Autism Spectrum Disorder Detection Using Skeleton-Based Body Movement Analysis via Dual-Stream Deep Learning
Abstract
1. Introduction
- Dual-Stream Framework for ASD Detection: We adapt and optimise the dual-stream architecture for ASD detection, integrating both spatial and temporal features from video data. The framework combines image classification and skeleton recognition models, allowing for comprehensive feature extraction while building upon previous work;
- Stream-1: Skepxels for Aggregated Spatial Feature Extraction: The first stream processes Skepxels—a spatial representation derived from skeleton sequences—using ConvNeXt-Base, an advanced image classification model. We replace the previous model (ViT) with ConvNeXt-Base to explore the potential benefits of CNN-based architectures for extracting robust spatial embeddings. During training, we employ pairwise Euclidean distance loss to ensure consistency and improve feature representation;
- Stream-2: Angular Features for Temporal Dynamics: The second stream encodes relative joint angles into the skeleton data and utilises MSG3D, which combines GCN and TCN, to extract detailed spatiotemporal dynamics. This stream is designed to capture intricate motion patterns that can be indicative of ASD;
- Experiments and Model Evaluation: The proposed architecture was evaluated on two publicly available datasets—Gait Fullbody and DREAM—achieving classification accuracies of 95.4% and 70.19%, respectively. These results demonstrate the framework’s ability to handle diverse ASD datasets effectively, confirming the potential of this approach for autism detection;
- Practical Impact and Advantages: This study contributes to advancing ASD detection by utilising complementary spatial and temporal features to improve the understanding of ASD-specific traits such as gait and gesture irregularities. The framework is scalable, adaptable, and suitable for real-world applications in healthcare, offering a robust tool for early detection and intervention. By addressing the limitations of traditional methods, this approach provides a promising solution to enhance outcomes for individuals with ASD.
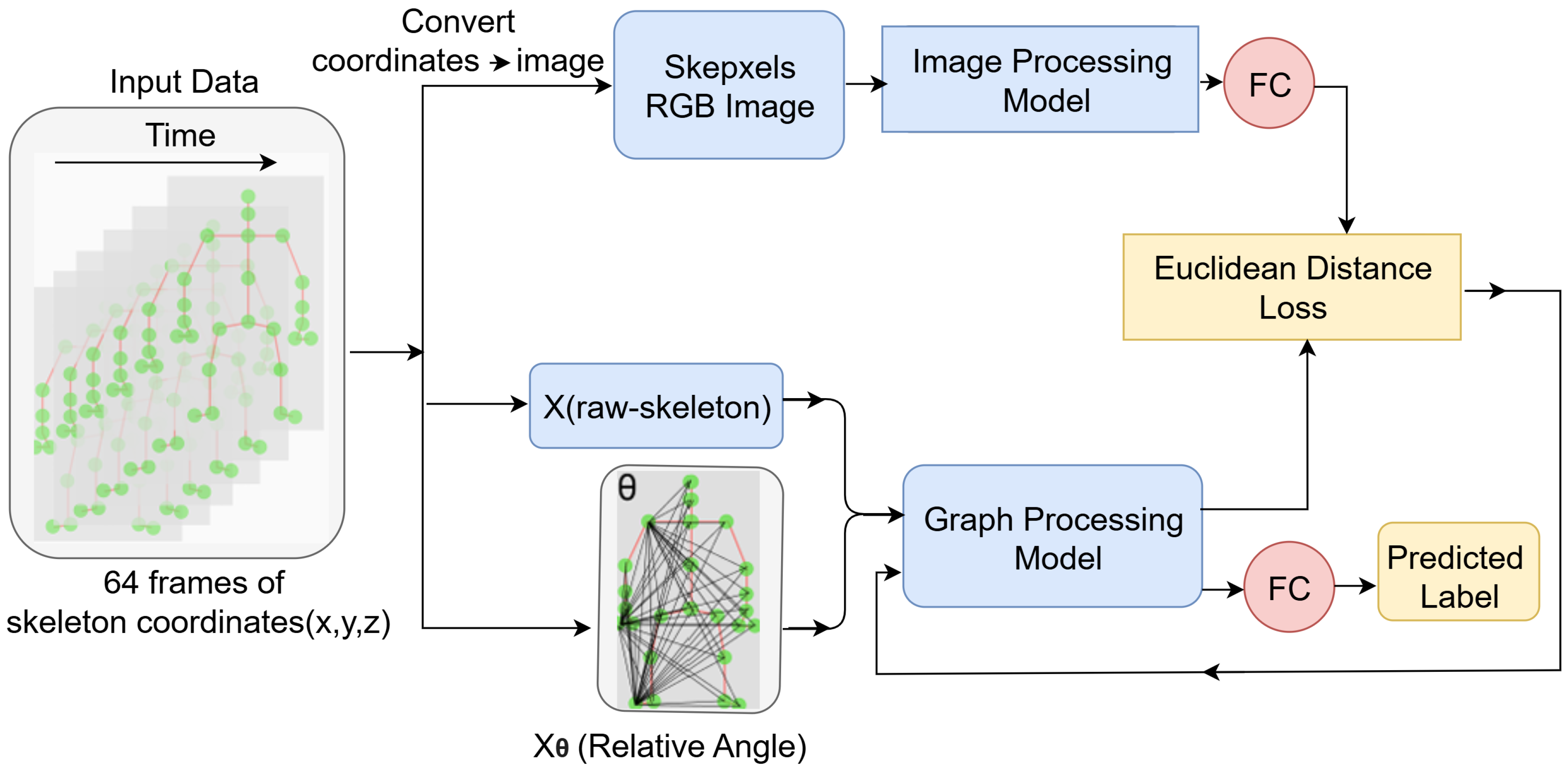
2. Proposed Methodology
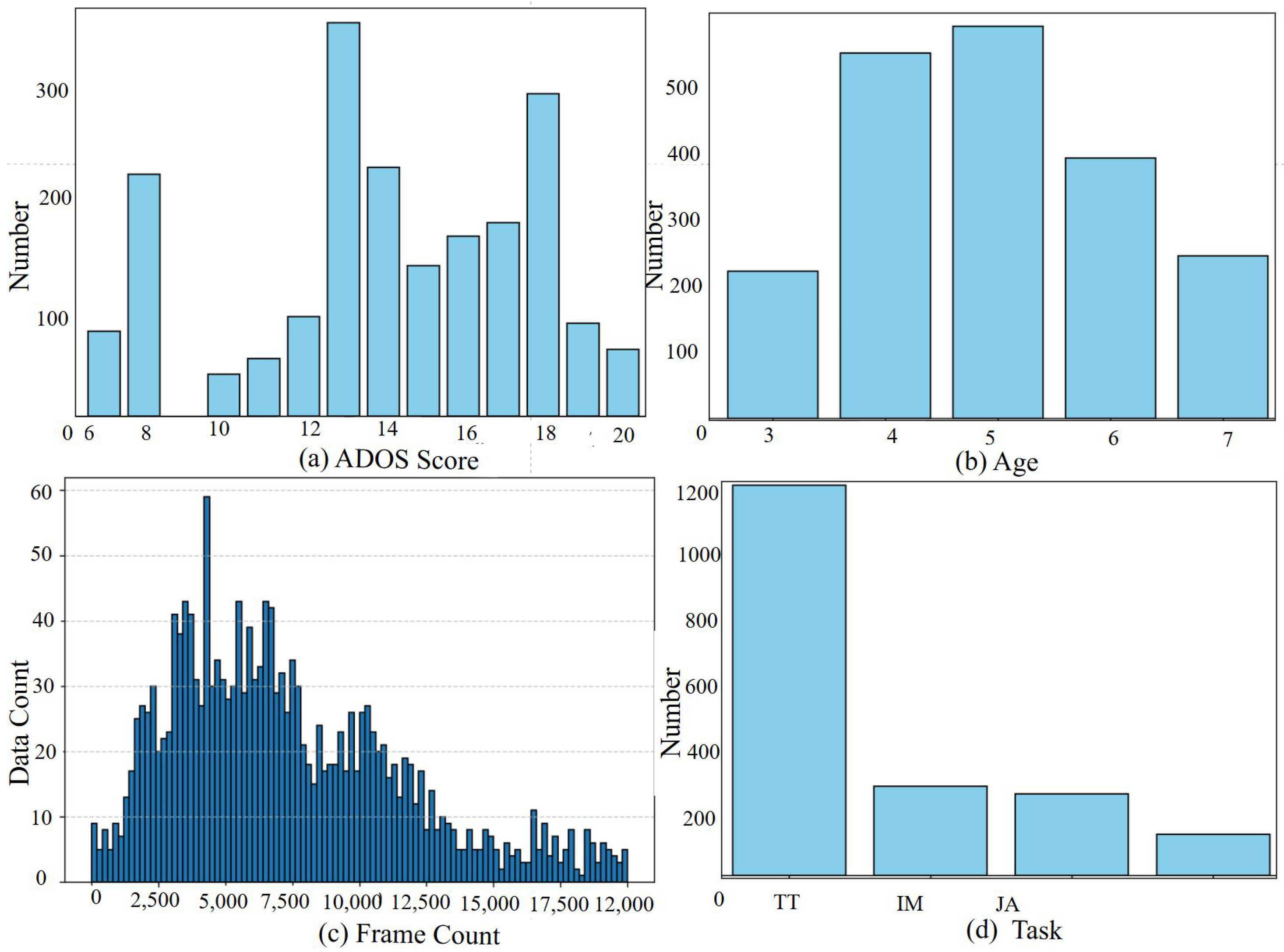
2.1. Dataset
2.1.1. Gait Fullbody Dataset
2.1.2. DREAM Dataset
2.2. Dataset Preprocessing
2.2.1. Joint Imputation
2.2.2. Dataset Labeling
2.3. Stream-1: Image Recognition Model Throw Skepxel
2.3.1. Skepxels
2.3.2. ConvNeXt-Base
- Feature Extraction Layer: ConvNeXt-Base begins with convolutional layers to extract spatial features from the input. Given an input , the convolutional layer produces output features Y, as described in Equation (1);
- Downsampling Layers: ConvNeXt incorporates downsampling operations to reduce the spatial resolution while preserving critical features. This is achieved using strided convolution operations, where the stride reduces the resolution:where s is the stride;
- Depthwise Convolution and LayerNorm: The model applies depthwise convolutions, followed by LayerNorm (Equation (2)), to improve efficiency and training stability;
- Fully Connected (FC) Layer: The final stage involves flattening the features and passing them through a fully connected layer for dimensionality reduction and final representation. If the extracted features are , the FC operation iswhere W is the weight matrix and b is the bias vector;
- ReLU Activation: Non-linearities are introduced using the ReLU activation function, defined as
2.4. Stream-2: Graph CNN Throw Relative Angle Information
- Angular Matrix
- Angle Embedding
- Graph-CNN on the Relative Angle Features
2.5. Model Integration and Training
3. Experimental Result
3.1. Environmental Setting
3.2. Training and Testing Configuration
3.2.1. Reason for Individual Streams Tests
Understanding Individual Stream Contributions
Performance Comparison
Better Interpretability and Debugging
Insights for Future Improvements
3.2.2. Euclidean Distance Loss and Its Benefits for Individual Stream Testing
Enhanced Feature Representation
Informed Individual Stream Evaluation
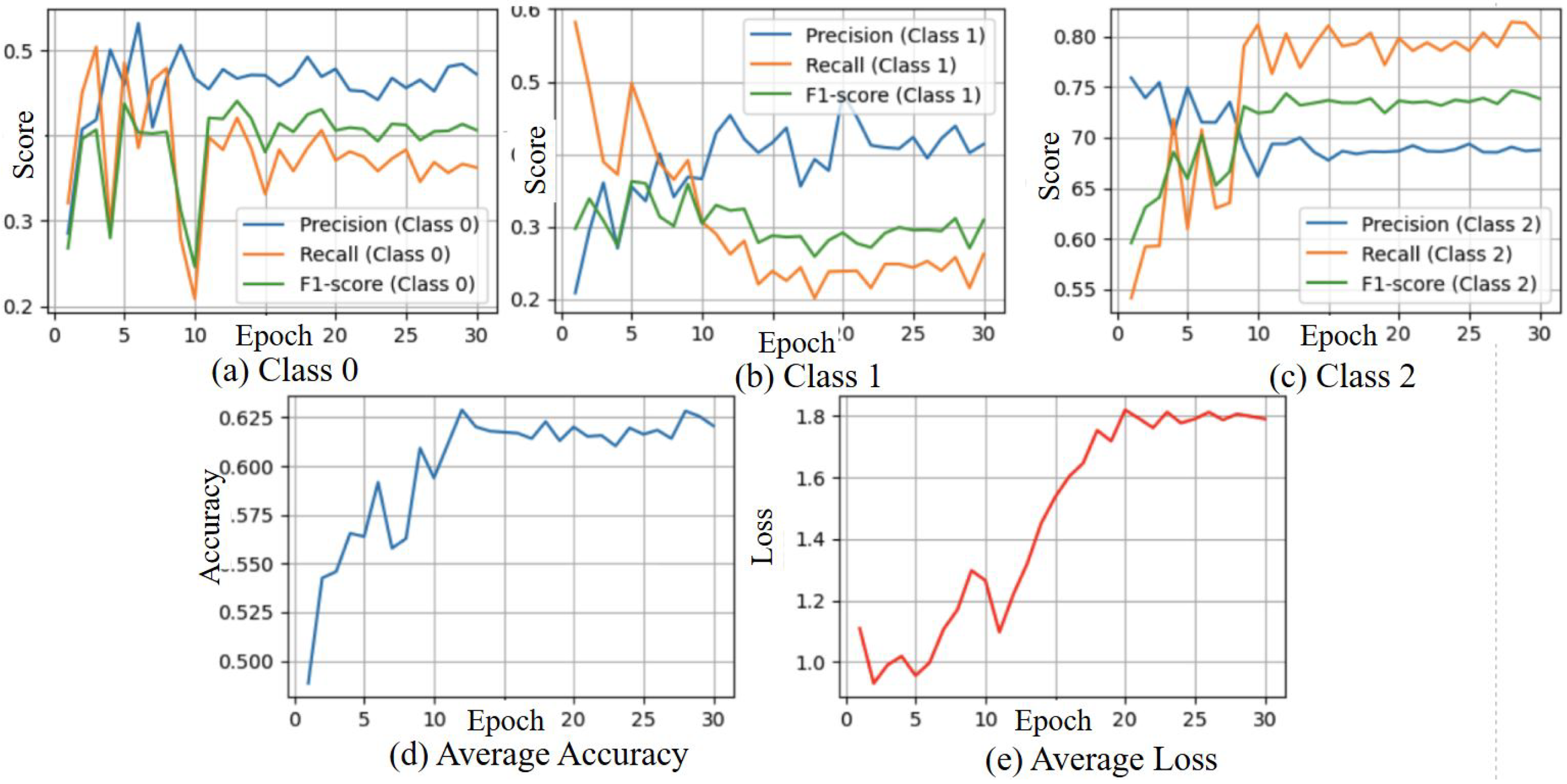
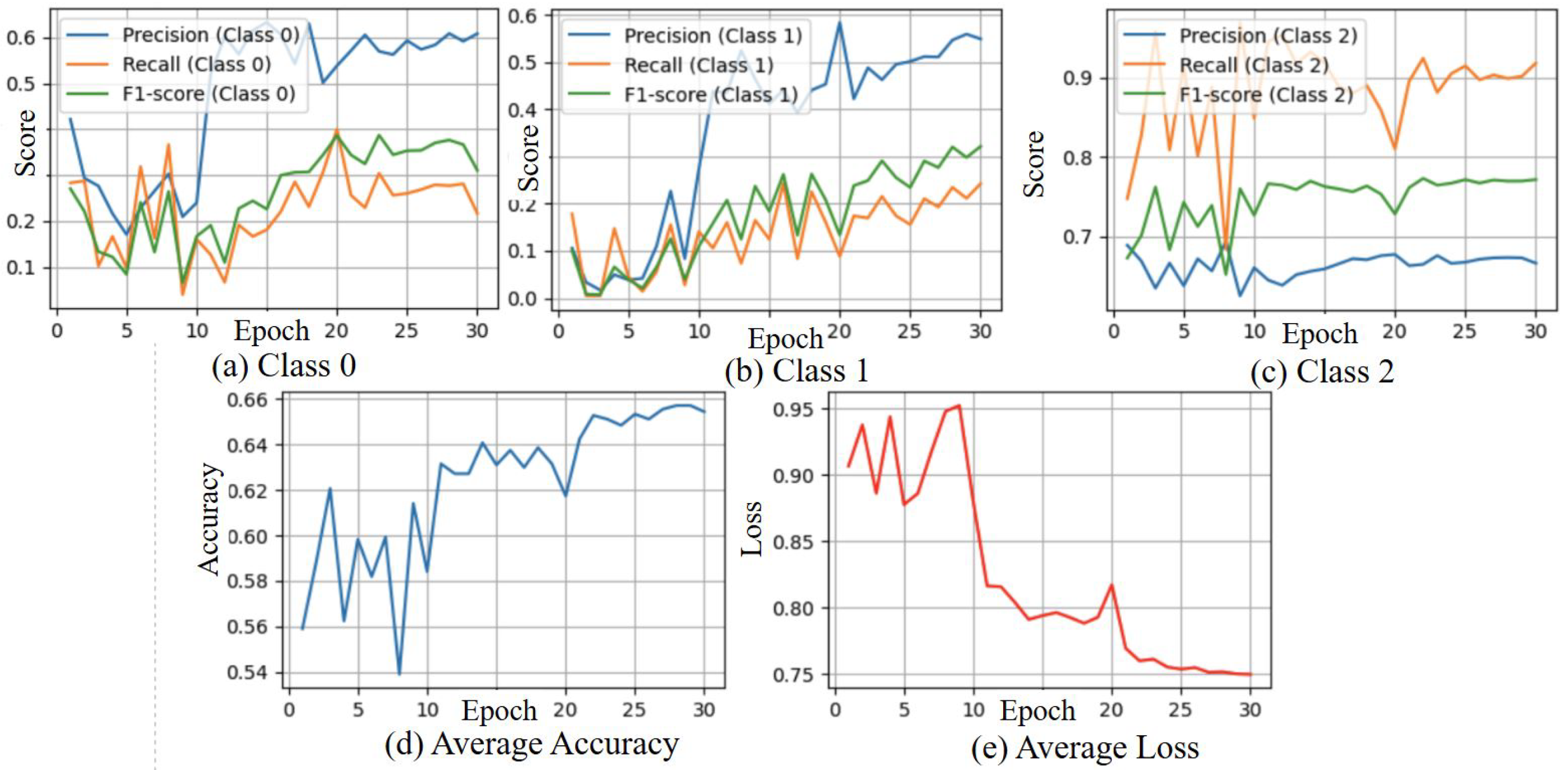
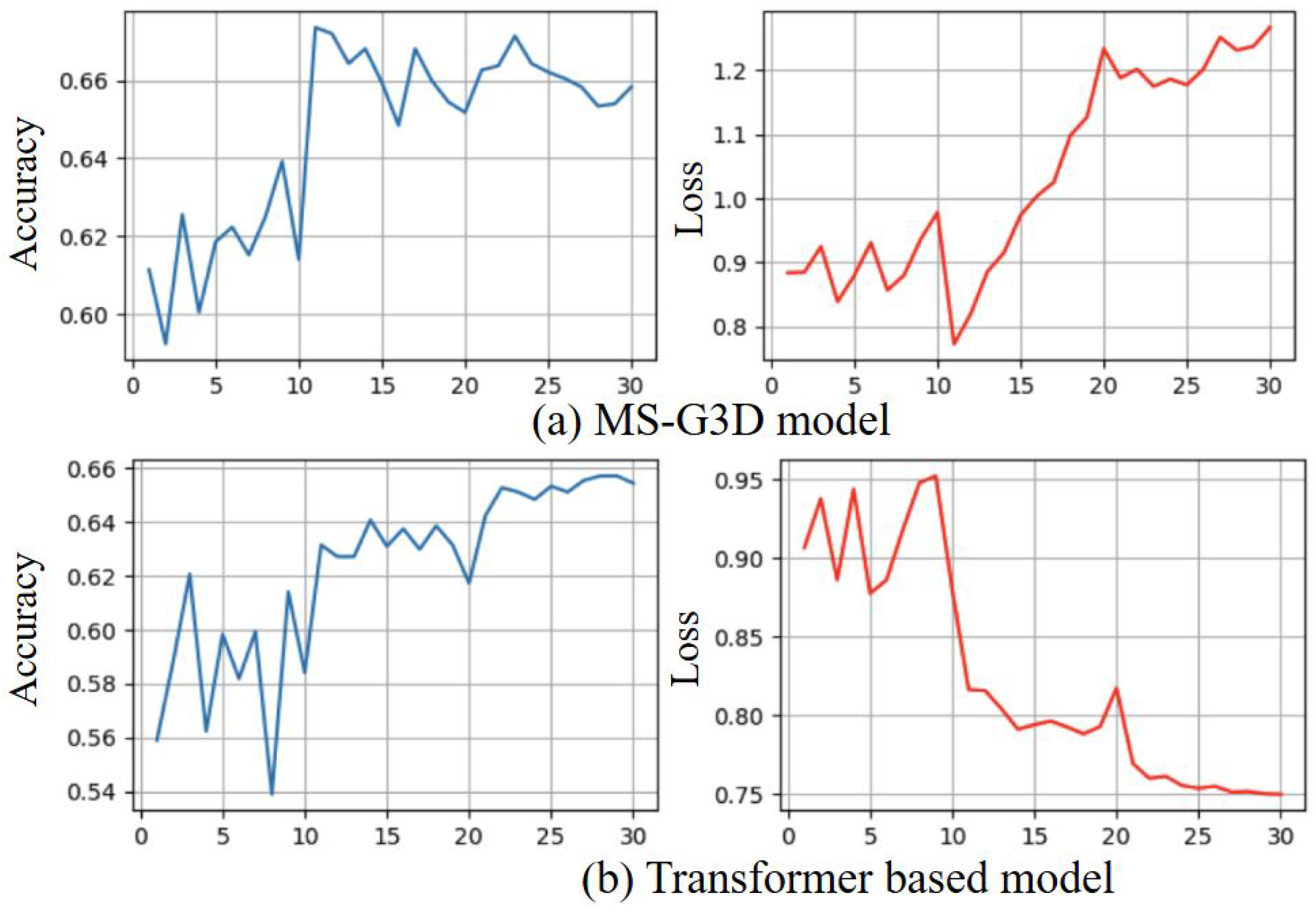
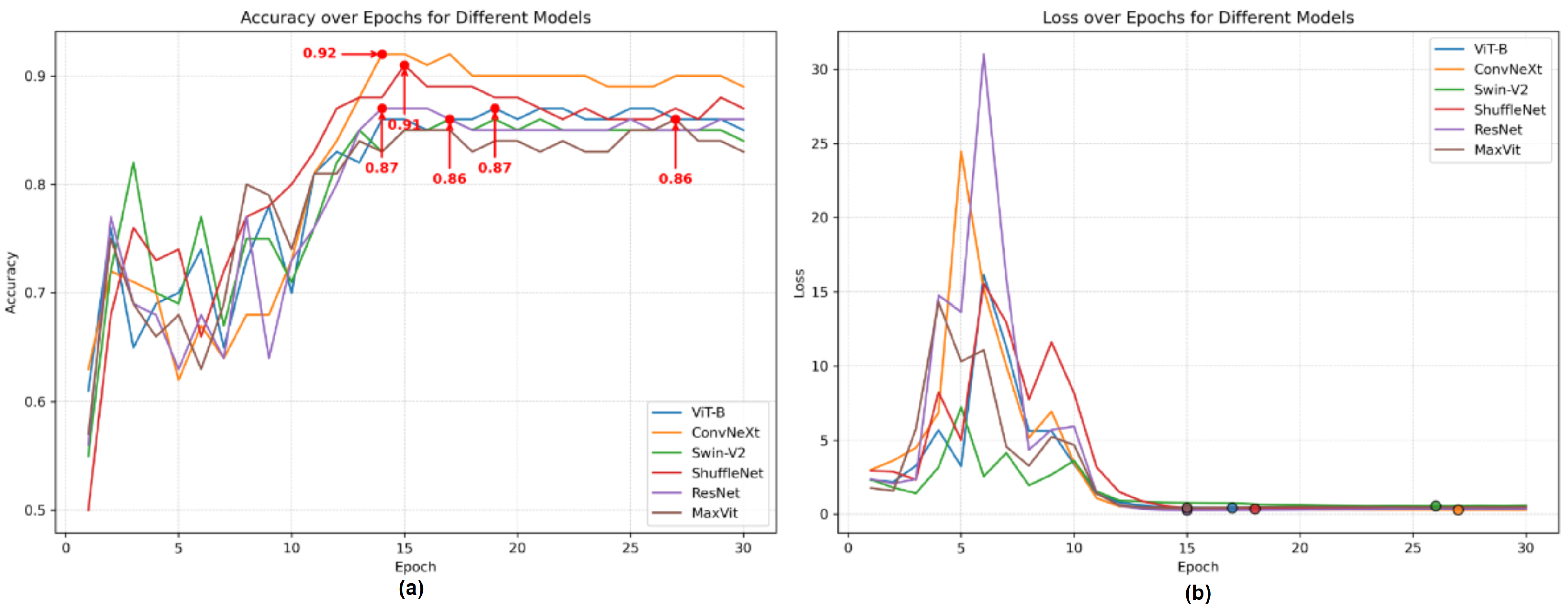
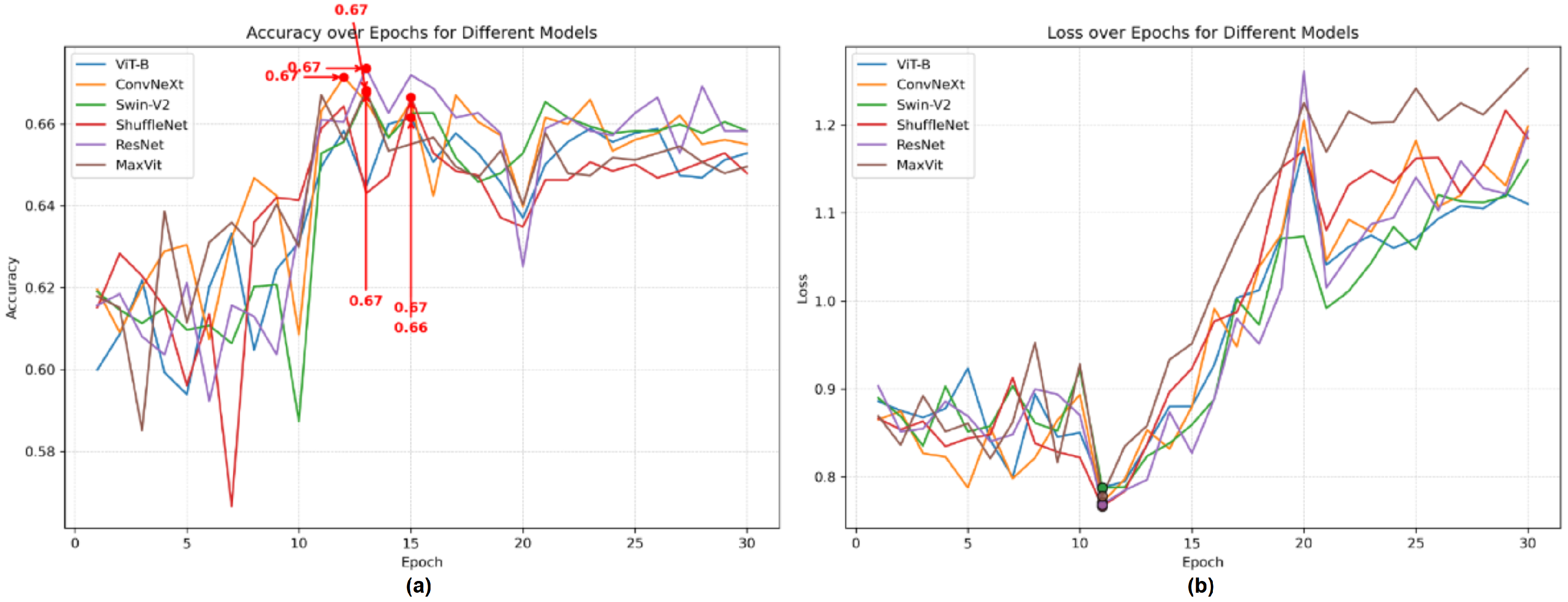
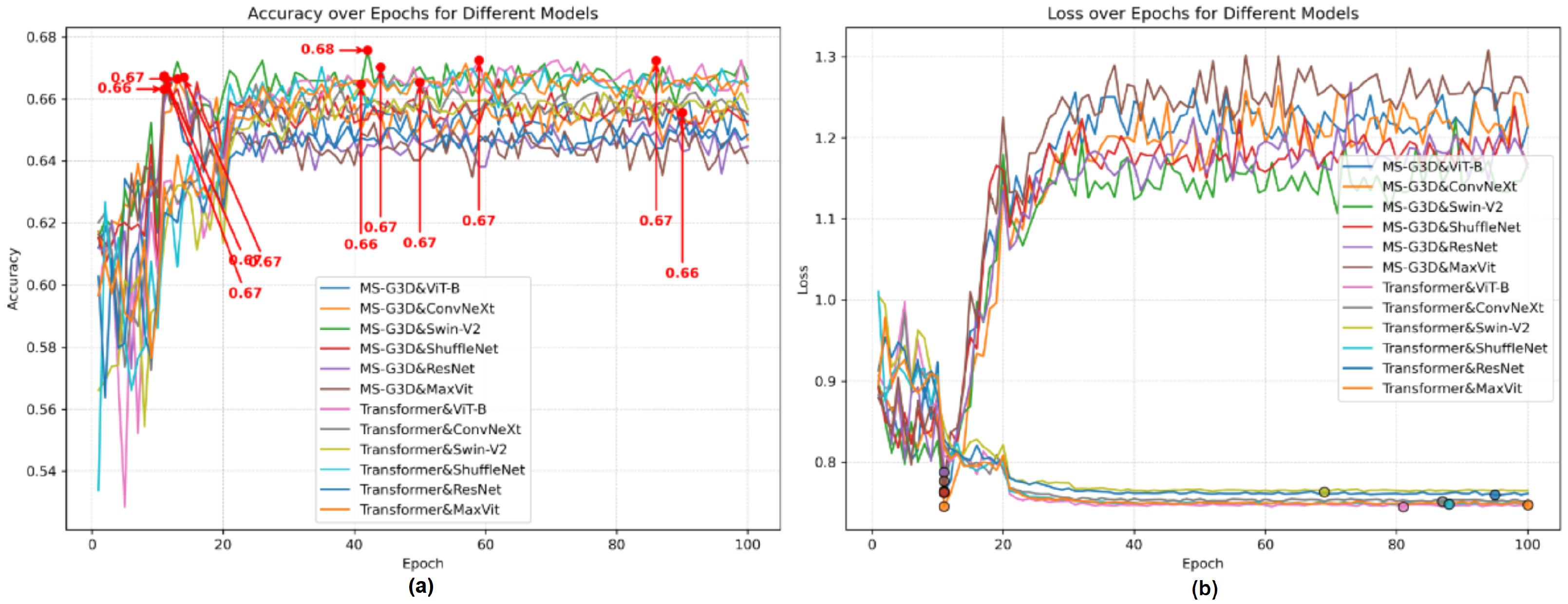
3.3. Ablation Study
3.3.1. Ablation Study with Gait Fullbody Dataset
3.3.2. Ablation Study with DREAM Dataset
3.3.3. Ablation Study with Data Augmentation of DREAM Dataset
3.3.4. Ablation Study of Skeleton Coordinate Recognition Models
3.4. Result from Stream 1 Ske-Conv Model with Loss from Both Streams
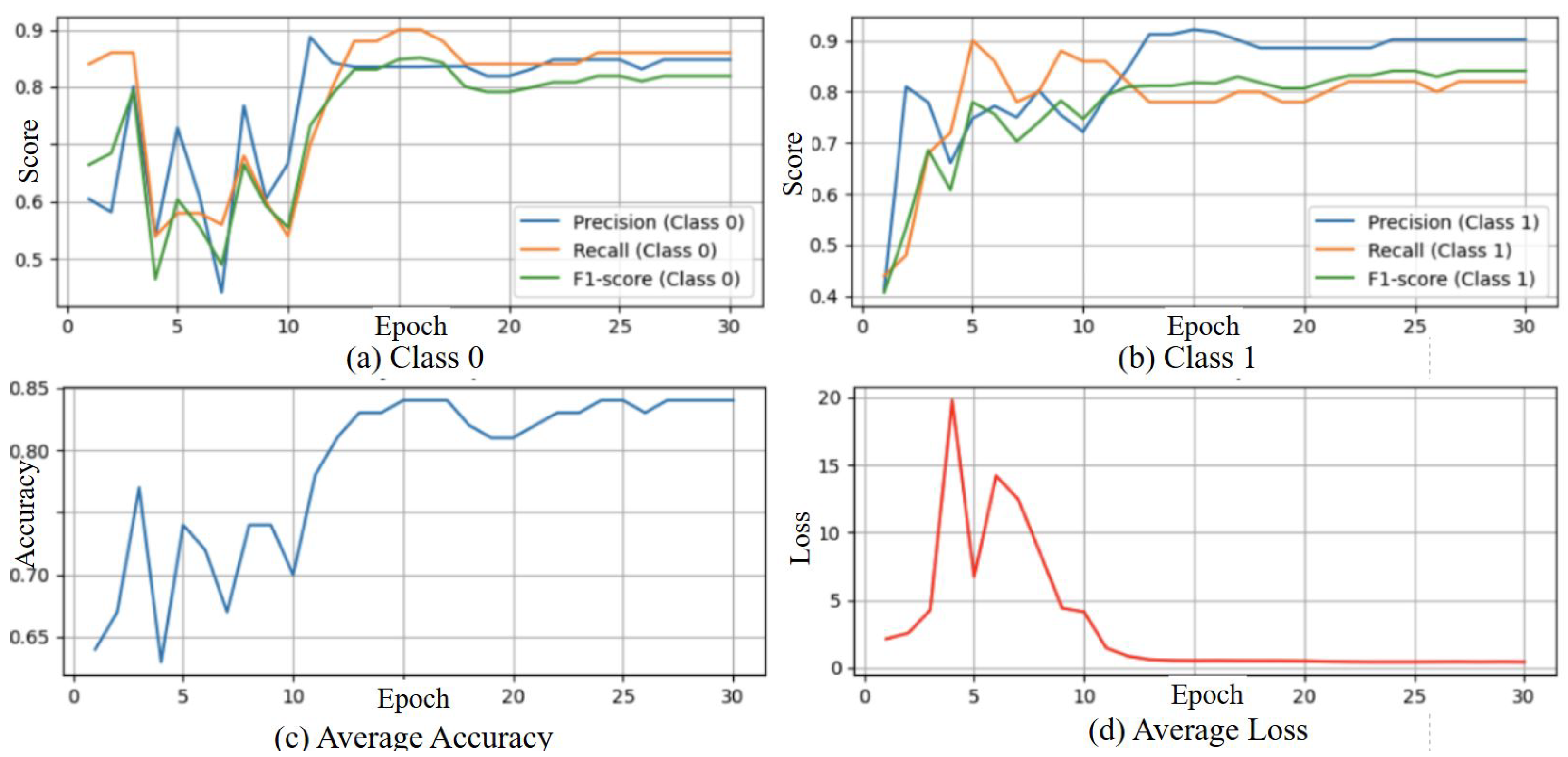
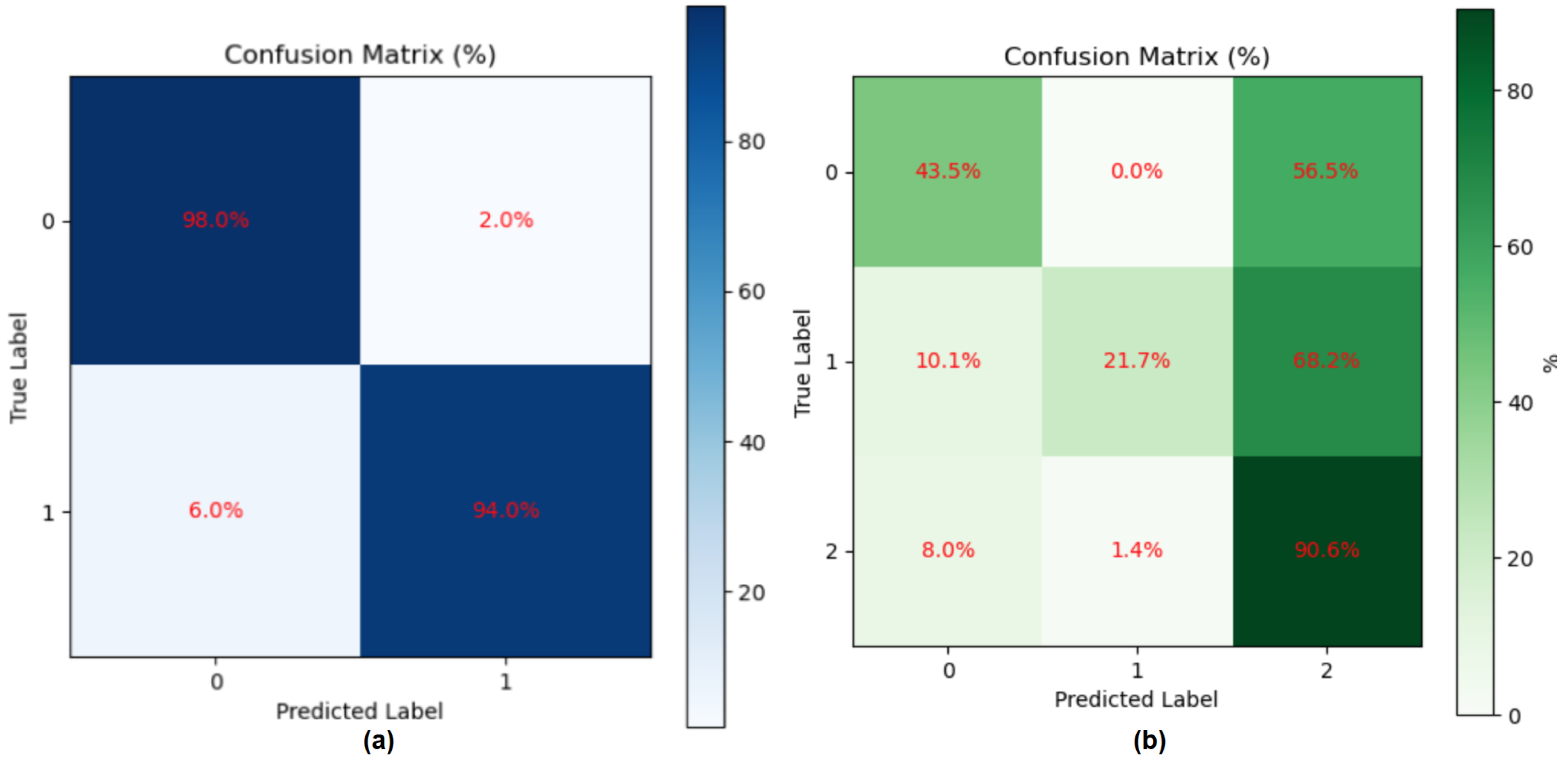
3.4.1. Result of Gait Fullbody Dataset Experiment
3.4.2. Results of DREAM Dataset Experiment
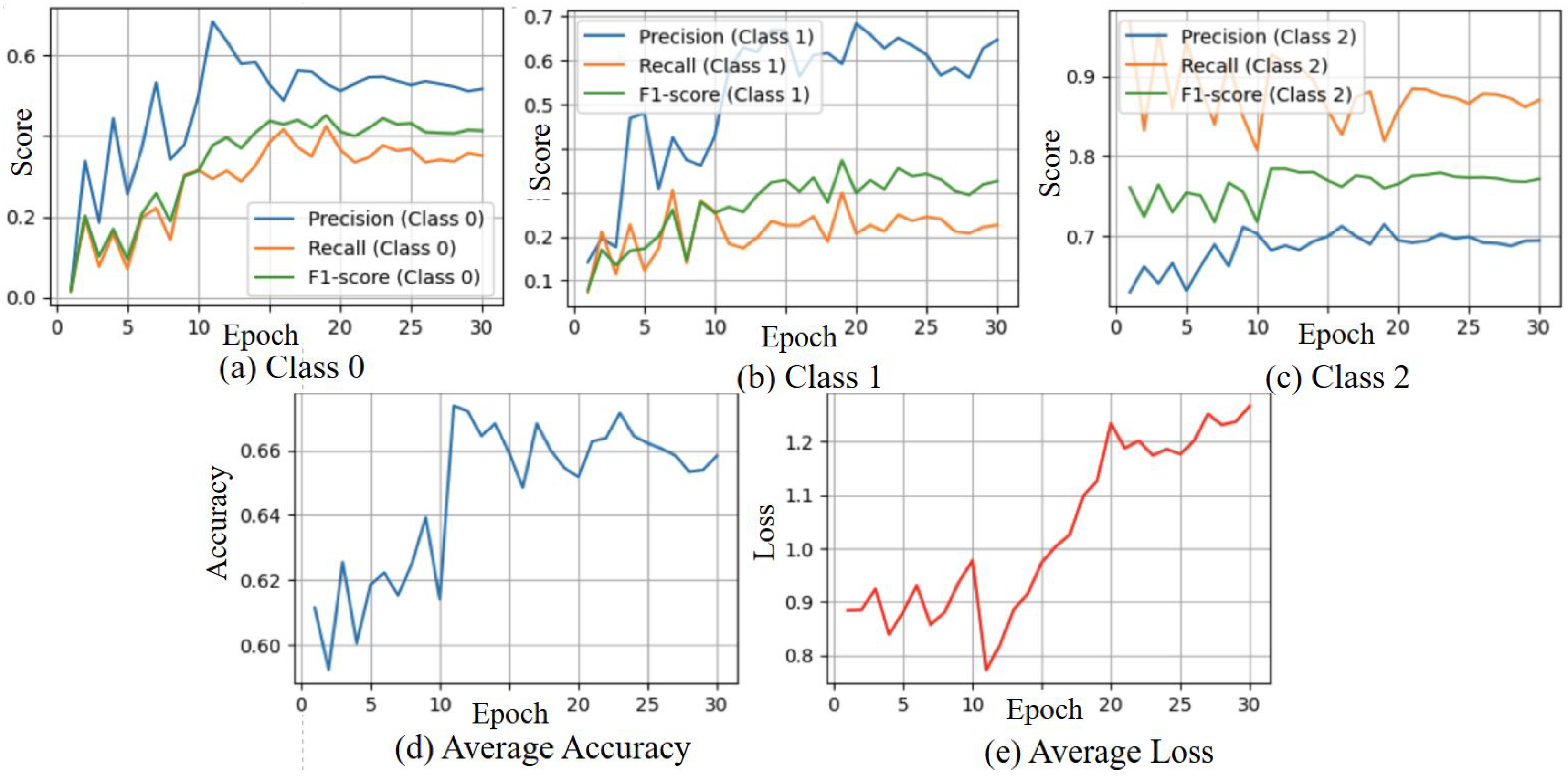
3.5. Results from Stream 2 Angle Skeleton-Based Model with Loss from Both Streams
3.6. State of the Art Comparison for Gait Fullbody Dataset
3.7. State of the Art Comparison for DREAM Dataset
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Joon, P.; Kumar, A.; Parle, M. What is autism? Pharmacol. Rep. 2021, 73, 1255–1264. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Autism Spectrum Disorders. 2024. Available online: https://www.who.int/news-room/questions-and-answers/item/autism-spectrum-disorders-(asd) (accessed on 22 December 2024).
- Smith, I.C.; Reichow, B.; Volkmar, F.R. The Effects of DSM-5 Criteria on Number of Individuals Diagnosed with Autism Spectrum Disorder: A Systematic Review. J. Autism Dev. Disord. 2015, 45, 2541–2552. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Autism Data and Statistics. 2024. Available online: https://www.cdc.gov/autism/data-research/index.html (accessed on 22 December 2024).
- Dawson, G.; Jones, E.J.H.; Merkle, K.; Venema, K.; Lowy, R.; Faja, S.; Kamara, D.; Murias, M.; Greenson, J.; Winter, J.; et al. Early behavioral intervention is associated with normalized brain activity in young children with autism. J. Am. Acad. Child Adolesc. Psychiatry 2012, 51, 1150–1159. [Google Scholar] [CrossRef] [PubMed]
- Rogers, S.J.; Vismara, L.A. Evidence-based comprehensive treatments for early autism. J. Clin. Child Adolesc. Psychol. 2008, 37, 8–38. [Google Scholar] [CrossRef] [PubMed]
- Zwaigenbaum, L.; Bauman, M.L.; Stone, W.L.; Yirmiya, N.; Estes, A.; Hansen, R.L.; McPartland, J.C.; Natowicz, M.R.; Choueiri, R.; Fein, D.; et al. Early Identification of Autism Spectrum Disorder: Recommendations for Practice and Research. Pediatrics 2015, 136, S10–S40. [Google Scholar] [CrossRef] [PubMed]
- Oosterling, I.J.; Wensing, M.; Swinkels, S.H.; van der Gaag, R.J.; Visser, J.C.; Woudenberg, T.; Minderaa, R.; Steenhuis, M.P.; Buitelaar, J.K. Advancing early detection of autism spectrum disorder by applying an integrated two-stage screening approach. J. Child Psychol. Psychiatry 2010, 51, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Al-Jubouri, A.A.; Ali, I.H.; Rajihy, Y. Gait and Full Body Movement Dataset of Autistic Children Classified by Rough Set Classifier. J. Phys. Conf. Ser. 2021, 1818, 012201. [Google Scholar] [CrossRef]
- Zahan, S.; Khan, M.; Islam, M.S. Human Gesture and Gait Analysis for Autism Detection. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Vancouver, BC, Canada, 17–24 June 2023; pp. 1413–1422. [Google Scholar]
- Serna-Aguilera, M.; Nguyen, X.B.; Singh, A.; Rockers, L.; Park, S.W.; Neely, L.; Seo, H.S.; Luu, K. Video-Based Autism Detection with Deep Learning. arXiv 2024, arXiv:2402.16774v2. [Google Scholar]
- Cook, A.; Mandal, B.; Berry, D.; Johnson, M. Towards Automatic Screening of Typical and Atypical Behaviors in Children With Autism. In Proceedings of the 2019 IEEE International Conference on Data Science and Advanced Analytics (DSAA), Washington, DC, USA, 5–8 October 2019; pp. 504–510. [Google Scholar] [CrossRef]
- Zhang, Y.; Tian, Y.; Wu, P.; Chen, D. Application of Skeleton Data and Long Short-Term Memory in Action Recognition of Children with Autism Spectrum Disorder. Sensors 2021, 21, 411. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Akhtar, N.; Mian, A. Skepxels: Spatio-temporal Image Representation of Human Skeleton Joints for Action Recognition. arXiv 2017, arXiv:1711.05941. [Google Scholar]
- Kohane, I.S.; McMurry, A.; Weber, G.; MacFadden, D.; Rappaport, L.; Kunkel, L.; Bickel, J.; Wattanasin, N.; Spence, S.; Murphy, S.; et al. The co-morbidity burden of children and young adults with autism spectrum disorders. PLoS ONE 2012, 7, e33224. [Google Scholar] [CrossRef] [PubMed]
- Al-Jubouri, A.A.; Hadi, I.; Rajihy, Y. Three Dimensional Dataset Combining Gait and Full Body Movement of Children with Autism Spectrum Disorders Collected by Kinect v2 Camera; Dryad Digital Repository: Davis, CA, USA, 2020. [Google Scholar] [CrossRef]
- Billing, E.; Belpaeme, T.; Cai, H.; Cao, H.L.; Ciocan, A.; Costescu, C.; David, D.; Homewood, R.; Garcia, D.H.; Esteban, P.G.; et al. The DREAM Dataset: Supporting a data-driven study of autism spectrum disorder and robot enhanced therapy. PLoS ONE 2020, 15, e0236939. [Google Scholar] [CrossRef] [PubMed]
- Gotham, K.; Pickles, A.; Lord, C. Standardizing ADOS scores for a measure of severity in autism spectrum disorders. J. Autism Dev. Disord. 2009, 39, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Mao, H.; Wu, C.Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A ConvNet for the 2020s. arXiv 2022, arXiv:2201.03545. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer using Shifted Windows. arXiv 2021, arXiv:2103.14030. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2015, arXiv:1512.03385. [Google Scholar]
- Shi, L.; Zhang, Y.; Cheng, J.; Lu, H. Two-stream adaptive graph convolutional networks for skeleton-based action recognition. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 12026–12035. [Google Scholar]
- Egawa, R.; Miah, A.S.M.; Hirooka, K.; Tomioka, Y.; Shin, J. Dynamic fall detection using graph-based spatial temporal convolution and attention network. Electronics 2023, 12, 3234. [Google Scholar] [CrossRef]
- Miah, A.S.M.; Hasan, M.A.M.; Jang, S.W.; Lee, H.S.; Shin, J. Multi-Stream General and Graph-Based Deep Neural Networks for Skeleton-Based Sign Language Recognition. Electronics 2023, 12, 2841. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, H.; Chen, Z.; Wang, Z.; Ouyang, W. Disentangling and unifying graph convolutions for skeleton-based action recognition. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; pp. 143–152. [Google Scholar]
- Hirooka, K.; Miah, A.S.M.; Murakami, T.; Akiba, Y.; Hwang, Y.S.; Shin, J. Stack Transformer Based Spatial-Temporal Attention Model for Dynamic Multi-Culture Sign Language Recognition. arXiv 2025, arXiv:2503.16855. [Google Scholar]


| Author(s) | Dataset | Algorithm | Accuracy | Year | Ref. |
|---|---|---|---|---|---|
| Ahmed A. Al-Jubouri et al. | Gait and full-body movement | Rough set classifier | 92% | 2021 | [9] |
| Sania Zahan et al. | Gait and full-body movement | Vit_b & MSG3D | 93% | 2023 | [10] |
| DREAM dataset (non NS) | Vit_b & MSG3D | 78.6% | 2023 | [10] | |
| Manuel Serna-Aguilera et al. | UTSA & UARK | EfficientNet & ResNet | 81.48% | 2024 | [11] |
| Andrew Cook et al. | YouTube video data | Decision Tree | 71% | 2019 | [12] |
| Yunkai Zhang et al. | Own dataset | LSTM | 71% | 2021 | [13] |
| Feature | Gait Fullbody Dataset | DREAM Dataset |
|---|---|---|
| Number of Participants | 100 children (50 ASD, 50 typical) | 61 children |
| Devices Used | Microsoft Kinect v2, Samsung Note 9 | Kinect v1 |
| Data Types | 3D joint places (25 keypoints) | 3D joint places (10 keypoints) |
| Task (Action) | Walking toward the camera | 3 tasks (TT, IM, and JA) |
| Label Name | Module-1 | Module-2 | ||
|---|---|---|---|---|
| Age | ADOS Score | Age | ADOS Score | |
| NS | ADOS ≤ 10 | 6 = ADOS | ||
| ADOS ≤ 6 | ||||
| ASD | 10 < ADOS ≤ 15 | 8 ≤ ADOS ≤ 9 | ||
| ADOS = 8 | ||||
| AUT | 15 ≤ ADOS | 9 < ADOS | ||
| 8 < ADOS | ||||
| ModelName | AveAcc (%) | MaxAcc (%) | MinAcc (%) | SD | Architecture Type |
|---|---|---|---|---|---|
| ViT-B(ref) | 94.8 | 96.0 | 94.0 | 0.83 | Transformer |
| ConvNeXt-Base | 95.0 | 96.0 | 94.0 | 1.00 | CNN |
| Swin-V2-B | 93.2 | 94.0 | 92.0 | 0.83 | Transformer |
| ShuffleNetV2-X2 | 95.4 | 97.0 | 94.0 | 1.14 | CNN |
| ResNet-152 | 94.0 | 95.0 | 93.0 | 0.71 | CNN |
| MaxViT-T | 95.4 | 97.0 | 93.0 | 1.82 | CNN + Transformer |
| ModelName | AveAcc (%) | MaxAcc (%) | MinAcc (%) | SD | Architecture Type |
|---|---|---|---|---|---|
| ViT-B(ref) | 69.34 | 69.87 | 68.61 | 0.65 | Transformer |
| ConvNeXt-Base | 70.19 | 70.80 | 69.43 | 0.69 | CNN |
| Swin-V2-B | 69.28 | 69.75 | 68.39 | 0.77 | Transformer |
| ShuffleNetV2-X2 | 68.96 | 69.54 | 68.45 | 0.54 | CNN |
| ResNet-152 | 69.24 | 69.43 | 69.15 | 0.15 | CNN |
| MaxViT-T | 68.92 | 69.81 | 67.90 | 0.96 | Transformer |
| Skeleton Classification Model | Image Classification Model | Architecture Type | Accuracy (%) | Ave Acc (%) |
|---|---|---|---|---|
| MSG3D (GCN-based) | ViT-B (ref) | Transformer | 68.94 | 69.39 |
| ConvNeXt-Base | CNN | 69.26 | ||
| Swin-V2-B | Transformer | 70.52 | ||
| ShuffleNetV2-X2 | Lightweight CNN | 69.38 | ||
| ResNet-152 | CNN | 69.27 | ||
| MaxViT-T | CNN + Transformer | 68.99 | ||
| Transformer-based [27] | ViT-B (ref) | Transformer | 68.77 | 68.08 |
| ConvNeXt-Base | CNN | 68.23 | ||
| Swin-V2-B | Transformer | 67.62 | ||
| ShuffleNetV2-X2 | Lightweight CNN | 68.39 | ||
| ResNet-152 | CNN | 66.81 | ||
| MaxViT-T | CNN + Transformer | 68.66 |
| Method | Data | Subject Selection | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) |
|---|---|---|---|---|---|---|
| Ahmed et al. [9] | Skeleton | Random | 92.00 | – | – | – |
| Sania et al. [10] | Skeleton | Random Average | 92.33 | – | – | – |
| Proposed (Ours) | Skeleton | Random Average | 95.00 | 97.92 | 94.00 | 95.92 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, J.; Miah, A.S.M.; Kakizaki, M.; Hassan, N.; Tomioka, Y. Autism Spectrum Disorder Detection Using Skeleton-Based Body Movement Analysis via Dual-Stream Deep Learning. Electronics 2025, 14, 2231. https://doi.org/10.3390/electronics14112231
Shin J, Miah ASM, Kakizaki M, Hassan N, Tomioka Y. Autism Spectrum Disorder Detection Using Skeleton-Based Body Movement Analysis via Dual-Stream Deep Learning. Electronics. 2025; 14(11):2231. https://doi.org/10.3390/electronics14112231
Chicago/Turabian StyleShin, Jungpil, Abu Saleh Musa Miah, Manato Kakizaki, Najmul Hassan, and Yoichi Tomioka. 2025. "Autism Spectrum Disorder Detection Using Skeleton-Based Body Movement Analysis via Dual-Stream Deep Learning" Electronics 14, no. 11: 2231. https://doi.org/10.3390/electronics14112231
APA StyleShin, J., Miah, A. S. M., Kakizaki, M., Hassan, N., & Tomioka, Y. (2025). Autism Spectrum Disorder Detection Using Skeleton-Based Body Movement Analysis via Dual-Stream Deep Learning. Electronics, 14(11), 2231. https://doi.org/10.3390/electronics14112231









