1. Introduction
Graphs are a powerful and basic type of data structure, commonly used to describe different relationships (referred to as edges) between objects (referred to as vertices). Most real-world data may be represented using graphs, which can be utilized to describe the in between data relationship [
1,
2]. For example, graphs can represent the nanotubes used in nanoelectronics [
3]. Additionally, chemical graph theory formulates chemical structures as a molecular graph, where the graph’s edges represent the chemical bonds that connect the compound’s atoms, and the graph’s nodes represent the atoms themselves [
4]. In bioinformatics, biological networks make a significant addition to the molecular structure of proteins, genetic disease association networks, and protein interaction networks [
5]. In social networks, graphs are employed to implement newsfeeds, determine page rank, and make online recommendations [
6]. The functionality of human brains is examined using graphs in the discipline of neuroscience [
7]. There are numerous other applications in various fields. These show why working with graphs is important.
In recent years, we have seen a tremendous increase in the size of graph data as well as the rising importance of graph data mining [
8]. The field of graph data mining, which deals with processing, analyzing, and extracting valuable information from real-world graph data, has recently seen a rise in interest [
9]. Real-world problems, including web document classification, a recommendation on a streaming service, and fraud detection in banking, are all solved using graph data mining [
10,
11]. Some of the common tasks involved in graph mining are community detection, node classification, graph classification, link prediction, and graph embedding [
12].
Graph classification, a significant subfield of graph mining, has been used in numerous real-world applications, including bioinformatics, chemoinformatics, social network analysis, etc. In bioinformatics, it is used for determining enzymatic proteins, determining whether cells are malignant, and determining altered DNA protein sequences. In the field of chemistry, it predicts the toxicity and anti-cancer effectiveness of various chemicals. Furthermore, social network applications and websites utilize graph mining to provide users with recommendations for pages or friends according to the user’s connections [
13,
14]. As friends on the social network are treated as nodes in the graph, the connections between people who are friends or followers are represented by graph edges [
15]. Graph classification is used to identify the most likely class labels for unseen graphs based on the constructed graph classification model [
16].
Recent years have seen a considerable increase in research interest in deep learning on graphs because graphs have a significant function and are frequently used in the real world. DNN provides powerful tools for graph classification, allowing for the development of complex models with high accuracy [
14]. Subsequently, deep learning research has numerous innovative applications in a variety of domains, including social networks, e-commerce networks, computer vision, speech recognition, object detection, and NLP [
17,
18]. CNN, a type of deep learning model, demonstrated remarkable performance in numerous computer vision and machine learning problems [
19]. It is a type of feedforward neural network that uses convolutional architecture to extract features from data [
20].
Graphs are not only used to store information in a structured way but they are also an important part of machine learning algorithms and data mining applications [
21]. Effectively learning the complicated non-Euclidean structure of graph data is a significant challenge [
22]. Since graph data cannot be directly classified by traditional classification algorithms, finding a way to include details about the graph’s structure in the machine learning model is the main problem when working with graphs [
23]. Consequently, embedding-based classifiers have gained interest because they allow us to perform node, graph, and subgraph mining tasks using traditional machine learning methods. In order to achieve this, a graph can be represented as a feature vector that can be used for community detection, node classification, and graph classification [
24,
25].
Graph classification presents unique challenges due to the inherent structural complexities and irregularities present in graph data. Therefore, numerous approaches have been proposed to address graph classification, including graph kernel methods, frequent subgraph mining methods, and deep learning methods. However, these methods suffer from various limitations. It is challenging for graph kernel methods to identify an appropriate kernel function that effectively captures the semantics of the structure while remaining computationally tractable. The frequent subgraph mining approach encounters complexities with implementation and the high computational costs associated with the mining stage. Moreover, this approach becomes increasingly time-consuming as the amount of data to be mined grows. On the other hand, deep learning approaches that act directly on graphs can overfit easily and fail to achieve good accuracy in graph classification tasks.
To overcome these limitations, in this paper, we propose two supervised embedding-based deep neural network (EBDNN) and embedding-based convolutional neural network (EBCNN) classifiers. The basic idea of our methods is to represent each graph or subgraph with an embedding vector, which automatically captures relevant features in the data, as opposed to relying on manually crafted features found in graph kernels.
To this end, we begin by comparing various graph embedding methods. Each embedding technique captures different aspects of graph structures, allowing for the extraction of diverse and complementary features. The first method involves extracting a set of rooted subgraphs to build a vocabulary of subgraphs, while the second method focuses on generating a subgraph vocabulary that preserves the neighborhood and structural properties of each subgraph. Then, to learn graph embedding, after constructing the vocabulary, a neural network is used to generate an embedding representation of each graph or subgraph. Subsequently, both DNN and CNN classifiers are trained for graph classification tasks, aiming to improve the accuracy of graph classification. These classifiers utilize the learned graph embeddings as input, enabling them to leverage the rich information encoded within the embeddings.
The main contributions of our work are summarized as follows:
Inspired by the Doc2Vec [
26] models in NLP, we designed a neural network for automatically learning graph and subgraph embeddings. These embeddings representation learning techniques aim to learn a mapping that embeds graphs or subgraphs as points in a low-dimensional vector space. The embedding representation of a graph or subgraph reflects its content and perhaps some latent semantic relationships within.
We evaluated the performance when utilizing different graph and subgraph embeddings, with the goal of achieving more efficient results in graph classification based on embedding.
Additionally, we developed two efficient supervised classifiers, EBDNN and EBCNN, to enhance classification accuracy.
By conducting experiments on benchmark graph classification, we provided empirical evidence showcasing the strong competitiveness of EBDNN and EBCNN classifiers in comparison to traditional classification methods and numerous deep learning methods for the graph classification task, particularly on large datasets.
The rest of this paper is organized as follows. The related work is presented in
Section 2, the details of the proposed approach are introduced in
Section 3, and the experimental results and implementation details are discussed in
Section 4. Finally,
Section 5 presents the conclusions and discusses future work.
2. Related Work
Graph classification aims to create a model that can correctly predict the target class for unseen graphs [
27]. To solve the graph classification problem, many different strategies have been presented, which generally fall into two categories: graph embedding methods and Graph Neural Network (GNN) methods. Approaches to learning the low-dimensional representations or embeddings that encode the properties of the graphs have become increasingly popular in recent years [
28]. Many graph-based tasks, including classification, clustering, and link prediction, can be performed using the embedding vector, as processing a whole graph is more difficult than working with vectors. In this section, we review some of the previous work used for graph classification.
DeepWalk [
29] was one of the earliest approaches that has been widely applied to graph learning techniques. It used a graph as input, generated a feature representation as an output, and then fed this representation into a traditional classifier such as Logistic Regression (LR). It performed short random walks on the graphs to generate sequences of nodes from the network and then passed the generated sequence through the skip-gram model to generate embedding vectors.
Graph2vec [
30], a graph embedding technique that is an unsupervised technique that generates embeddings using the skip-gram model, was proposed. Then, data were classified using the kernel algorithm as a Support Vector Classifier (SVC). Graph2vec views rooted subgraphs as the elements that compose a particular graph. Sub2vec [
25], a subgraph embedding technique that aims to learn subgraph embeddings while maintaining structural and neighborhood features, was introduced. Then, it classified with traditional classifiers such as SVC. However, these methods only used traditional classifiers after embedding.
A Deep Divergence Graph Kernels method [
31], which first learns the structure of a graph by training an encoder that is implemented as a fully connected DNN capable of reconstructing its structure given partial or distorted information, was proposed. Then, it trained a cross-graph attention mechanism that establishes a divergence score for each pair of graphs and measures how well one graph’s representations can encode another. It used these pairwise divergence scores to create an embedding space for all graphs and then used SVC to classify them.
A new graph classification method using graph structure learning that automatically learns hidden representations of substructures to help in resolving the subgraph isomorphism testing issue was designed [
32]. It built a vocabulary of rooted subgraphs for each graph and extracted topological attribute vectors to be added to the output layer to help the model learn the general information of the graph. Then, a graph embedding neural network was trained to obtain the embedding vector, which was then used in SVC for graph classification.
A graph classification method based on feature construction was applied [
24]. The topological feature vector of the graph, including node-level and global attribute features for the complete graph, was extracted. It constructed a vocabulary of subgraphs using a four-layer subgraph mining process and a subgraph isomorphism test to filter out subgraphs, which were then used as input in training graph-embedded neural networks. Finally, it classified graphs using two layers of DNN. The embedding not only reflected the features of each graph itself but also included the relative relationships among the graphs.
A two-stage classification algorithm was illustrated [
33]. First, it trained a classifier and then aggregated, rather than the other way around. It used a multi-layer perceptron classifier (MLP) to produce a predicted label vector using node features and then performed multihop aggregation of the predicted labels to improve accuracy and handle larger graphs. When employing node embedding for a graph classification task, all node embeddings could be averaged to produce the graph vector, but information about the general structure of the graph was missing.
GNNs have become a potential new learning framework that could bring the power of deep learning to graphs. They are an example of a neural network model that can act on graph structure data to express graph information. They efficiently mine the information included in the graph data using the node and structure information of a graph [
34]. The fundamental concept behind GNNs is to compute a state for each node in a graph, which is then repeatedly updated based on the states of the neighboring nodes. GraphSAGE, an inductive framework that trains a collection of aggregator functions rather than individual embeddings for every node to figure out the topology of each node’s neighborhood and the distribution of the node attributes within it, was introduced [
35].
DIFFPOOL, a graph pooling module that stacks GNNs and pooling layers to generate a hierarchical representation of graphs, was proposed [
36]. At each layer of a deep GNN, it mapped nodes to a set of clusters based on their learned embeddings, forming the coarsened input for the next GNN layer.
A study to analyze the expressive power of GNNs was proposed [
37]. The authors considered the relationship between GNNs and the Weisfeiler-Lehman graph isomorphism test in identifying graph structures. They developed a straightforward neural architecture called the Graph Isomorphism Network (GIN) to overcome the drawbacks associated with various GNN architectures in terms of GNN expressive capacity for capturing graph structures. The GIN employed neighbor aggregation and graph readout functions that operate on a multiset of node features. It demonstrated that readout operations with an MLP and summation can improve graph classification performance. When stacking more GNN layers, the majority of GNNs experienced drawbacks including noise from neighbor nodes and over-smoothing problems.
The Graph-of-Graph Neural Network Framework [
38] was introduced to alleviate the graph imbalance issue; it was composed of two modules, one from a global viewpoint and one from a local viewpoint. First, it constructed a graph of graphs (GoG) based on kernel similarity to establish a K-nearest neighbor graph and performed GoG propagation to aggregate neighboring graph representations. Then, topological augmentation was locally used by masking node features or removing edges using self-consistency regularization to generate stochastic augmentations of each graph to make the model more general and enhance graph classification.
CNN is one of the best-known DNNs and has been successful in a variety of domains; it began with images and then expanded to work on graphs. CNNs have proven to be extremely effective at extracting complex information. A CNN architecture was developed that was directly applied to graphs to learn the feature representations that can be used for classification [
39]. The authors extracted locally connected regions from graphs by choosing a group of nodes to represent a sizable portion of the graph before creating normalized neighborhood representations for each node.
A study using CNN’s architecture [
40] for graph classification was presented. It received graph data as input without converting it into tensors and designed a SortPooling layer to sequentially read graph vertices in a meaningful and consistent order instead of summing them up.
Two spatial CNN architectures [
41] were applied, the first of which was a simple CNN structure with two convolutional layers: a dense layer and a SoftMax layer. The other one used the first CNN as the first layer followed by a channel concatenation layer to determine the classification result of the entire graph according to the category of the neighborhood graphs. The weighting method was used according to the central node sequence and a combination of the local and global features of the vertices to construct the neighborhood graphs. Then, each graph corresponded to a fixed-size tensor that acted as an input to the CNN for graph classification.
An end-to-end deep graph CNN model for graph classification (up to 32 layers) was designed to overcome the shallow structure of most graph convolution network models and solve the over-smoothing problem when stacking too many graph convolution layers [
42]. It was divided into four parts. The first stage used 16 layers of graph convolutional layers to extract comprehensive structural information from the input graph, along with non-local structural information from the input node. The second-stage graph convolutional layers, which had another 16 layers, concatenated the node features extracted by the first stage and the initial low-dimensional features as input to obtain deeper structural information and node features and then defined a consistent vertex ordering. The third stage was the SortPooling layer, which organized the node features discovered by the second stage and unified the number of nodes as the input to the stage after it. The final stage was one-dimensional convolution layers and dense layers that read the sorted continuous node features for graph classification.
A subgraph CNN method [
43] was proposed for predicting the mutability of molecular graphs. This graph was divided by a graph partitioning algorithm according to its structure and features. The partitioned and original graphs were converted into embedding vectors. Finally, the embedding vectors were combined into one vector that goes to a classifier to predict the mutagenicity of the graph.
The method proposed in this paper is based on Graph2vec [
30] and Sub2vec [
25] for (sub)graph embedding. These methods utilize the (sub)graph neighbors as well as the graph structure for generating the graph embedding vectors. Then, the embedding vector is used as input for deep learning classifiers. The main aim of this method is to evaluate the performance of utilizing graph and subgraph embeddings, with the goal of achieving more efficient results in graph classification based on embedding. Many existing methods heavily rely on manual feature extraction, which can be time-consuming and labor-intensive and may fail to capture the complete representation of graph structures. Additionally, some methods encounter challenges in effectively scaling with the increasing size and complexity of graph data. As a result, when the volume of graph data is large, substituting the traditional classifier with a deep learning model can increase classification accuracy, as deep learning models can learn patterns on their own and can be applied to different fields.
3. Proposed Graph Classification Algorithm
Embedding-based deep neural network (EBDNN) and embedding-based convolutional neural network (EBCNN) classifiers are introduced to learn a model from labeled graphs and use that model to classify unseen graphs. This is inspired by Doc2Vec [
26], which uses neural networks to learn document embedding. In the same manner, the Doc2Vec model for documents can also be modified for graph data to train graph embedding. The text is very similar to the graph data. The graph is viewed as a document, and the subgraphs that surround each node in the graph are viewed as words. In addition, when training graph embedding, it is more suitable to use the structure of the PV-DBOW model in Doc2vec. In the graph data, the subgraphs are viewed as words in the document; therefore, there is no sequence relationship between the subgraphs, as it only considers which words a document contains but not the order of words in the document.
In the following subsections, we first explain the method of graph embedding using rooted subgraphs. Then, we explain the method of subgraph embedding with the goal of preserving neighborhoods and the structural properties of subgraphs. Finally, the graph classification framework is given. To boost the performance of graph classification, we replace traditional classifiers with deep learning classifiers.
3.1. Problem Formulation
Graph classification is the problem of determining graph classes. Consider a collection of undirected graphs, G = G1, G2, G3, …, GN, where N represents the number of graphs and each graph, G = (V, E), is composed of a set of vertices (V) and a set of edges (E) as well as their target labels. Our goal is to train a deep learning model that accepts the graph embedding vector of the training graph and its corresponding class as an input to predict the correct class label for unseen graphs; a previously unseen graph is one that was not used for training and validating the model.
Accordingly, efficient supervised DNN and CNN embedding-based classifiers on large-scale graphs are proposed. The fundamental concept is to represent each graph or subgraph with an embedding vector, which is then fed into an embedding layer as inputs in the graph classification framework using DNN and CNN.
Definition 1 (Graph Classification). Given a collection of graphs with their target labels, G = {G1, G2, G3, …, GN}, where each Gi = (Vi, Ei) has Vi vertices and Ei edges; the goal is to learn a function μ: G → L, where G is the input graphs, and L is the set of graph labels.
Definition 2 (Graph Embedding). Given a set of undirected graphs {G1, G2…, GN} and a positive integer d that determines the dimensionality of embedding, the goal is to learn a representation vector for every graph. The matrix of the vectors of all graphs is denoted as φ ∈ R|G|×d. Learning an embedding func-tion {f(G1), f(G2) …, f(GN)} that satisfies the following condition: as two graphs, Gi and Gj, are semantically more similar, f(Gi) and f(Gj) become closer in the d-dimensional vector space.
Definition 3 (Subgraph Embedding). Given a set of undirected graphs and a positive integer d that determines the dimensionality of embedding, we extract a set of subgraphs S = {g1, g2, …, gn} from the same graph where gi (vi, ei) is a subgraph of the graph if vi ⊆ V and ei ⊆ E. Our goal is to learn a representation vector for every subgraph, with the probability of maintaining the neighborhood and structural features of each subgraph.
3.2. Graph Embedding
Graph embedding is a significant approach used for graph classification; it transforms graph-structured data into a fixed-length feature vector. There are several embedding approaches available, but we use Graph2vec and Sub2vec for our experiments [
25,
30].
Since a graph is complex and non-linear, we need to extract features from it that capture information. Graph embeddings can be primarily divided into two categories. One emphasizes node embedding while the other concentrates on whole graph embedding. To accomplish classifications, we focus on graph and subgraph embeddings. Thus, we construct an embedding matrix and provide it as input to the classifier. The embedding matrix is a matrix with a row size equal to the number of unique graphs and a column size equal to the embedding vector dimension. Graph embedding [
30] is an unsupervised training technique that does not rely on the training dataset’s class label information but instead uses the graph’s information and features.
Assuming we have a set of undirected, unweighted graphs, G = (V, E), we first use the breadth-first search strategy to extract a set of rooted subgraphs (i.e., neighborhoods) around each node for each graph, and we treat these rooted subgraphs as the vocabulary of the graph. We consider each node in the graph as the root node, and then we find its neighborhood to a certain degree of the intended subgraph d (up to degree 4). After that, we aggregate all the rooted subgraphs of the four layers and eliminate the repeated subgraphs to construct a vocabulary. Since the method of extracting rooted subgraphs requires node labels, if the graph lacks any, we label the nodes using the Weisfeiler-Lehman relabeling approach based on their degrees, by assigning a unique label from the alphabet to every node.
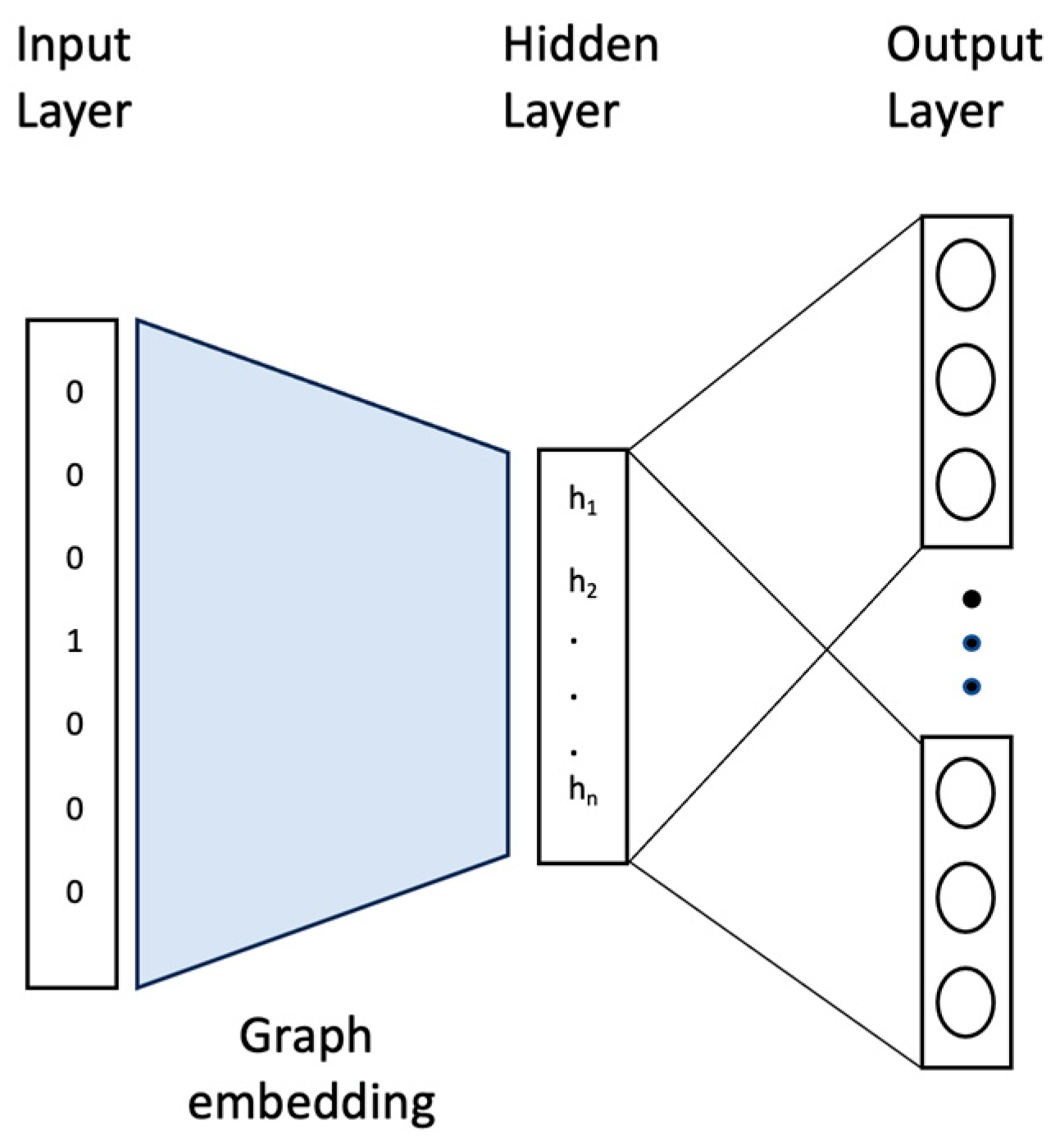
Based on the document embedding model Doc2vec, each graph is represented by a set of rooted subgraphs, just as each document is represented by a set of words. The vocabulary of the graph is obtained by summing these rooted subgraphs and giving each one of these rooted subgraphs a unique label. We train the Doc2vec model using the skip-gram algorithm to learn graph embedding after extracting the rooted subgraphs and constructing the vocabulary. It is a neural network with a single hidden layer. The architecture of the graph embedding neural network is shown in
Figure 1. The input layer serves as the first layer in which the graphs (or rooted subgraphs) are inputs. It takes a one-hot vector of the graphs, where the length of the vector is equal to the total number of training and test graphs in the dataset. Then, the number of neurons in the hidden layer corresponds to the dimensionality of the fixed-length feature vectors obtained after training the graph embedding. The parameters between the input layer and the hidden layer of the trained neural network are used as the embedding of the graphs we need to train. Finally, the output layer corresponds to the vocabulary of the subgraphs. Since there are a lot of rooted subgraphs, we train the skip-gram model using the negative sampling technique to speed up the training process.
The graph embedding procedure using a rooted subgraph is shown in Algorithm 1. Algorithm 1 shows the process of learning graph embedding for all the graphs in the dataset. R
sub(i) represents the extracted rooted subgraphs around each node, v; V
sub(i) represents the vocabulary of rooted subgraphs after aggregation; R
wl(i) represents the nodes after relabeling in the case of unlabeled nodes; and φ represents the embedding of each rooted subgraph. Then, these embedding vectors are used to classify the graphs.
| Algorithm 1 Graph Embedding with Rooted Subgraph Extraction and Skip-Gram Model (Graph2vec) |
Input: G = {G1, G2, G3, …., Gn}: Set of graphs such that each graph gi = (V, E) be a graph, where V
represents the set of vertices and E represents the set of edges in the graph.
n: number of graphs
k: desired embedding size
Output: A matrix of vectors representing the embedded graph.
1 Begin
2 For each gi in G:
3 Rsub(i) ← Extract rooted subgraph by performing a breadth-first search around each node v in
V of gi.
Rsub(i) = {Rv(i) | v ∈ V}, where Rv represents the rooted subgraph extracted for node v.
4 Vsub(i) ← Construct the vocabulary of rooted subgraphs considering a maximum degree of d.
Vsub(i) = {v’ | v’ is a rooted subgraph with maximum degree d}
5 IF gi does not have node labels:
6 Rwl(i) ← Apply the Weisfeiler-Lehman relabeling method to assign labels to the nodes.
Rwl(i)={λ(v) | v ∈ V}, where λ(v) represents the relabeled label for node v in graph gi.
7 φ ← Generate embedding vectors, using the skip-gram model.
φ = {φV | v ∈ V}, where φV represents the embedding vector for each rooted sub-
graph.
8 End for
9 Return the matrix of vectors, φ, as an n × k matrix.
10 END |
3.3. Subgraph Embedding
Subgraph embedding methods are simple but highly effective for converting graphs into the optimal format for a machine learning task. They are an unsupervised training approach to learning feature representations of arbitrary subgraphs.
Assuming we have a collection of undirected, unweighted graphs, we generate a set of subgraphs, S = {g
1, g
2…, g
n}, from the same graph using two different frameworks to preserve different properties of each subgraph. One framework preserves the neighborhood features of each subgraph, and the other preserves the structural aspects of each subgraph [
25].
In the neighborhood framework, the neighborhood information for each node inside a subgraph is captured. Thus, we establish a set of ID-paths annotated by the IDs of the nodes that represent important connectivity data in each subgraph. As there are unlimited paths in each subgraph, we use a random walk of fixed length L for each subgraph to effectively produce samples of the pathways. Each series of nodes in the ID paths illustrates how the subgraph’s neighborhood is changing.
In the structural framework, we create Degree-paths that capture the subgraph’s overall structure, as opposed to neighborhood properties, which only focus on local connectivity data. First, we assign a degree to each node based on the number of edges adjacent to the node in each subgraph. Then, the ratio of the degree of a node to the total number of nodes in the subgraph is captured for the Degree-paths. Afterward, based on the ratio of degrees, we assign a label to each node. After generating the labels for each node, we do a random walk on each subgraph and generate a sequence of characters for the degree paths.
After generating the samples of the ID paths/Degree-paths in each subgraph by running random walks, the vocabulary of the subgraphs is obtained, with each subgraph having its own ID within the larger graph. Then, we feed this vocabulary into Doc2vec, an unsupervised model that produces vector representations for subgraphs. It is a neural network with a single hidden layer to obtain the embedding for each subgraph. This neural network is similar to the PV-DBOW model in Doc2vec. Our main focus lies in the embedding matrix between the input layer and the hidden layer in Doc2vec because they are the subgraph embeddings/vectors that we need to use. Finally, the output layer corresponds to the vocabulary of the subgraphs. The intention is to ensure that the resulting embeddings effectively preserve the neighborhood information or the inherent structural relationships among nodes within the subgraphs. After the training converges, subgraphs with similar structures are mapped to a similar position in the vector space.
The subgraph embedding procedure to preserve different properties of the subgraphs is shown in Algorithm 2. Algorithm 2 shows the process of learning the subgraph embedding for all the graphs in the dataset. E represents a walk set of all subgraphs in each graph, and P represents samples of the ID paths/Degree-paths in each subgraph after running random walks; then the walk list E is updated with P, and φ represents the embedding of each subgraph. Then, these embedding vectors are used to classify the graphs.
| Algorithm 2 Sub-graph Embedding (Sub2vec) |
Input: Graph G, subgraph set S = {g1, g2, …, gn},
d: Dimension,
L: Walk length,
w: Window size
output: A matrix of vectors representing the embedded subgraph.
1 Begin
2 E ← Initialize an empty dictionary for the walk set, where the key represents the subgraph id and
the value represents ID-paths/Degree-paths of each subgraph.
3 for gi in S do
4 P ← Random Walk (gi, L)
E[gi] ← P
5 end for
6 φ ← Doc2vec (E, d, w)
7 Return the matrix of vectors, φ.
9 END |
3.4. Graph Classification Framework
The input to the classification algorithm consists of a set of embedding vectors and labels (xi, yi) for i ∈ T, where T ⊆ V is the set of training (sub)graphs. By stacking all the (sub)graphs’ embedding vectors and labels, we form an embedding matrix X and a label matrix Y.
The results obtained from the embedding neural network are combined with the final classifier to form a complete graph classification framework based on graph embedding. We develop DNN and CNN models for this study. A DNN is created with inspiration from the neurons in the human brain. It is an extension of an artificial neural network (ANN) with multiple hidden layers, enabling it to handle complex tasks. In a DNN, each layer is exclusively connected to the previous one and is only connected to the next layer within the sequential architecture. A CNN is one of the most common types of DNN architecture. It utilizes convolutional layers to automatically learn the spatial hierarchies of features from input data. It demonstrates exceptional proficiency in capturing local patterns by means of convolutional operations and achieving downsampling through pooling layers. A CNN’s architecture consists of a variety of building components, including convolutional layers, pooling layers, and fully connected layers.
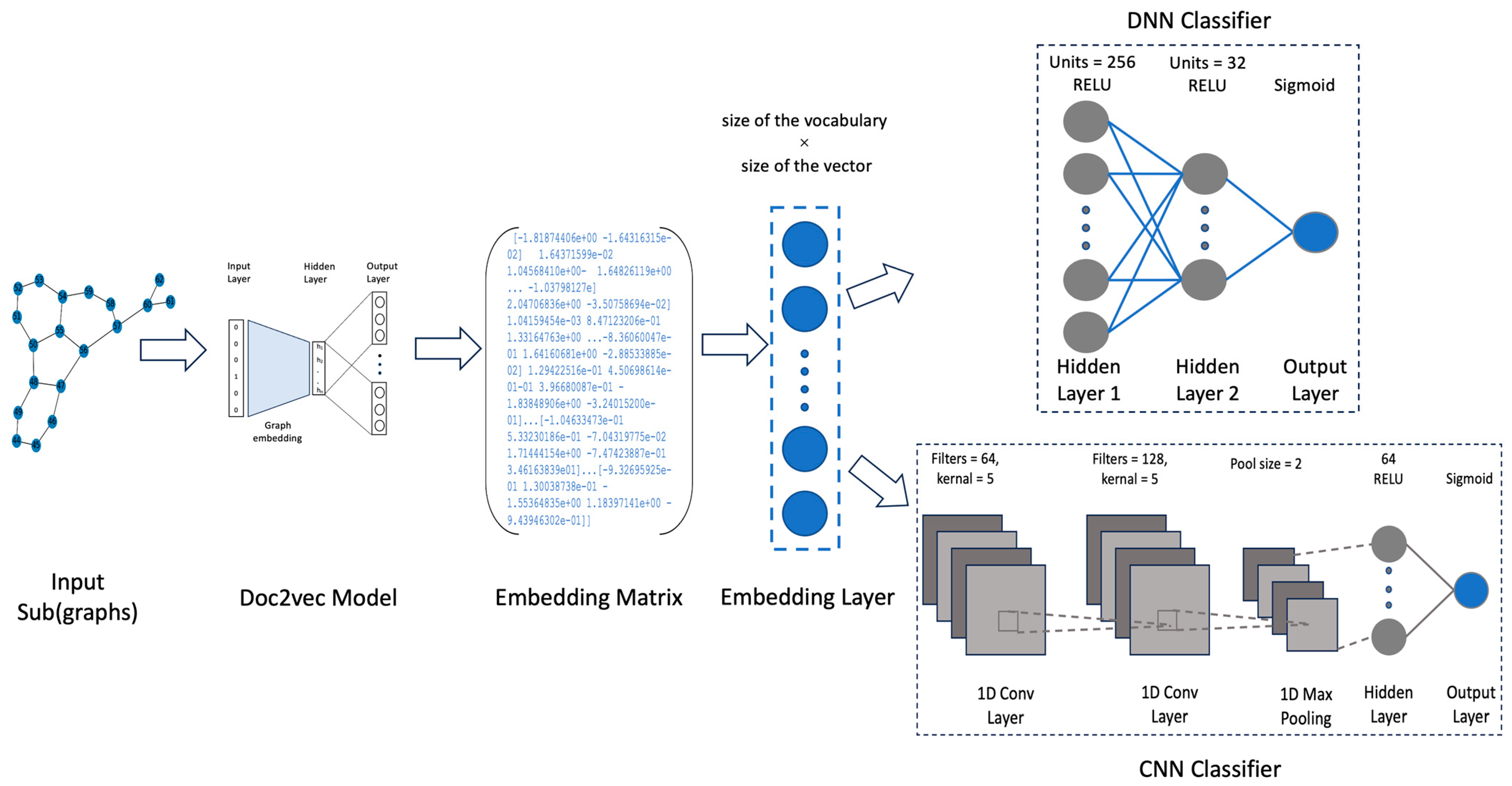
In the graph classification process based on graph embedding, first, we extract a vocabulary of subgraphs, which are then used as input to the graph embedding neural network. The parameters between the input layer and the hidden layer of the trained neural network are taken out as the embedding representation of the training graph and the test graph. Each subgraph corresponds to a fixed-length embedded vector. Finally, the embedding of the training graph and its corresponding class are input into DNN and CNN classifiers for training. An overview of the architecture of the proposed graph classification framework is shown in
Figure 2.
Next, when making the proposed DNN and CNN models, it is important to choose what the model’s architecture is. A DNN model has an embedding layer used as the first layer of the model, which takes the embedding matrix as input. There are three necessary arguments used to construct the embedding layer, which is the first layer of the DNN. First, the size of the vocabulary, which refers to the total number of graphs in the dataset. Second, the size of the vector space corresponds to the dimensionality of the output vector in which graphs or subgraphs are embedded. Finally, the input length describes the length of the input sequences. The embedded vectors are employed as data in the embedding layer for the DNN and CNN models. Then, there are two hidden layers and the output layer.
In a CNN model, the embedding matrix is initially utilized by the embedding layer, similar to a DNN model. Subsequently, two one-dimensional convolution layers are employed to effectively learn features from the shorter segments of the entire dataset. Max-pooling layers are incorporated to prevent overfitting of the learned features by selecting the maximum value among several features within a predefined sliding window. Following that, a final dense layer, also known as a fully connected layer, integrates the learned features from all the combinations of the previous layers. Finally, there is an output layer consisting of two neurons, along with their associated probabilities. As the amount of graph data is large, replacing the traditional classifier with a deep learning model is also helpful to improve the classification accuracy.
4. Experiments
We conduct our experiments on benchmark datasets to evaluate the performance of EBDNN and EBCNN. First, we examine the impact of applying subgraph and graph embedding. Then, the graph classification algorithm presented in this paper is compared with graph classification methods based on traditional classifiers and methods based on deep learning in terms of classification accuracy.
4.1. Datasets
We evaluate our proposed method using five bioinformatic benchmark graph datasets with node labels: MUTAG, PTC, PROTEINS, NCI1, and NCI109. The chemical compounds of each dataset are transformed into graphs, where nodes stand in for atoms and edges for chemical bonds.
MUTAG [
44] is a dataset of 188 mutagenic aromatics. The aromatic compounds in the graph set can be divided into two classes based on whether they have a mutagenic effect on bacteria or not.
PTC [
45] is a set of 344 chemical compounds that are presented as graphs with 19 distinct node labels that detail each compound’s ability to cause cancer in rats.
PROTEINS [
46] is a collection of 1113 protein structures represented as graphs, where the nodes are secondary structural elements (SSEs), and the edges are neighborhoods in either a 3D space or an amino acid sequence. The graph’s nodes have three different types of labels. Protein structures can be divided into two classes: enzymes and non-enzymes.
NCI1 [
47] is a large-scale balanced dataset of chemical compounds and has more than 4000 graphs in it. Each of the NCI anti-cancer screens poses a classification problem. The chemical compounds in this collection are evaluated as either active or inactive for lung cancer cells.
NCI109 [
47] is a large balanced dataset composed of substances that inhibit the activity of ovarian cancer cell lines that were published by the National Cancer Institute (NCI). The information from the above five datasets is shown in
Table 1.
4.2. Experimental Settings
This study proposes graph classification methods based on graph embedding that utilizes the training of a neural network for graph embedding. We evaluate the model based on the experimental results. Finally, we compare our graph classification methods with traditional classifiers and other deep learning methods.
All experiments are developed in a hardware environment of NVIDIA V100 Tensor Core GPUs with 16 GB RAM. We use the PyTorch library to implement the skip-gram model for Graph2vec and the Gensim library to implement the Doc2vec model for Sub2vec. The DNN and CNN classifiers are built using the Keras library. It is a high-level neural network library that functions on top of TensorFlow, a Google open-source library primarily created for deep learning applications.
We apply 5-fold cross-validation to every dataset, and we present the average accuracies achieved to validate the performance of our methods in graph classification. First, the training set is randomly divided into five folds of equal size. As a result, four out of five folds of the training set are utilized to train, while the remaining fold of the training set is used to validate the training model’s performance. Hyper-parameter tuning is the process of fine-tuning the best setting values to obtain the lowest generalization error and modify the model’s practical capacity in accordance with computational resources. We configure the optimal hyper-parameters via random search. We tune the number of hidden units in each layer ∈ {256, 128, 64, 32}, learning rate ∈ {0.01, 0.001, 0.001}, batch size ∈ {8, 64}, optimization methods {SGD, Adam}, dropout ratio after the dense layer ∈ {0, 0.5}, and activation function in the hidden layers and the output layer for all models and report the best parameters to achieve the best accuracy.
Our methods use the neural network in both the training graph embedding and the classifier, where the parameter updating process is time-consuming. However, with the acceleration of the GPU, the overall time has not increased much. Since the first approach uses an embedding vector of size 1024, the other method uses an embedding vector of size 512, and the training time increases as the size of the embedding vector does, it is difficult to compare the training times for graph and subgraph embeddings.
Additionally, by comparing three datasets of various sizes, including MUTAG (188 graphs), PROTEINS (1113 graphs), and NCI1 (4110 graphs), the experimental results demonstrate that the classification time needed by the suggested approaches grows linearly O(n) with the linear growth in data size. This is due to the methods used in this research, which generate a different sub(graph) “vocabulary” for each graph. As a result, the suggested methods have excellent stability, and the expansion of the dataset does not result in a major rise in classification time.
A DNN classifier with two hidden layers is used; an embedding layer is used as the first layer, in which the embedding matrix is passed as input. This embedding layer is configured with specific parameters. Initially, it requires the input dimensions, representing the total number of graphs in the dataset. For instance, in the case of the MUTAG dataset, which contains 188 graphs, the embedding layer has an input dimension of 188. Then, the output dimension corresponds to the size of the embedding vector. In the subgraph embedding method (Sub2vec), the output dimension is set to 512, while in the graph embedding method (Graph2vec), it is set to 1024. Finally, the input length, which represents the length of the input sequences, is set to the same value as the output dimension.
After the embedding layer, a flatten layer is employed to transform the two-dimensional vector into a one-dimensional vector, which is then used as an input to the next layer. Then, two dense layers are followed by an output layer. The number of dense layers’ units is 256 and 32. A dropout layer with dropout rates (0.2 and 0.5) is applied after the dense layer to prevent overfitting. Rectified Linear Units (ReLU) are used as an activation function in the hidden layers, as they facilitate faster learning and better performance compared to other activation functions. In the output layer, the Sigmoid activation function is used. Sigmoid is preferred in binary classification tasks, as it updates more quickly when using a single output unit.
The learning rate is chosen from (0.001, 0.0001) for different datasets, and the number of epochs is set to 100 in all datasets. Stochastic gradient descent (SGD) is used for optimization. Binary Cross-Entropy is used as a loss function.
In the CNN classifier, we have the embedding layer as the first layer, in which the embedded vectors are employed as data. The embedding layer of CNN shares the same parameters as the DNN classifier. The embedding layer is followed by two one-dimensional convolution layers; each convolutional layer has 64 and 128 filters of size 5, respectively. After the convolutional layers, a maximum pooling layer with pool size of 2 is applied. Next, there is a dense layer with 64 hidden units, followed by the output layer.
The parameters of the model are optimized by minimizing the difference between the predicted outputs and the ground truth labels using an SGD algorithm. This optimization process aims to improve the model’s performance and accuracy. As a loss function, Binary Cross-Entropy is employed.
4.3. Experimental Results and Discussion
Through graph embedding training, every graph of the five datasets is converted into a 512-dimensional embedding vector in the subgraph embedding method (Sub2vec), and we set the length of the random walk to 100,000 and the default window size to two. In the graph embedding method (Graph2vec), a 1024-dimensional embedding vector is used. In addition, the learning rate is set to 0.0001 in the skip-gram model, and the number of epochs in all datasets is set to 20.
We transform each graph in the graph dataset into a fixed-length feature vector. To observe the embedding result, we use the t-distributed Stochastic Neighbor Embedding (t-SNE) approach to carry out nonlinear dimensionality reduction on graph embedding to make it easier to observe the changed vector effect.
Figure 3 shows the 2D visualization results after the graph embeddings (Graph2vec) of the five datasets, and different colors indicate different classes of graphs in the dataset.
As demonstrated in
Figure 3, the visualization outcomes of the different classes of graph embedding are clearly distinguishable in the two datasets of MUTAG and PROTEINS and are well-separated in each embedding space. However, the PTC_MR dataset, consisting of two distinct classes, exhibits a complex and intertwined distribution of data, lacking a clear clustering structure. On NCI1 and NCI109, even though there is not a clear line between the two classes of graphs, they are not all mixed up together. Instead, one class can be observed to be divided into multiple subclusters, and there is a noticeable distinction between the subclusters belonging to different classes.
In summary, the training of graph embedding primarily relies on the substructure and feature information within the graphs. Consequently, graph data with similar substructures tend to be clustered together, while graph data with significant differences in substructures are distanced from one another.
While this approach demonstrates the potential of training and visualizing graph-level embeddings, it is not practical in its current state. However, it highlights that a well-trained unsupervised embedding can effectively separate graphs based on a specific training task, which can then be visualized for further analysis.
We evaluate the model based on the experimental results and mainly compare our graph classification methods in two aspects. On the one hand, we compare our proposed method with traditional classifiers. On the other hand, we compare it with other deep learning approaches. The abbreviations of the proposed and compared methods are reported in
Table 2. The abbreviations of the proposed methods are in bold.
4.3.1. Comparisons with Traditional Classifiers for Graph Classification
We compare our proposed methods with the following traditional classifiers methods:
Graph2vec [
30]: extracted rooted subgraphs and the neural network are used to train the embedding of graphs and use SVC to classify graphs.
Sub2vec (Sub2vec-N, Sub2vec-S) [
25]: specialize in subgraph embeddings by generating a set of subgraphs from the same graph based on structural and neighborhood features and then classifying with traditional classifiers such as SVC.
Spectral Features (SF-RFC) [
48]: assume the graph is connected; if it is not, find the largest connected graph, obtain its normalized Laplacian, and use Random Forest (RF) as a classifier.
Learning Graph Representations for Deep Divergence Graph Kernels (DDGK) [
31]: create an encoder using DNN to learn a graph’s structure and the cross-graph attention mechanism to calculate a divergence score for each pair of graphs that are used to produce an embedding space and use SVC as a classifier.
Graph Classification via Graph Structure Learning (GC-GSL) [
32]: build a vocabulary of rooted subgraphs for each graph and extract topological attribute vectors to be added to the output layer; then, a graph embedding neural network is trained to obtain the embedding vector and use SVC to classify graphs.
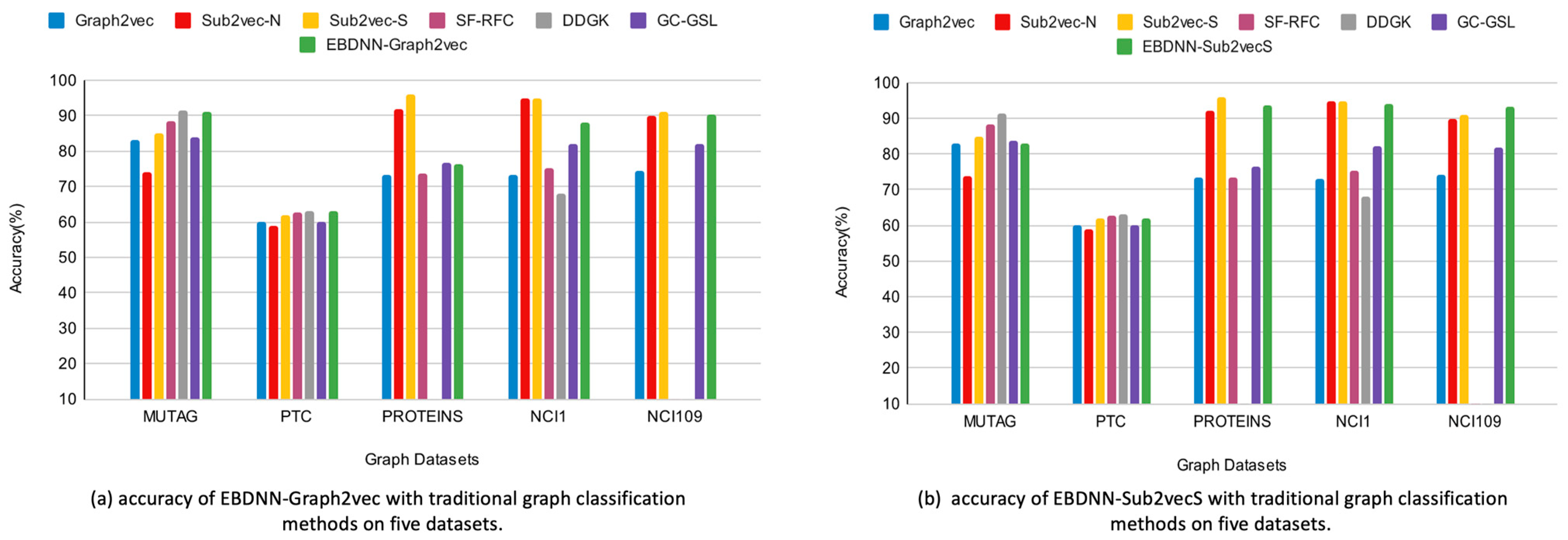
The average classification accuracy of the proposed methods and the traditional graph classification methods mentioned above on five datasets are shown in
Table 3. For the other methods, we take the reported accuracy directly from their papers (“--” means not available). The best results are shown in bold.
It can be seen from
Table 3 that the classification results of the EBDNN-Graph2vec model proposed in this paper demonstrate similar accuracy levels as the top-performing method on the MUTAG and PTC datasets, which have smaller graph sizes. Additionally, when comparing EBDNN-Graph2vec with the other methods, its classification performance is always in the top 2 across all datasets. On the other hand, EBCNN-Graph2vec achieves comparable classification results on all datasets except for the PROTEINS dataset.
Replacing traditional classifiers with deep learning classifiers using the Graph2vec embedding technique enhances the accuracy of all datasets. However, on larger datasets such as PROTEINS, NCI1, and NCI109, traditional classifiers with Sub2vec-N outperform EBDNN-sub2vecN and EBCNN-sub2vecN.
Although the EBDNN-sub2vecS and EBCNN-sub2vecS models do not achieve the highest accuracy on the PROTEINS and NCI1 datasets, they still achieve accuracy rates above 90%, which is 9–20% higher than other graph classification methods.
On the other hand, all the proposed methods achieve the highest accuracy on the NCI109 dataset when compared to traditional classifiers. Additionally, the EBDNN model also shows a slight improvement in accuracy over the EBCNN model, except for the NCI109 dataset.
The PROTEINS dataset is characterized by its large size and more complex internal structure within each graph. Consequently, using the EBDNN-Graph2vec and EBCNN-Graph2vec methods, which employ the rooted subgraph mining technique along with the Weisfeiler-Lehman relabeling method and only focus on learning local graph information and structures while neglecting the global graph structure, leads to the loss of significant information. This, in turn, affects the subsequent classification results.
Analyzing the NCI1 and NCI109 datasets reveals that, despite having the largest number of subgraphs, they also possess a higher number of node labels compared to the average number of subgraph nodes in other datasets. As a result, numerous subgraphs in NCI1 and NCI109 do not fully contain all types of nodes. Although the proposed methods improve the accuracy of graph classification on these datasets, they do not achieve the highest accuracy, except for the EBDNN-Sub2vecS and EBCNN-Sub2vecS models when applied to the NCI109 dataset.
4.3.2. Comparisons with Deep Learning Approaches for Graph Classification
We compare our proposed methods with the following deep learning approaches for graph classification:
PATCHY-SAN (PSCN) [
39]: chooses a node sequence from a graph; neighborhoods are assembled and normalized for each node in the chosen sequence, and then the convolutions are applied to the normalized neighborhoods graph.
Deep Graph Convolution Neural Network (DGCNN) [
40]: classifies graphs in three stages: first, a convolutional layer that directly reads graphs to obtain multi-scale vertex features, then a SortPooling layer that sorts the vertex features, and, finally, convolutional and dense layers that classify the graphs.
Capsule Graph Neural Network (CapsGNN) [
49]: proposes a new vector propagation by generating graph capsules and class capsules on the basis of node capsules that are extracted from GNN; then, dynamic routing is applied to update weights over graph capsules to generate final class capsules for graph classification.
Graph Isomorphism Network (GIN) [
37]: proposes a readout function that uses the embeddings of individual nodes to produce the embedding of the entire graph and then uses MLP and summation for graph classification.
Graph Embedding (GE) [
24]: builds a vocabulary of node-level and global attribute features for the whole graph; then, a neural network is used to train the embedding of graphs, and a DNN is used as the classifier.
Imbalanced Graph Classification via Graph-of-Graph Neural Network (G2 GNN) [
38]: constructs a graph of graphs based on kernel similarity, performs propagation to aggregate neighboring graph representations, and uses topological augmentation locally by removing edges to generate stochastic augmentations of each graph used for graph classification.
A Deep Graph Convolutional Neural Network architecture for Graph Classification (DGCNNII) [
42]: designs a very deep graph CNN up to 32 layers to extract the graph’s structure information and the nodes’ non-local structure information, concatenates the node features and the initial low-dimensional features, and then uses the SortPooling layer, 1D convolution layers, and dense layers to read the sorted continuous node features for graph classification.
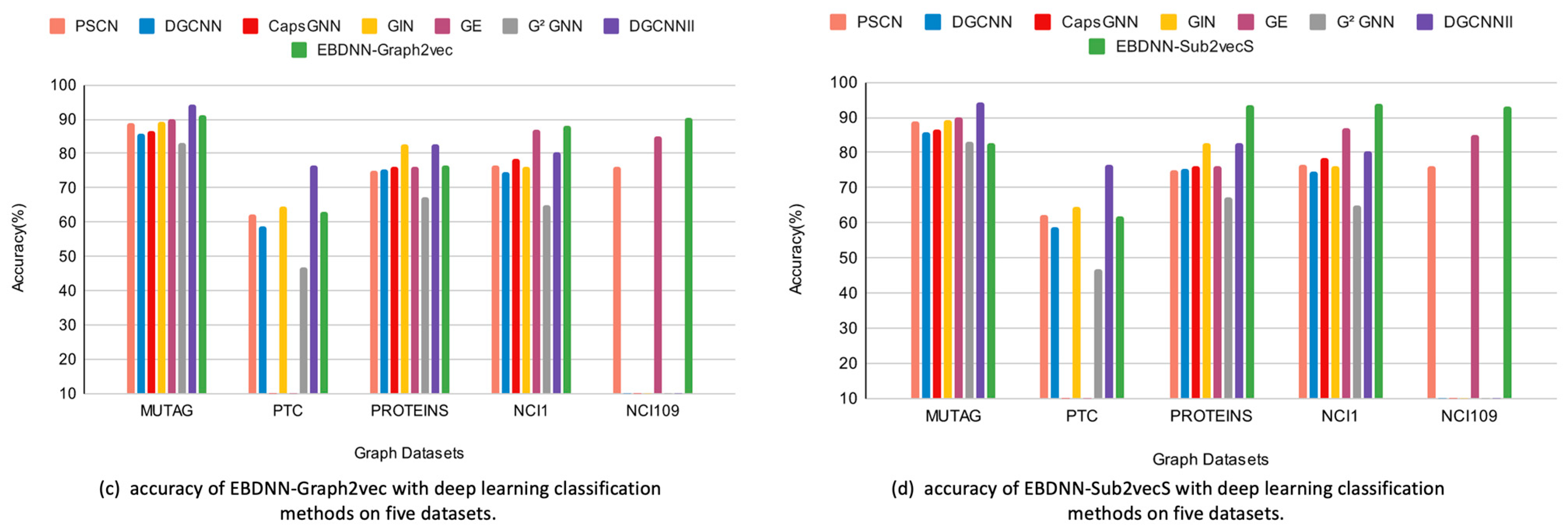
The average classification accuracy of the proposed methods and the graph classification based on deep learning are shown in
Table 4. The best results on each dataset are shown in bold.
From the results in
Table 4, we can see that the proposed EBDNN-Sub2vecS and EBCNN-Sub2vecS achieve the highest classification accuracy on the PROTEINS, NCI1, and NCI109 datasets when compared to deep learning methods.
However, the accuracy of EBDNN-Sub2vecS and EBDNN-Sub2vecN is relatively low on small-size datasets such as MUTAG. This can be attributed to the limited size of the dataset, which consists of only 188 graphs. The small dataset size results in an imbalanced class distribution and relatively simple structural complexity, which impacts the accuracy of the models in comparison with deep learning methods.
Comparing EBDNN-Graph2vec and EBCNN-Graph2vec with other methods, their classification performance is always in the top 2 across all datasets except for PTC. However, the accuracy rate of EBDNN-Graph2vec on the PROTEINS dataset is limited to 76.5%. This can be attributed to the complex internal structure within each graph and the lack of node labels, which impact the effectiveness of the embedding approach that employs node labels, consequently affecting the subsequent classification results.
EBDNN-Sub2vecN and EBCNN-Sub2vecN show the lowest accuracy across all datasets, except for the PROTEINS dataset where they achieve the highest accuracy compared to other methods. This can be attributed to the PROTEINS dataset having a high number of classes, making the neighborhood property more significant in determining the graph class.
On the PROTEINS dataset, EBDNN-Sub2vecS and EBCNN-Sub2vecS outperform other baseline approaches. EBDNN-Sub2vecS achieves an accuracy of 93.6%, with an average improvement of 10.8% to 26.21% in accuracy rates. Similarly, EBCNN-Sub2vecS achieves an accuracy of 92.2%, with an average improvement of 9.4% to 24.8% in accuracy rates. This indicates that utilizing structural subgraphs, particularly for datasets with complex internal structures such as PROTEINS, enhances classification accuracy.
On the NCI1 dataset, EBDNN-Sub2vecS achieves an accuracy of 94%, while EBCNN-Sub2vecS achieves 91.3%, surpassing the performance of all other baseline methods, none of which have achieved an accuracy rate of 90%. The highest accuracy is also obtained on the NCI109 dataset, where EBDNN-Sub2vecS achieves 93.1%, and EBCNN-Sub2vecS achieves 93.5%.
For large datasets such as NCI1 and NCI109, using EBDNN-Graph2vec and EBDNN-Sub2vecS achieves the highest accuracy when compared with other deep learning methods. This can be attributed to the large number of subgraphs present in the datasets, a significant number of node labels, and both datasets being balanced.
Comparing the proposed methods, EBDNN-Graph2vec and EBDNN-Sub2vecS, it becomes apparent that the latter performs better on large datasets, indicating that the classification results of structural subgraphs have a greater influence on classification accuracy compared to using a rooted subgraph representation for each graph.
To evaluate the impact of graph classification, we conduct an analysis of the classification results obtained by EBDNN-Graph2vec and EBDNN-Sub2vecS in comparison to previous methods across five datasets. The comparison results are presented in
Figure 4 and
Figure 5.
Figure 4a,b showcase the comparison of EBDNN-Graph2vec and EBDNN-Sub2vecS, respectively, with traditional classifiers. It can be observed that the classification accuracy of EBDNN-Graph2vec is about the same as that of the top-performing method on the MUTAG and PTC datasets, which have smaller graph sizes. However, the improvement in classification accuracy is not remarkable for the PROTEINS and NCI1 datasets. EBDNN-Sub2vecS achieves the lowest accuracy with the small datasets, MUTAG and PTC. However, it demonstrates high accuracy on PROTEINS and NCI1, although not the highest accuracy. Both EBDNN-Graph2vec and EBDNN-Sub2vecS achieve the highest accuracy on the NCI109 dataset.
Figure 5c,d, on the other hand, depict the comparison of EBDNN-Graph2vec and EBDNN-Sub2vecS, respectively, with deep learning classifiers.
Figure 5c,d demonstrate that EBDNN-Graph2vec and EBDNN-Sub2vecS outperform other deep-learning-based approaches for graph classification on three datasets. This can be attributed to the fact that in the PROTEINS, NCI1, and NCI109 datasets, the local structural features, such as the active compounds in cells, are more obvious and easier to extract. Additionally, the larger size of these datasets also contributes to the superior performance of EBDNN-Graph2vec and EBDNN-Sub2vecS in classification tasks.
Overall, the experimental results highlight the effectiveness of EBDNN-Graph2vec and EBDNN-Sub2vecS in graph classification, particularly on datasets with larger graph sizes and distinct local structural features.