Abstract
This paper presents LUVS-Net, which is a lightweight convolutional network for retinal vessel segmentation in fundus images that is designed for resource-constrained devices that are typically unable to meet the computational requirements of large neural networks. The computational challenges arise due to low-quality retinal images, wide variance in image acquisition conditions and disparities in intensity. Consequently, the training of existing segmentation methods requires a multitude of trainable parameters for the training of networks, resulting in computational complexity. The proposed Lightweight U-Net for Vessel Segmentation Network (LUVS-Net) can achieve high segmentation performance with only a few trainable parameters. This network uses an encoder–decoder framework in which edge data are transposed from the first layers of the encoder to the last layer of the decoder, massively improving the convergence latency. Additionally, LUVS-Net’s design allows for a dual-stream information flow both inside as well as outside of the encoder–decoder pair. The network width is enhanced using group convolutions, which allow the network to learn a larger number of low- and intermediate-level features. Spatial information loss is minimized using skip connections, and class imbalances are mitigated using dice loss for pixel-wise classification. The performance of the proposed network is evaluated on the publicly available retinal blood vessel datasets DRIVE, CHASE_DB1 and STARE. LUVS-Net proves to be quite competitive, outperforming alternative state-of-the-art segmentation methods and achieving comparable accuracy using trainable parameters that are reduced by two to three orders of magnitude compared with those of comparative state-of-the-art methods.
1. Introduction
Eyes are the principle visual sensory organs for humans. Their vulnerability to a number of pathologies and ailments with aging is a serious concern around the world. This concern stems from the fact that, according to the World Health Organization (WHO), close to 2 billion people are facing some form of visual impairment globally [1]. Some of the leading diseases include trachoma, glaucoma, diabetic retinopathy (DR), hypersensitivity retinopathy (HR), cataract and age-related macular degeneration (AMD).
Since many of these ailments cause irreversible damage that may even lead to blindness, a large body of research is dedicated to development of techniques and technologies for the early diagnosis of eye diseases. To this end, visual computing and machine learning have played central roles in driving a significant volume of the current research pertaining to eye health [2,3,4].
The automatic detection of eye disease has many benefits, including being fast and having low costs and a low probability of misdiagnosis. This facilitates accurate diagnosis at an early stage. Consequently, there is a significant incentive to design a non-invasive automated diagnostic system to solve the difficulties that ophthalmologists encounter during routine patient examinations [5,6,7]. The retinal blood vessels in the fundus are extremely delicate and, hence, are more prone to eye diseases. There is a strong correlation between deterioration in the fundus vasculature and occurrence of the aforementioned diseases. DR can be discovered by detecting abnormalities in the shape and composition of the retinal vessels that are caused by diabetes [5].
Glaucoma causes particular type of changes in the retinal vascular structure that can be detected using computerized methods to diagnose the disease [6]. HR is a retinal disease driven by high blood pressure, where patients may have increased vessel stenosis or arterial blood pressure as a result of the curvature of the vessels [7]. AMD causes the deterioration of the central region of the retina (the macula) resulting in blurry vision and difficulty focusing. If not detected early on, these diseases can advance to more serious stages that may even lead to complete loss of eyesight. By studying changes in the retinal vascular structures in the retina, these diseases can be diagnosed, and their progression can be monitored [8].
Traditionally, eye specialists diagnose eye diseases by studying retinal fundus images without any automatic diagnostic aid. However, more recently, computer-vision techniques are playing an increasingly significant role in the field of medical imaging due to advancements in the areas of machine learning and digital image processing [9,10,11]. Features in the retinal blood vessels visible in fundus images were identified as markers indicative of a variety of eye diseases as well as other ailments (diabetes, cardio-vascular, etc.) [12].
It is essential for any visual computing platform to effectively extract the images of blood vessels (extraction/segmentation) from retinal fundus images for the accurate detection of pathological symptoms. Consequently, a large volume of current research is devoted to improving the precision and accuracy of existing techniques for the retinal vascular segmentation procedure [12,13]. Other research involves the development of a pipeline of operations targeted at the improved detection of eye diseases [13], and more is being done to improve noise-removal methods [14,15,16].
Deep-learning-based techniques have shown very promising results when it comes to detecting complex patterns in a wide variety of medical images generally including those in retinal fundus images [17,18,19]. Numerous procedures to segment retinal blood vessels automatically from digital fundus images have been presented in the literature [20]. The segmentation of an image is the procedure of dividing a digital image into a number of segments of interest. Recent approaches leverage segmentation as a classifier. Segmentation can be used as classifier for any number of things in images of a wide variety.
For instance, Tsai et al. [21] demonstrated the efficacy of using a segmentation network to segment out the shape of a hand in an image at a state-of-the-art efficiency. Similarly, Wang et al. [22] proposed an advanced multi-scale segmentation technique to detect defects in a subway tunnel. This research used segmentation to distinguish whether individual pixels in a fundus image are a component of a vessel or not. Using segmentation to classify artifacts as being either vessels or not improves the precision of vascular analysis compared to other classification techniques. Consequently, a large volume of recent research is concerned with the optimization of retinal vascular segmentation algorithms [23].
While deep-learning-based techniques produce highly accurate results, they are constrained by their heavy computational requirements. There is potential in the literature to expand the capacity of medical image analyses to platforms with relatively low computational capabilities (mobile phones and embedded systems). Significant work is being done to develop “lightweight” neural networks that are able to perform on limited hardware comparable to high-end computing platforms.
This work presents the Lightweight U-Net Vessel Segmentor (LUVS-NET) for retinal vasculature detection in fundus images. The main contribution of this work lies in removing redundant layers/operations, which add to the computational cost without notably contributing to the overall performance. To this end, we suggest an optimized framework that can extract deep features required to segment retinal vessels. For fair analysis, we compare the proposed LUVS-Net with the versions of U-Net while also stating the structural differences with the latter. LUVS-Net is trained and tested with the STARE, DRIVE and CHASE_DB1 datasets. The efficacy of LUVS-Net is demonstrated by benchmarking it relative to contemporary networks.
2. Related Works
Convolutional neural network (CNN)-based approaches are by far the most popular learning techniques that are used for medical image analysis and semantic segmentation today [23,24,25]. In visual computing, semantic segmentation is considered to be a fundamental technique where artifacts in an image are classified based on a pixel-wise discrimination process The segmentation algorithm is able to differentiate between any object of interest and the background of an image on a pixel level.
This means that bone fractures, implants, deformities, carcinogenic growth and other things of interest that may be present in any form of medical image can automatically be identified. The technology today is able to detect and identify minute regions of interest in an image. This makes semantic segmentation ideally suitable to detect a vasculature network inside a retinal fundus image by teaching the network to analyze the vascular structure.
When it comes to image segmentation, the seminal U-Net developed by Ronneberger et al. [26] and ResNet presented by He et al. [27] form the backbone of most new developments in the area of segmentation today, specifically those of medical images. Several recent works augmented and expanded the capabilities of U-Net and ResNet. For instance, Zhuang et al. [28] used residual blocks to stack a pair of U-Nets that resulted in an increase in the number of information flow channels (LadderNet). Alom et al. [29] used an optimal combination of U-Net, RCNN and ResNet to develop a highly efficient network (R2U-Net) for the segmentation of medical images.
A great deal of recent work has focused on modifying and upgrading these “industry standard” networks to produce lightweight variants. Laibacher et al. [30] presented a modified light U-Net that has components of MobileNetV2 encoder that performs real-time inference on ARM-based embedded platforms. Khan et al. showed that computational requirements can be reduced by minimizing the pooling layers, reducing the number of feature channels and skipping connections between the encoder and decoder to produce pathology classification comparable to a traditional deep network [31].
Li et al. [32] augmented a U-Net with an attention module that aids in image acquisition and feature fusion for light enhancement called IterNet. It was presented as a lightweight network for image segmentation that is able to find obscure artifacts from a segmented image instead of the heavy raw image. Jiang et al. [33] modified a U-Net by down-scaling it to three layers with only five dense convolutional blocks, reduced feature channels and the introduction of dropout layers to effectively detect laser scars left after ophthalmic treatment. Kamran et al. [34] presented a lightweight generative adversarial network (GAN) for retinal vessel segmentation that uses two generators with two auto-encoding discriminators for improved performance.
A great deal of recent development in terms of segmentation networks has been towards producing lightweight networks presenting lower than normal computational requirements. Galdran et al. [35] demonstrated that, if a U-Net is trained with precision and tested rigorously, its performance does not deteriorate even if the parameters are reduced by many orders of magnitude. More recently, ref. [36] presented several retinal vessel segmentation models that were minimized and modified to produce good and comparable results to their heavier counterparts.
Moreover, ref. [37] was able to improve performance while decreasing the number of parameters by using prompt blocks to extract useful features. Khan et al. presented a novel protocol to use image complexity as a factor to determine how various macro-level design aspects can be modified to achieve optimal reduction in computational requirements [38]. Lastly, an attention block was employed in [39] to improve the segmentation of the encoder–decoder design, coupled with the use of adversarial learning.
3. Proposed Methodology
Most of the popular deep-learning-based methods used for retinal vessel segmentation are not robust enough for minor vessels [40]. The identification of tiny vessels is critical for accurate and precise disease analysis from medical images. In terms of traditional segmentation algorithms, this translates into deep networks with many convolution and pooling layers. This leads to vanishing gradient issues. The overall performance of pixel-wise classification suffers as a result of the lack of spatial information.
This work presents the Lightweight U-Net Vessel Segmentor (LUVS-Net) architecture, a retinal vessel segmentation technique based on U-Net. It introduces an elegant architecture that uses group convolutions to extend the network width enabling it to learn a larger volume of low- and intermediate-level features. Group convolution allows the reduction of inter-layer convolutional filters to further minimize the network complexity. Furthermore, it integrates aspects of both supervised and unsupervised learning to create a hybrid learning system. These augmentations allow for a lightweight network compared to a fully convolutional network. Lastly, this work demonstrates its efficacy in training very high-resolution images and producing segmentation with efficiency that is competitive compared to other existing methods.
3.1. LUVS-Net Architecture
The proposed LUVS-Net is illustrated in in Figure 1 where the core components of the network are summarized. The LUVS-Net can be divided into two parts:
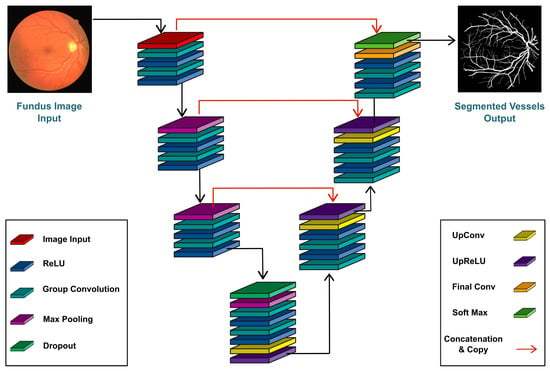
Figure 1.
Lightweight U-Net Vessel Segmentor (LUVS-Net) architecture.
- The encoder, also known as a contracting encoder, is located on the left side of the proposed network and is used for feature extraction.
- The decoder, also known as the expanding decoder, is towards the right of the proposed network and restores the features extracted at the encoder side.
Both the encoder and the decoder stages of LUVS-Net have the same number of convolutional layers as proposed in the base U-Net architecture; however, in this case, group-convolution is performed. Padding is used to keep the output image’s scale consistent with the input image. The use of padding ensures that the output image is the same size as the input image. A 1 × 1 convolutional layer is applied to convert the final output channel number into the number of divided categories.
3.1.1. Encoder
LUVS Net is a fully connected CNN with six dense stages: three stages for each the encoder and decoder. In simple terms, encoding is the conversion of one type of data to another type of data for the ease of transmission, communication and storage. The LUVS-Net encoder also acts as a contracting encoder. The contracting encoder in the proposed network has three stages, each with two convolutional layers of kernel size followed by a ReLU (rectified linear unit) activation function and a max-pooling layer of pool size.
In the first step, the input image (dimensions: ) is passed through a series of two convolutional layers proceeded by a pair of ReLU activation function layers. The second and third stages take in the feature maps generated in the preceding step as input and pass them through the max-pooling layer to minimize the size of the feature maps. The bottleneck used in each stage reduces the number of channels after concatenation to minimize issues related to memory and space.
The overall number of convolutional filters is reduced in consecutive layers. Furthermore, grouped convolutions are employed to reduce the complexity of the network even further. During the encoding phase, while convoluting for image segmentation, there is some spatial loss where useful information is lost. This loss can be covered by deep-feature concatenation. The final feature map is for a input image. Each dense stage in the LUVS-Net encoder structure provides feature empowerment, while the bottleneck layer reduces the feature map depth.
3.1.2. Bridge
The LUVS-Net Bridge takes the convoluted image from the encoder as input and passes it to the decoder to reconstruct the image with the original dimensions with minimum information loss. In the bridge phase, the layers used are a dropout layer, max-pooling layer, convolutional layer, ReLU layer, convolutional layer, ReLU layer, dropout layer, up-convolutional layer and up-ReLU layer. The function of the dropout layer is to reduce the risk of over-fitting by dropping the node connections in the dense layers randomly during the back-propagation phase.
The max-pooling layer minimizes the feature map further by only considering the largest value pixels in the patch to pass a sparse feature map, thereby reducing the computational costs. The connecting convolutional layers use the ReLU activation function to generate feature maps. The up-convolutional layers are used along with the up-ReLU activation functions to revive the original feature map dimensions with the help of padding around the feature map before convoluting with the kernels.
3.1.3. Decoder
The output from the bridge is passed into the LUVS-Net decoder for reconstruction of the image, where the dimensions progressively increase until they reach the original size. The LUVS-Net decoder as shown in Figure 1, performs the inverse operation of the encoder by reviving the original dimensions of the feature map reduced by the corresponding encoder. The decoder has three stages, each stage starting with an un-pooling layer followed by a series of up-convolution layers with up-ReLU activation function. The output at the third stage is the reconstructed and segmented image result.
The depth-wise concatenation layers of each decoder stage are driven by three inputs. These comprise the first and second convolution in addition to direct information from the outer dense connection of the corresponding encoder block. The concatenation of feature maps helps to give localization information. This approach improves the latency by leveraging only the edge data from the encoder to decoder. The edge data are provided by the outer dense paths. The decoder receives an pixel input from the encoder. This input is then used to build the (final) feature map, the size of which is equal to the input image provided at the beginning of the encoder phase.
The last bottleneck layer is configured with two channels that serve to reduce the feature map depth. The bottleneck layer doubles up as a class mask layer with the two classes; “Retinitis-pigmentosa” and “Background”, corresponding to the two channels. The third and concluding layer of the decoder before the image output uses the Softmax activation function for predicting the classes.
Table 1 and Figure 1 depict the proposed network in detail. Blood vessels are the key components of fundus retinal images. The vessels are automatically distinguished from the digital color images of the fundus. The main goal of this study is to classify individual pixels in the image as either belonging to a vessel or not. The two classes of pixels, vessel and non-vessel pixels, are considered in this segmentation process, and each function vector belongs to one of these two classes. The purposed model focuses on the following aspects:

Table 1.
Proposed LUVS-Net filter details.
- The use of a shallow network with one or two layers in each stage, respectively.
- The optimization in the number of filters throughout the network.
The shallow network enables the use of a minimum number of filters that preserve the high-frequency pixels. Usually, four to five filters or max-pooling layers need to be utilized for a basic U-Net architecture. Since this research deals with a one-class problem, using more filters increases the overlaps, which, in turn, decreases the performance. Hence, utilizing more filters complicates the learning process.
3.2. Architectural Differences with U-Net
The proposed LUVS-Net significantly reduces the computational cost by removing redundant layers/ operations without compromising the performance of the network. Each stage in LUVS-Net consists of an input layer, three contracting encoder layers, a bridge layer, three expanding decoder layers and an output layer. The activation layer is the ReLU activation function. The layers in LUVS-Net are summarized and compared to the classical U-Net in Table 2. The number of convolutional layers in LUVS-Net is significantly less than in U-Net.

Table 2.
Layers in LUVS-Net.
The number of filters in LUVS-Net starts from 24 for every convolutional layer at every level and doubles as they are downsampled, going up to a maximum of 96 (per convolutional layer) as shown in Table 1. In U-Net, for comparison, the number of filters starts from 64 (per convolutional layer) in the first level, doubling with each instance of downsampling to a total of 1024 filters per convolution. LUVS-Net is further showcased in terms of the number of parameters and model size in comparison with both U-Net and Segnet in Table 3. LUVS-Net represents close to a 98% decrease in the number of parameters and close to a 99.5% decrease in the model size compared to U-Net.

Table 3.
A comparison of the number of parameters and model size.
LUVS-Net is, arguably, computationally much lighter when compared to the classical U-Net. This conclusion can be drawn by considering the relatively lower number of convolutional layers, lower number of filters and drastically lower number of total parameters compounded by the much smaller relative model size in LUVS-Net compared to U-Net. This leads to the implicit conclusion that LUVS-Net has massively reduced computational requirements relative to U-Net. These lowered requirements come without any degradation in the relative performance (as demonstrated in the following sections), lending merit to LUV-Net’s credentials as a lightweight alternative to U-Net that is well suited for mobile and embedded computational platforms.
4. Experimental Results
This section presents a comparative analysis of the proposed LUVS-Net using state-of-the-art methods. The following three fundus retinal image datasets, all of which are available in the public domain, were used for the performance analysis:
- STARE (Structured Analysis of the Retina): A collection of twenty fundus images gathered in the United States [41].
- CHASE DB1 ( Child Heart and Health Study in England): A collection of retinal fundus images based on fourteen pediatric subjects [42].
- DRIVE (Digital Retinal Images for Vessel Extraction): A set of 40 fundus images of patients with diabetic retinopathy based in the Netherlands [43].
Among these three datasets, only the DRIVE dataset is available with a binary mask that reveals the field of view (FOV). Blood vessels were segmented from the retinal images in DRIVE manually. In the case of STARE and CHASE_DB1, binary masks were manually generated using standard procedures [44]. While DRIVE and CHASE_DB1 are available with their training and testing sub-sets already bifurcated, the STARE dataset was divided into training and testing subsets using the “leave-one-out” strategy [44].
In this strategy, training starts with samples, that are tested on samples, where m are the remaining samples. This process is repeated for n iterations in a manner that ensures that each individual sample in the dataset is left out at least once throughout the entire run of iterations. Table 4 summarizes the distribution of the training and testing subsets for each of the three selected datasets in the benchmarking performed in this research.

Table 4.
Sample size and distribution of the experimental dataset.
4.1. Performance Parameters
We used adaptive moment estimation (Adam) with a fixed learning rate of as the optimization solver. A weighted cross-entropy loss was used throughout as an objective function for training as we observed that, for each retinal image’s vessel segmentation instance, the non-vessel pixels outweighed the pixels of the vessels by a significant difference. We used median frequency balancing to determine the class association weights [45]. The network batch size was 8.
With a 10 + 10 split between training and testing images, the “leave-one-out” strategy was used. Furthermore, the number of images was increased to a sufficient number for training by employing data augmentation. Augmentation was achieved through both contrast enhancement and one-degree rotations. Additionally, image brightness and contrast were manipulated using randomized factors. Lastly, the network was trained and tested on a platform with an Nvidia GeForce GTX2080TI GPU, Intel(R) Xeon(R) W-2133 3.6 GHz CPU and 96 GB RAM.
4.2. Evaluation Criteria
Most neural network models that are used to segment vessels from retinal images are essential two-class classifiers (vessels and background). These segmentation classifiers were evaluated using ground truth photos for their effectiveness. These images were examined and graded by ophthalmologists.
For the performance evaluation of the proposed system, we used the three metrics described in Equations (1)–(3) based on the following four parameters.
- True Negative (TN): Correctly classified as a background pixel.
- False Positive (FP): Incorrectly classified as a vessel pixel.
- True Positive (TP): Correctly classified as a vessel pixel.
- False Negative (FN): Incorrectly classified as a background pixel.
The sensitivity, specificity and accuracy are, henceforth, referred to as Se, Sp and Acc, respectively. Additionally, the ROC-AUC (receiver operating characteristic area under the curve) was used to assess the quality of classification in case of a dataset with distribution imbalance. The PR-AUC (precision–recall area under the curve) was used to measure the efficiency of classification.
4.3. Comparison with the State-of-the-Art
Figure 2, Figure 3 and Figure 4 depict the perceptible outcomes of the simulation on the three datasets. Each row in the figures shows a tested image, its corresponding ground truth and the segmented outcome of the proposed network from left to right.
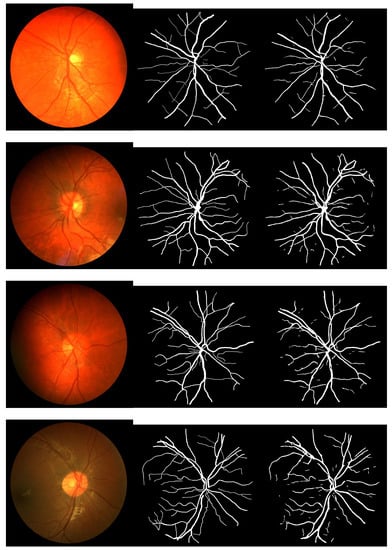
Figure 2.
Sample images, their corresponding ground truth and the outcome of the proposed network while testing the CHASE_DB1 dataset (left to right).
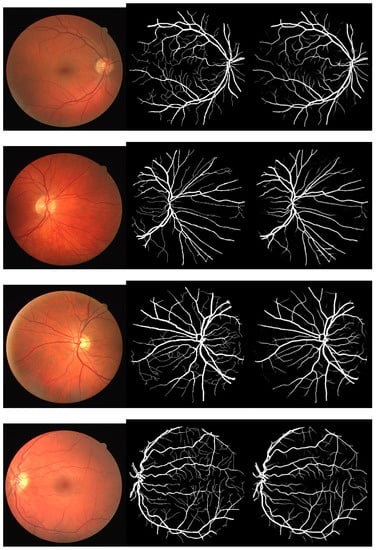
Figure 3.
Sample images, their corresponding ground truth and the outcome of the proposed network while testing the DRIVE dataset (left to right).
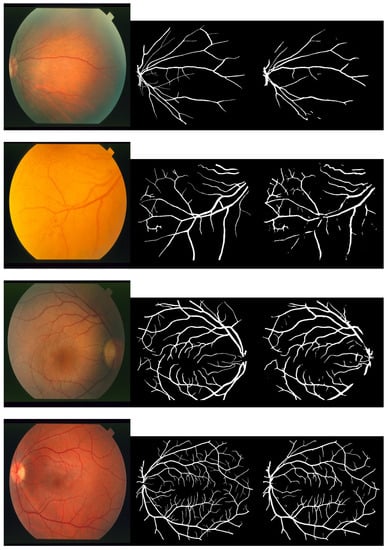
Figure 4.
Sample images, their corresponding ground truth and the outcome of the proposed network while testing the STARE dataset (left to right).
The data are presented and summarized in tabular form to evaluate and compare them to state-of-the-art models. In Table 5, the results obtained from simulations of the proposed LUVS-Net and other models on CHASE_DB1 are compared. The Se, Sp and Acc of the proposed LUVS-Net are 0.8269, 0.9846 and 0.9738, respectively.

Table 5.
LUVS-Net compared to existing models as tested (dataset: CHASE_DB1).
The results of the proposed LUVS-Net model compared to the state-of-the-art models using the DRIVE dataset can be seen in Table 6 where best performance metrics in each column are highlighted in bold. Clearly, Se, Sp and Acc of the proposed LUVS-Net are 0.8258, 0.983 and 0.9692, respectively. Similarly, Table 7 compares the outcomes achieved by all models, including the proposed LUVS-Net using the STARE dataset. The Se, Sp and Acc of the proposed LUVS-Net are 0.8133, 0.9861 and 0.9733, respectively.

Table 6.
LUVS-Net compared to existing models as tested (dataset: DRIVE).

Table 7.
LUVS-Net compared to existing models as tested (dataset: STARE).
On the basis of analyses using industry-standard metrics, LUVS-net can clearly be seen to outperform existing cutting-edge models.
The summary shown in Figure 5 shows the consistency of LUV-Net’s performance across all three datasets that were used for training and validation.
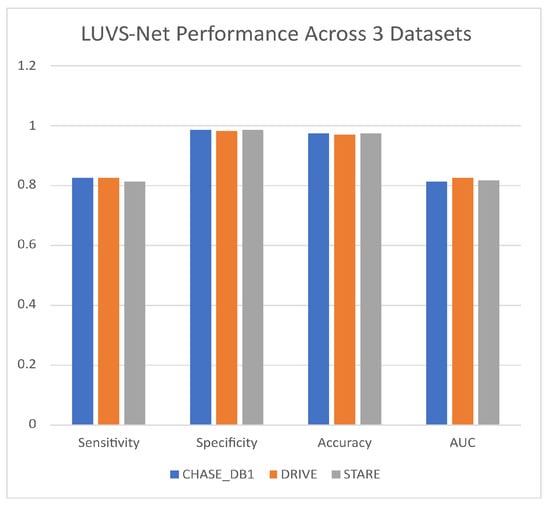
Figure 5.
Lightweight U-Net Vessel Segmentor (LUVS-Net) architecture performance.
5. Conclusions
One of the most prevalent ophthalmic ailments afflicting people with diabetes is DR, which has the potential to lead to complete blindness. While screening and detecting diabetic retinopathy, an ophthalmologist can greatly benefit from the precise segmentation of blood vessels in the retina. This work presented LUVS-Net for the segmentation of blood vessels in the retina, which can greatly aid in the diagnosis of this disease. As a result of the rich feature set in the dense block, LUVS-Net is able to attain and transfer the spatial information of the image. The transfer from the encoder to the decoder of data from the edges allows for fast convergence of the network.
LUVS-Net has three main design attributes: First, feature concatenation improves the feature quality, whereas the bottleneck layers in dense blocks control the memory requirements. Secondly, the number of convolution layers in each of the network’s six blocks is reduced to minimize spatial information loss, and group convolution is used to enhance the network width. Finally, LUVS-Net leverages dense paths for feature empowerment that aids in extracting very fine information from images.
Three publicly available datasets were used to demonstrate LUVS-Net’s ability to surpass the accuracy and computational efficiency of existing state-of-the-art methods for each of the three datasets it was tested on, thus proving its efficacy.
In conclusion, LUVS-Net can be used as a lightweight segmentor network to supplement the analysis and eventual diagnosis of diabetic retinopathy from fundus images with, arguably, state-of-the-art performance. With the potential for inference using hand-held smart platforms, this network has the potential to be employed in an app for the automated diagnosis of diabetic retinopathy. Such a system could be used as a platform to provide alternative opinions to assist medical doctors and ophthalmologists in the diagnosis of DR. Future work will focus on increases in the accuracy of blood-vessel segmentation considering the fact that this can aid in the diagnosis of a variety of retinal diseases in addition to DR.
Author Contributions
Conceptualization, K.N., M.T.I., A.N., H.A.K. and S.M.G.; Investigation, K.N., M.T.I. and A.N.; Writing—original draft, M.T.I., K.N., H.A.K., A.N. and S.M.G.; Writing—review and editing, A.N., H.A.K., K.N. and S.W.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported in part by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2021R1A6A1A03039493) and in part by the NRF grant funded by the Korea government (MSIT) (NRF-2022R1A2C1004401).
Data Availability Statement
All the datasets used in this study are freely available online.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bourne, R.R.; Flaxman, S.R.; Braithwaite, T.; Cicinelli, M.V.; Das, A.; Jonas, J.B.; Keeffe, J.; Kempen, J.H.; Leasher, J.; Limburg, H.; et al. Magnitude, temporal trends, and projections of the global prevalence of blindness and distance and near vision impairment: A systematic review and meta-analysis. Lancet Glob. Health 2017, 5, e888–e897. [Google Scholar] [CrossRef] [PubMed]
- Malik, S.; Kanwal, N.; Asghar, M.N.; Sadiq, M.A.A.; Karamat, I.; Fleury, M. Data driven approach for eye disease classification with machine learning. Appl. Sci. 2019, 9, 2789. [Google Scholar] [CrossRef]
- Armstrong, G.W.; Lorch, A.C. A (eye): A review of current applications of artificial intelligence and machine learning in ophthalmology. Int. Ophthalmol. Clin. 2020, 60, 57–71. [Google Scholar] [CrossRef] [PubMed]
- Murtagh, P.; Greene, G.; O’Brien, C. Current applications of machine learning in the screening and diagnosis of glaucoma: A systematic review and Meta-analysis. Int. J. Ophthalmol. 2020, 13, 149. [Google Scholar] [CrossRef] [PubMed]
- Lieth, E.; Gardner, T.W.; Barber, A.J.; Antonetti, D.A. Retinal neurodegeneration: Early pathology in diabetes. Clin. Exp. Ophthalmol. Viewp. 2000, 28, 3–8. [Google Scholar] [CrossRef]
- Abdullah, F.; Imtiaz, R.; Madni, H.A.; Khan, H.A.; Khan, T.M.; Khan, M.A.; Naqvi, S.S. A review on glaucoma disease detection using computerized techniques. IEEE Access 2021, 9, 37311–37333. [Google Scholar] [CrossRef]
- Moyer, J.H.; Heider, C.; Pevey, K.; Ford, R.V. The effect of treatment on the vascular deterioration associated with hypertension, with particular emphasis on renal function. Am. J. Med. 1958, 24, 177–192. [Google Scholar] [CrossRef]
- Dougherty, G.; Johnson, M.J.; Wiers, M.D. Measurement of retinal vascular tortuosity and its application to retinal pathologies. Med. Biol. Eng. Comput. 2010, 48, 87–95. [Google Scholar] [CrossRef]
- Currie, G.; Hawk, K.E.; Rohren, E.; Vial, A.; Klein, R. Machine learning and deep learning in medical imaging: Intelligent imaging. J. Med. Imaging Radiat. Sci. 2019, 50, 477–487. [Google Scholar] [CrossRef]
- Iqbal, S.; Khan, T.M.; Naveed, K.; Naqvi, S.S.; Nawaz, S.J. Recent trends and advances in fundus image analysis: A review. Comput. Biol. Med. 2022, 151, 106277. [Google Scholar] [CrossRef]
- Kim, M.; Yun, J.; Cho, Y.; Shin, K.; Jang, R.; Bae, H.j.; Kim, N. Deep learning in medical imaging. Neurospine 2019, 16, 657. [Google Scholar] [CrossRef]
- Imran, A.; Li, J.; Pei, Y.; Yang, J.J.; Wang, Q. Comparative analysis of vessel segmentation techniques in retinal images. IEEE Access 2019, 7, 114862–114887. [Google Scholar] [CrossRef]
- Khan, T.M.; Khan, M.A.; Rehman, N.U.; Naveed, K.; Afridi, I.U.; Naqvi, S.S.; Raazak, I. Width-wise vessel bifurcation for improved retinal vessel segmentation. Biomed. Signal Process. Control. 2022, 71, 103169. [Google Scholar] [CrossRef]
- Naveed, K.; Abdullah, F.; Madni, H.A.; Khan, M.A.; Khan, T.M.; Naqvi, S.S. Towards automated eye diagnosis: An improved retinal vessel segmentation framework using ensemble block matching 3D filter. Diagnostics 2021, 11, 114. [Google Scholar] [CrossRef]
- Khawaja, A.; Khan, T.M.; Naveed, K.; Naqvi, S.S.; Rehman, N.U.; Nawaz, S.J. An improved retinal vessel segmentation framework using frangi filter coupled with the probabilistic patch based denoiser. IEEE Access 2019, 7, 164344–164361. [Google Scholar] [CrossRef]
- Naveed, K.; Ehsan, S.; McDonald-Maier, K.D.; Ur Rehman, N. A multiscale denoising framework using detection theory with application to images from CMOS/CCD sensors. Sensors 2019, 19, 206. [Google Scholar] [CrossRef] [PubMed]
- Raza, M.; Naveed, K.; Akram, A.; Salem, N.; Afaq, A.; Madni, H.A.; Khan, M.A.; Din, M.Z. DAVS-NET: Dense Aggregation Vessel Segmentation Network for retinal vasculature detection in fundus images. PLoS ONE 2021, 16, e0261698. [Google Scholar] [CrossRef]
- Naqvi, S.S.; Fatima, N.; Khan, T.M.; Rehman, Z.U.; Khan, M.A. Automatic optic disk detection and segmentation by variational active contour estimation in retinal fundus images. Signal Image Video Process. 2019, 13, 1191–1198. [Google Scholar] [CrossRef]
- Khan, T.M.; Alhussein, M.; Aurangzeb, K.; Arsalan, M.; Naqvi, S.S.; Nawaz, S.J. Residual connection-based encoder decoder network (RCED-Net) for retinal vessel segmentation. IEEE Access 2020, 8, 131257–131272. [Google Scholar] [CrossRef]
- Asgari Taghanaki, S.; Abhishek, K.; Cohen, J.P.; Cohen-Adad, J.; Hamarneh, G. Deep semantic segmentation of natural and medical images: A review. Artif. Intell. Rev. 2021, 54, 137–178. [Google Scholar] [CrossRef]
- Tsai, T.H.; Huang, S.A. Refined U-net: A new semantic technique on hand segmentation. Neurocomputing 2022, 495, 1–10. [Google Scholar] [CrossRef]
- Wang, A.; Togo, R.; Ogawa, T.; Haseyama, M. Defect detection of subway tunnels using advanced U-Net network. Sensors 2022, 22, 2330. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chuah, J.H.; Raza, A.; Wang, Y. Retinal vessel segmentation using deep learning: A review. IEEE Access 2021, 9, 111985–112004. [Google Scholar] [CrossRef]
- Kayalibay, B.; Jensen, G.; van der Smagt, P. CNN-based segmentation of medical imaging data. arXiv 2017, arXiv:1701.03056. [Google Scholar]
- Anwar, S.M.; Majid, M.; Qayyum, A.; Awais, M.; Alnowami, M.; Khan, M.K. Medical image analysis using convolutional neural networks: A review. J. Med. Syst. 2018, 42, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Zhuang, J. LadderNet: Multi-path networks based on U-Net for medical image segmentation. arXiv 2018, arXiv:1810.07810. [Google Scholar]
- Alom, M.Z.; Hasan, M.; Yakopcic, C.; Taha, T.M.; Asari, V.K. Recurrent residual convolutional neural network based on u-net (r2u-net) for medical image segmentation. arXiv 2018, arXiv:1802.06955. [Google Scholar]
- Laibacher, T.; Weyde, T.; Jalali, S. M2U-Net: Effective and efficient retinal vessel segmentation for resource-constrained environments. arXiv 2018, arXiv:1811.07738. [Google Scholar]
- Khan, T.M.; Robles-Kelly, A.; Naqvi, S.S. T-Net: A Resource-Constrained Tiny Convolutional Neural Network for Medical Image Segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–8 January 2022; pp. 644–653. [Google Scholar]
- Li, L.; Verma, M.; Nakashima, Y.; Nagahara, H.; Kawasaki, R. Iternet: Retinal image segmentation utilizing structural redundancy in vessel networks. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Snowmass Village, CO, USA, 1–5 March 2020; pp. 3656–3665. [Google Scholar]
- Jiang, Y.; Pan, J.; Yuan, M.; Shen, Y.; Zhu, J.; Wang, Y.; Li, Y.; Zhang, K.; Yu, Q.; Xie, H.; et al. Segmentation of Laser Marks of Diabetic Retinopathy in the Fundus Photographs Using Lightweight U-Net. J. Diabetes Res. 2021, 2021, 8766517. [Google Scholar] [CrossRef]
- Kamran, S.A.; Hossain, K.F.; Tavakkoli, A.; Zuckerbrod, S.L.; Sanders, K.M.; Baker, S.A. RV-GAN: Segmenting retinal vascular structure in fundus photographs using a novel multi-scale generative adversarial network. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin/Heidelberg, Germany, 2021; pp. 34–44. [Google Scholar]
- Galdran, A.; Anjos, A.; Dolz, J.; Chakor, H.; Lombaert, H.; Ayed, I.B. The little w-net that could: State-of-the-art retinal vessel segmentation with minimalistic models. arXiv 2020, arXiv:2009.01907. [Google Scholar]
- Galdran, A.; Anjos, A.; Dolz, J.; Chakor, H.; Lombaert, H.; Ayed, I.B. State-of-the-art retinal vessel segmentation with minimalistic models. Sci. Rep. 2022, 12, 6174. [Google Scholar] [CrossRef]
- Arsalan, M.; Khan, T.M.; Naqvi, S.S.; Nawaz, M.; Razzak, I. Prompt deep light-weight vessel segmentation network (PLVS-Net). IEEE/ACM Trans. Comput. Biol. Bioinform. 2023, 20, 1363–1371. [Google Scholar] [CrossRef]
- Khan, T.M.; Naqvi, S.S.; Meijering, E. Leveraging image complexity in macro-level neural network design for medical image segmentation. Sci. Rep. 2022, 12, 22286. [Google Scholar] [CrossRef]
- Naqvi, S.S.; Langah, Z.A.; Khan, H.A.; Khan, M.I.; Bashir, T.; Razzak, M.I.; Khan, T.M. GLAN: GAN Assisted Lightweight Attention Network for Biomedical Imaging Based Diagnostics. Cogn. Comput. 2023. [Google Scholar] [CrossRef]
- Haleem, M.S.; Han, L.; Van Hemert, J.; Li, B. Automatic extraction of retinal features from colour retinal images for glaucoma diagnosis: A review. Comput. Med. Imaging Graph. 2013, 37, 581–596. [Google Scholar] [CrossRef]
- Hoover, A.; Kouznetsova, V.; Goldbaum, M. Locating blood vessels in retinal images by piecewise threshold probing of a matched filter response. IEEE Trans. Med. Imaging 2000, 19, 203–210. [Google Scholar] [CrossRef]
- Fraz, M.M.; Remagnino, P.; Hoppe, A.; Uyyanonvara, B.; Rudnicka, A.R.; Owen, C.G.; Barman, S.A. An Ensemble Classification-Based Approach Applied to Retinal Blood Vessel Segmentation. IEEE Trans. Biomed. Eng. 2012, 59, 2538–2548. [Google Scholar] [CrossRef]
- Staal, J.; Abràmoff, M.D.; Niemeijer, M.; Viergever, M.A.; Van Ginneken, B. Ridge-based vessel segmentation in color images of the retina. IEEE Trans. Med. Imaging 2004, 23, 501–509. [Google Scholar] [CrossRef]
- Soares, J.V.; Leandro, J.J.; Cesar, R.M.; Jelinek, H.F.; Cree, M.J. Retinal vessel segmentation using the 2-D Gabor wavelet and supervised classification. IEEE Trans. Med. Imaging 2006, 25, 1214–1222. [Google Scholar] [CrossRef]
- Badrinarayanan, V.; Handa, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for robust semantic pixel-wise labelling. arXiv 2015, arXiv:1505.07293. [Google Scholar]
- Khawaja, A.; Khan, T.M.; Khan, M.A.; Nawaz, S.J. A multi-scale directional line detector for retinal vessel segmentation. Sensors 2019, 19, 4949. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Dashtbozorg, B.; Bekkers, E.; Pluim, J.P.; Duits, R.; ter Haar Romeny, B.M. Robust retinal vessel segmentation via locally adaptive derivative frames in orientation scores. IEEE Trans. Med. Imaging 2016, 35, 2631–2644. [Google Scholar] [CrossRef] [PubMed]
- Arsalan, M.; Owais, M.; Mahmood, T.; Cho, S.W.; Park, K.R. Aiding the diagnosis of diabetic and hypertensive retinopathy using artificial intelligence-based semantic segmentation. J. Clin. Med. 2019, 8, 1446. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Meng, Z.; Pham, T.D.; Chen, Q.; Wei, L.; Su, R. DUNet: A deformable network for retinal vessel segmentation. Knowl.-Based Syst. 2019, 178, 149–162. [Google Scholar] [CrossRef]
- Yin, P.; Yuan, R.; Cheng, Y.; Wu, Q. Deep guidance network for biomedical image segmentation. IEEE Access 2020, 8, 116106–116116. [Google Scholar] [CrossRef]
- Wang, D.; Haytham, A.; Pottenburgh, J.; Saeedi, O.; Tao, Y. Hard attention net for automatic retinal vessel segmentation. IEEE J. Biomed. Health Inform. 2020, 24, 3384–3396. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Yu, S.; Ma, K.; Wang, J.; Ding, X.; Zheng, Y. Multi-task neural networks with spatial activation for retinal vessel segmentation and artery/vein classification. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin/Heidelberg, Germany, 2019; pp. 769–778. [Google Scholar]
- Guo, S.; Wang, K.; Kang, H.; Zhang, Y.; Gao, Y.; Li, T. BTS-DSN: Deeply supervised neural network with short connections for retinal vessel segmentation. Int. J. Med. Inform. 2019, 126, 105–113. [Google Scholar] [CrossRef]
- Wu, Y.; Xia, Y.; Song, Y.; Zhang, D.; Liu, D.; Zhang, C.; Cai, W. Vessel-Net: Retinal vessel segmentation under multi-path supervision. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin/Heidelberg, Germany, 2019; pp. 264–272. [Google Scholar]
- Wang, B.; Qiu, S.; He, H. Dual encoding u-net for retinal vessel segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin/Heidelberg, Germany, 2019; pp. 84–92. [Google Scholar]
- Gu, Z.; Cheng, J.; Fu, H.; Zhou, K.; Hao, H.; Zhao, Y.; Zhang, T.; Gao, S.; Liu, J. Ce-net: Context encoder network for 2d medical image segmentation. IEEE Trans. Med. Imaging 2019, 38, 2281–2292. [Google Scholar] [CrossRef]
- Gargari, M.S.; Seyedi, M.H.; Alilou, M. Segmentation of Retinal Blood Vessels Using U-Net++ Architecture and Disease Prediction. Electronics 2022, 11, 3516. [Google Scholar] [CrossRef]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).