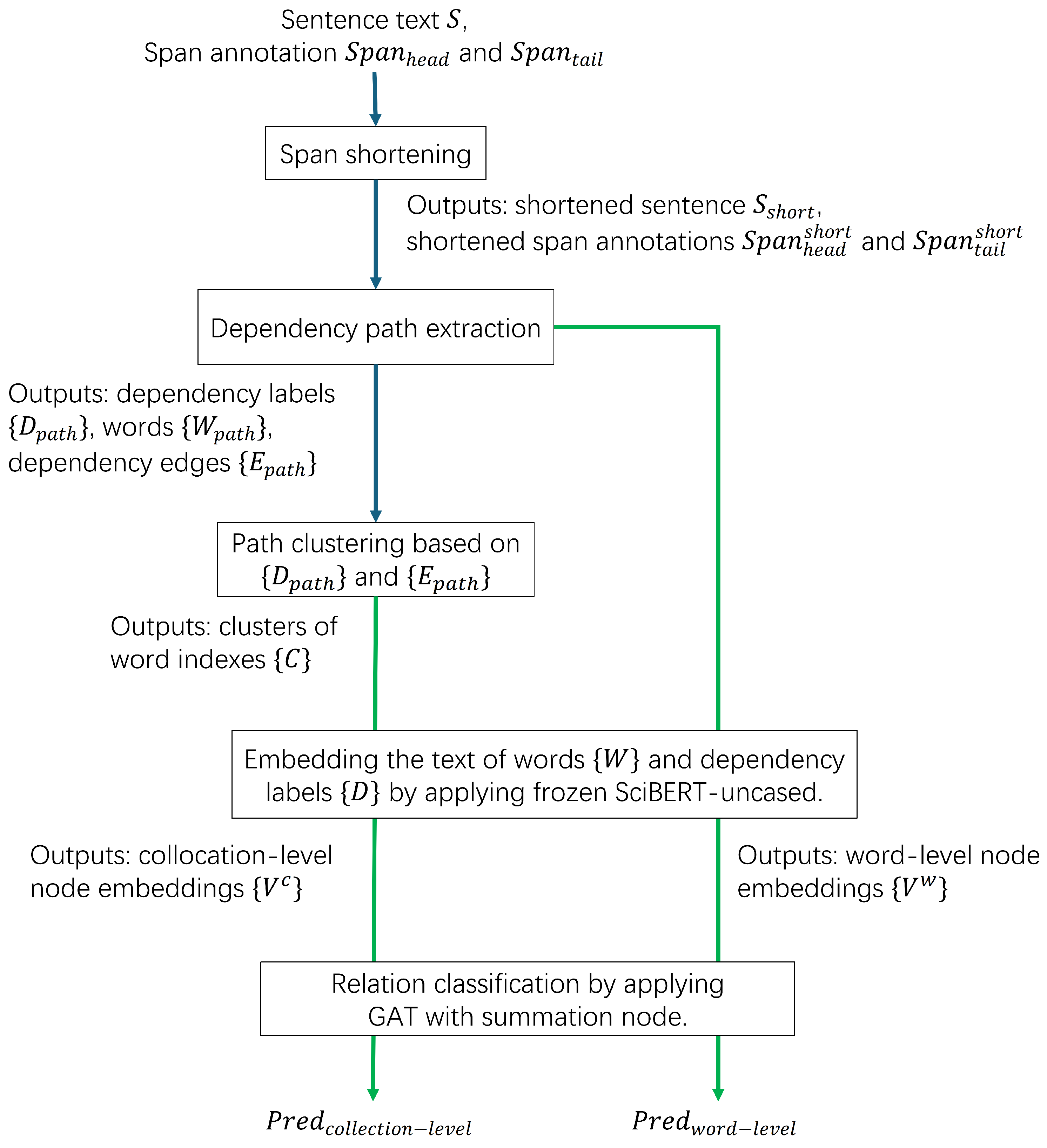
After extracting and clustering dependency paths, a GAT was applied for all experiments performing relation classification on extracted dependency paths. A brief overview of the proposed method is depicted in
Figure 2. Compared to one of its popular competitors GCNs, GATs could process graphs with variant number of nodes and, thus, further consideration of padding was not needed. Meanwhile, the attention mechanism of the GAT could highlight the importance of each input node from the view of classifier after training and, thus, had suitable explainability. All GATs experimented on were trained on the training set of SciERC and then tested on the test set. The GAT implementation from pyGAT) was adopted and modified for performing relation classification. Following the graph-specific design of GNNs described in [
16], the major input graph designs involved adding an extra node for summation, which was different from other nodes for words. In this design, the input nodes were linked directly to the added summation node. This input graph design adopting one summation node was adopted for two main reasons. First, this input graph design could reveal how much the word-level patterns within dependency paths could be used by GNN models for scientific relation classification. Simply linking the input nodes to the summation nodes means difficulty in explicitly utilizing the absolute positional information of words but easiness in reflecting the contribution of each input node’s content and, thus, it helped investigate word-level patterns and collocation-level patterns. Second, the adopted GAT architecture was the simplest case implementing graph pooling since it always tried to pool all the input nodes into one.
The modified GAT architecture was visualized in
Figure 3. First, for an input sequence with
N words, the embeddings of each word
(
) in the input sequence and its corresponding dependency label
are concatenated as in Equations (
2) and (
3). Equation (
2) is for investigating word-level semantic invariance, in which
denotes the
i-th word-level node embedding,
means embedding the input text, and
refers to the operation of concatenating multiple input embeddings; Equation (
3) is for collocation-level invariance, in which
denotes the
j-th collocation-level embedding (“c” here is short for “collocation”),
means summing the contextualized embeddings of words within word collocation
(“ctxt” is short for “contextualized”), and
means summing the static embeddings of dependency labels within word collocation
. All embeddings were generated by applying frozen (i.e., non-trainable) SciBERT-uncased, the embedding size of which is 768 (
). Therefore, the size of node embedding is 1536 (i.e.,
) after the concatenation. Second, the non-trainable embedding of the summation node
(“a” stands for “aggregated”) is appended to the end of the node embeddings
to form
, as in Equation (
4), where
is either
or
. The embedding of summation node had the same size as that of an input node but was initialized with zeros since it had no corresponding semantic content and dependency information.
is processed using two GAT layers in order, which are
(from
to
) and
(from
to
), as in Equation (
5). For relation classification, the output embedding of the summation node was applied with softmax to get the probability
for each class, as in Equation (
6). Specifically,
means selecting only the last element because
is always appended at the end. The left branch in in
Figure 3 depicts a GAT layer with multiple attention heads, where
denotes the first head (from
to
). Different with the one-head scenario shown as the right branch, the results of each head are concatenated and applied with ELU activation function.
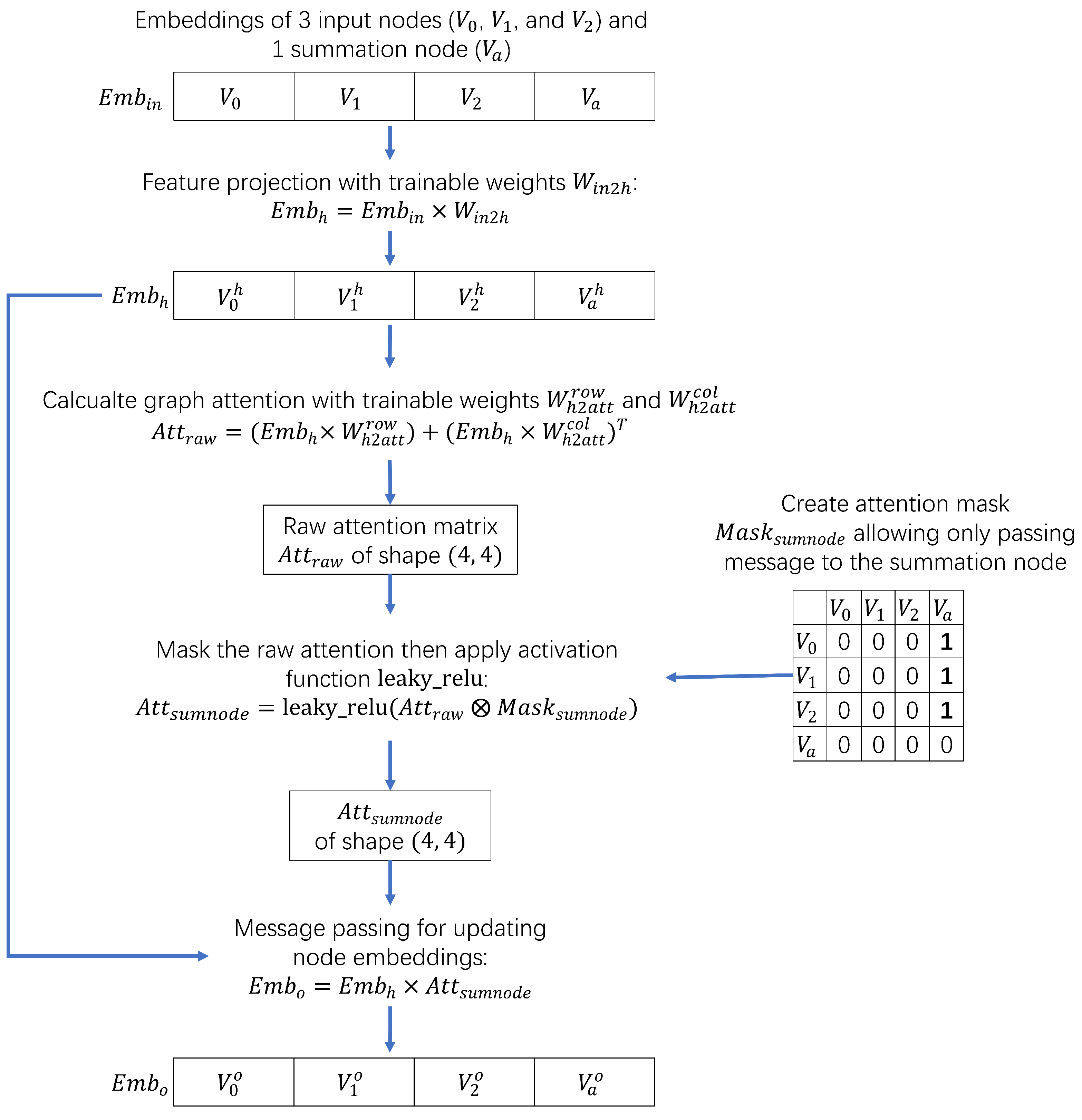
Within one GAT layer, the interactions between inputs and parameters are depicted in
Figure 4, where only the three weight matrices
,
, and
are trainable.
here contains four nodes, the last of which is the summation node. First, input embeddings transformed by applying
of shape
. Second, the raw attention scores
of shape
are calculated by applying
and
separately and then summing the rows and columns. Both
and
are of shape
. Third, the calculated
is masked by
via element-wise multiplication, denoted by ⊗.
only allows summing information from
to
by keeping only the edges from others to summation node. Finally, the updated embeddings
is obtained by
.
For revealing the contributions of word-level patterns, the embedding for each term in the SciBERT-uncased vocab file was collected from the last layer’s hidden state of SciBERT-uncased regardless of the context of input sentence to embed input words, which then generated static word embeddings. This static embedding method was adopted to ensure that only information within the extracted paths was used and, thus, ensured a focus on the word-level patterns for evaluating the potential of GNNs for scientific relation extraction. The embedding of a word was generated by first tokenizing it into SciBERT-uncased vocab terms and then performing a mean pooling on the static embeddings of these vocab terms (excluding the embeddings of [CLS] and [SEP]). To embed dependency labels, these labels were first verbalized following a bag-of-word idea, which used several words to express one label. The completed label verbalization scheme is provided in
Appendix A (
Table A1). Then, similar to embedding a single word previously, the static embeddings of the words describing a dependency label were collected and mean pooling was applied on. The embeddings of several words describing a label were averaged to simply ensure that similar labels had similar embeddings (e.g., in terms of cosine similarity). Since the dependency parsing results were generated following the idea of token classification, the embedding of a verbalized dependency label was concatenated to its corresponding word to form the embedding of a node in the input graph.
For revealing the contributions of collocation-level patterns, the embedding of each input node was a concatenation of two parts, which were the sum of contextualized vocab term embeddings of their corresponding words in the word collocation and the sum of dependency label embeddings of their corresponding words in the word collocation. The dependency label embedding of each word was obtained in the same way in the experiments for revealing word-level patterns. The contextualized embeddings of vocab terms for their corresponding words were first obtained in the context of the shortened sentences and then summed, which were generated after applying the span shortening described in
Section 3.3. Similarly, the embedding of [CLS] or [SEP] token were not included in the final calculation of node embeddings. Instead of using static embedding, the contextualized embeddings were adopted for performance optimization, since the semantics of a word collocation depended more on the word order.
3.5. Configurations of Experiments
The configuration of hyperparameters is provided in
Table 4. The hidden size was set to 200 following [
8,
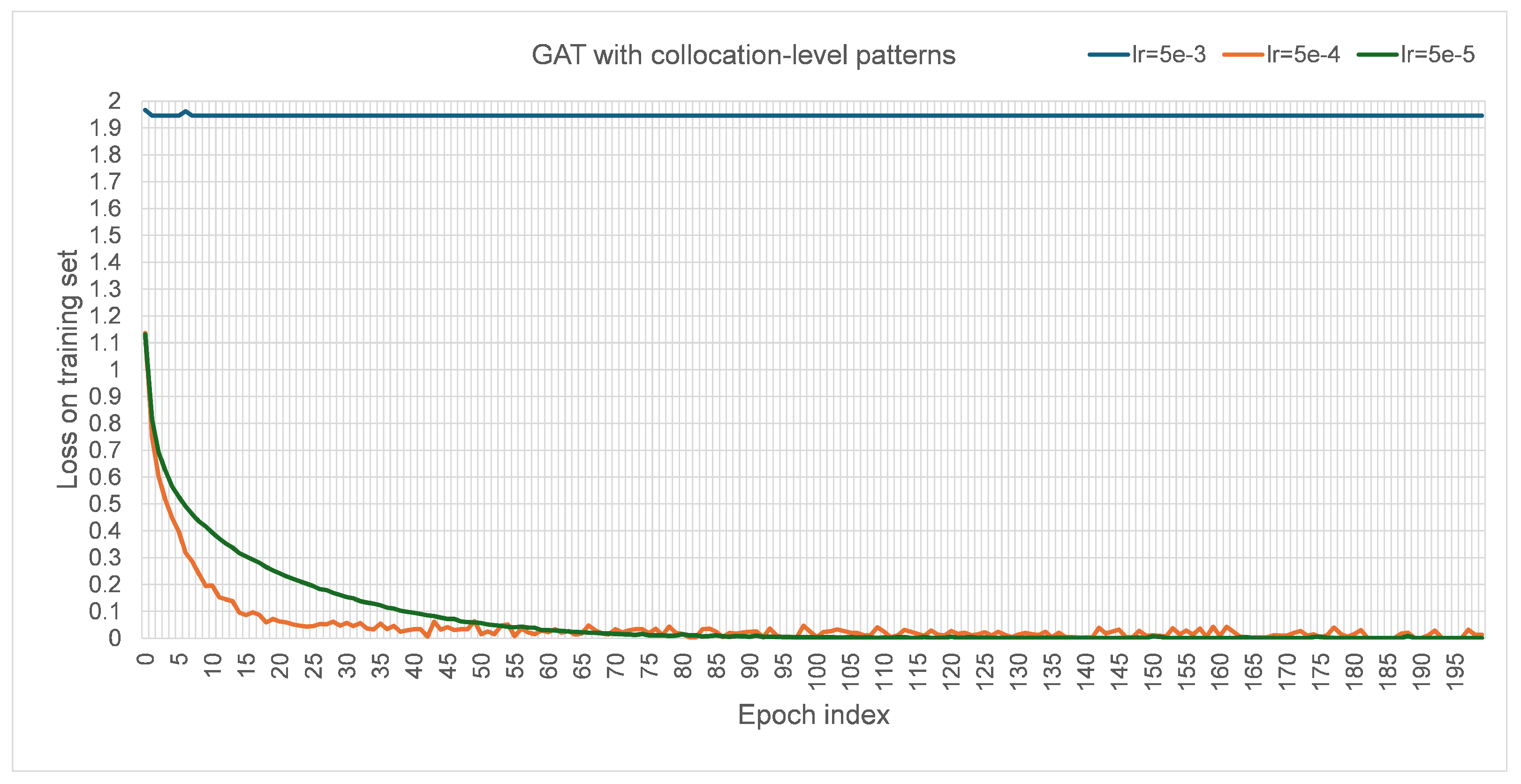
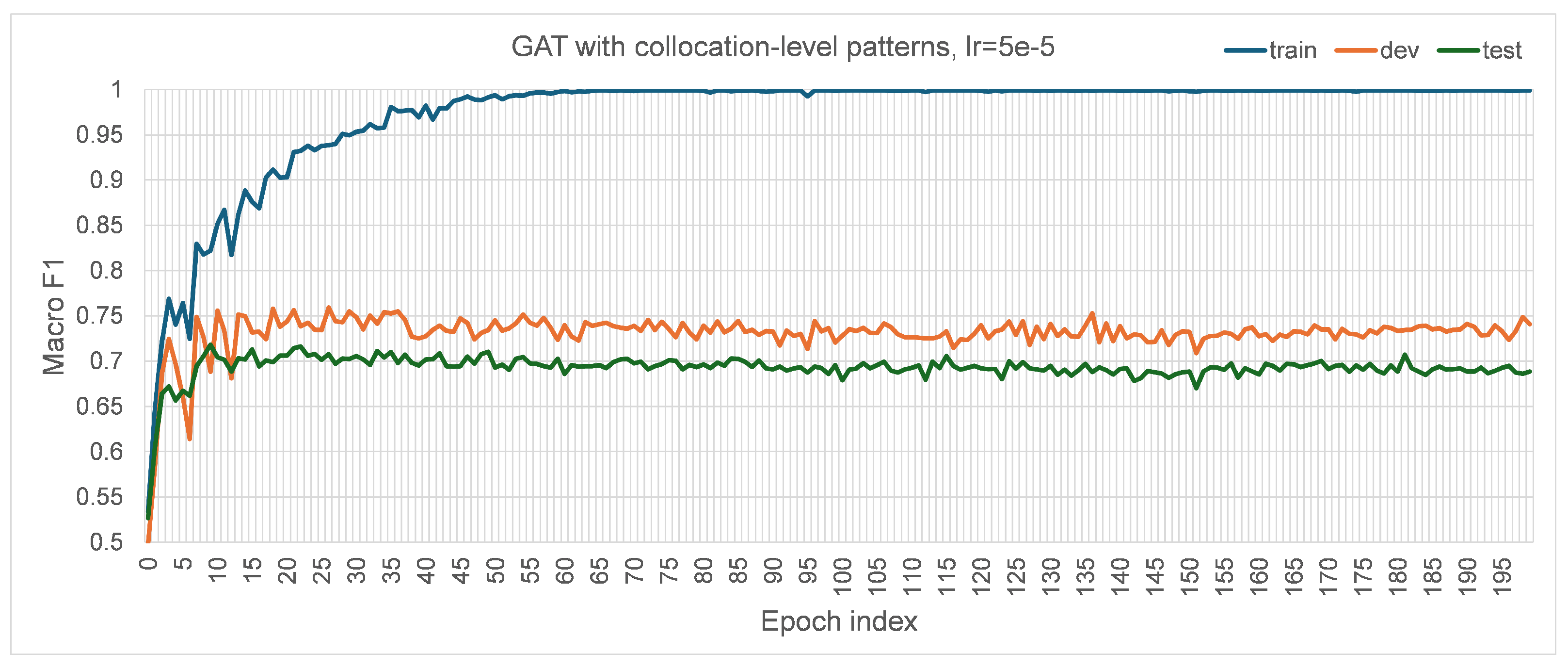
27] for convenient comparison. The number of heads was initially set to one. The dropout rate was set to zero to maintain all information currently. The negative slope of leaky ReLU was set to 0.01, following the default PyTorch 2.0.1 implementation; similarly, the alpha value for ELU was set to 0.2 following the default implementation. Every model was trained for 200 epochs to experiment with their ability to fit into the training set and generalization on the development set, based on which the models were considered if they were worth further training. The loss for training the designed GAT models was chosen to be cross-entropy loss. The learning rate was set to 0.0005 after some initial experiments trying {0.5, 0.05, 0.005, 0.00005}. When the learning rate was set to 0.5 and 0.05 with other hyperparameters unchanged, the losses were stuck at 1.9458. For 0.005, 0.0005, and 0.00005, their corresponding performance trends with the growth of training epochs in terms of macro F1 are visualized in
Appendix A (
Figure A2,
Figure A3,
Figure A4), correspondingly. Apparently, the best performance with the learning rate of 0.005 on the test set was lower than those with the learning rate of 0.0005 and 0.00005, so it was not used for experiments. For the learning rate of 0.00005, the best performance in macro F1 on the test set was 0.6544, which was slightly lower than the performance with the learning rate of 0.0005. Since using a small learning rate would usually slow the convergence during model training, the larger learning rate of 0.0005 was preferred and adopted. The default parameters of the Adam [
41] optimizer implemented in PyTorch 2.0.1 and a random seed value of 42 were used to train the model.
To evaluate the effectiveness of different models in the classification of scientific relations, four classification criteria including accuracy, precision, recall rate, and macro F1 score were used, which had also been used in previous studies. The macro F1 score was considered the main criterion as it takes into account both precision and recall rate, which under the multi-classification settings was calculated as the average of the F1 score of each class. Provided a sequence of model prediction results and their corresponding ground-truth values, precision, recall rate, basic F1 score, and macro F1 score are expressed in (
7)–(
11), where
is for the number of true positives (e.g., the samples both annotated and predicted as “USED-FOR”),
for the number of false positives (e.g., the samples both annotated and predicted as not “USED-FOR”),
for the number of false negatives (e.g., the samples annotated as “USED-FOR” but predicted as “USED-FOR”), and
is the enumeration of all natural numbers that are smaller than the number of relations.
The relation classification task on SciERC was formalized as two types: multi-classification and bi-classification. For multi-classification, the input samples covered all seven relation types, and a classification model was expected to choose the most suitable relation from the seven types for an input sample. This multi-classification design followed previous extraction studies on SciERC for convenient comparison. For bi-classification, input samples covered only two types indicating whether a sample was for a relation or not. Under this design, seven datasets were produced for each relation. This bi-classification formalization was designed to reveal the performance of the designed GAT for each relation type individually. To accommodate the two settings, the output size of the designed models was set to seven and two for multi-classification and bi-classification, respectively.
3.6. Comparative Methods
For demonstrating the ability and gaps of the experimented models, the benchmark model [
21] applied one linear layer on SciBERT-uncased and then finetuned it for classifying all seven relations. The architecture followed by the benchmark method is depicted in
Figure 5. Different from our classification design, the benchmark model feeds the embedding of [CLS] token to the linear classifier layer to obtain the for similar to [
8]. However, using [CLS] embedding for classification raises the problem of distinguishing different entity pairs in the same input sentence. Hence, to mark the head and tail entities, text symbols were used by the benchmark model to prompt the model. For example, in the sample “One is « string similarity » based on [[ edit distance ]]”, the symbol pair “[[” and “]]” is used to prompt the span within the pair as head entity; the symbol pair “«” and “»” is used to prompt the span within the pair as tail entity. This prompt design requires the user to rerun the whole language model each time when processing a new entity pair in the same sentence. For classifying one entity pair, the prompted text is first tokenized into input sequences and then fed to SciBERT-uncased to obtain contextualized embedding of [CLS] token (
), as in Equation (
12). In Equation (
12),
refers to the text function that mark entity pairs with the symbols;
is an indexing operation that return the first embedding of the input embedding sequence, which is normally the embedding of [CLS] token for SciBERT models. The
used for final classification is obtained by applying the linear classifier layer on the [CLS] embedding as in Equation (
13), where
is the linear classifier layer and matrix
of shape
and
of shape
are its parameters. Finally,
is applied with softmax to get the probabilities
for each class, as in Equation (
14).
The benchmark model was chosen since its architecture was simple enough to reflect the effectiveness of utilizing dependency graph information. Moreover, it was the only up-to-date work focusing on comprehensively discussing SciBERT scientific relation classification across each SciERC relation to our best knowledge, while other studies also considered extracting the expected span pairs as in
Section 3.2 task definition, which may add unnecessary complexity to reflect the effectiveness. The benchmark performance to compare for both of the task formalizations was obtained from [
21]. The performance of the benchmark on all relations is only provided in accuracy and macro F1, which are 0.8614 and 0.7949, correspondingly. The performance of the benchmark on each relation is provided in
Table 5. Provided the absence of bi-classification in the original work and our limited computation resources, the benchmark performance under multi-classification was also used in comparison with the designed graph-based models for subsequent analysis. The benchmark model achieved a performance suitable for application but required significantly more computation resources during training than the proposed method. A comparison of the finetuned/trained parameters between the benchmark model and the proposed method is shown in
Table 6, where “one-head” means using one attention head for GATs and “two-head” means using two.
In addition, a method named MRC was compared, which is short for multiple-relation-at-a-time classification. MRC modifies the self-attention mechanism of transformer architecture so that the representations of the relative positions of entities are considered [
21]. In other words, additional parameters are added to the SciBERT model while the pretrained parameters of SciBERT-uncased can still be directly loaded. Apart from this change, MRC adopts the same data and architecture as the benchmark model, which means it shares the Equations (
13) and (
14). Also using SciBERT-uncased, the performance of MRC on all relations is only provided in accuracy and macro F1, which are 0.8433 and 0.7744, correspondingly. The performance of MRC on each relation is provided in
Table 5. Similarly, the benchmark performance under multi-classification was also used in comparison with the proposed method.