1. Introduction
At the end of December 2019, some cases of novel coronavirus pneumonia of unknown origin were reported in Wuhan, Hubei province, which was officially named COVID-19 by the World Health Organization in February 2020 [
1]. COVID-19 has pandemic characteristics and spread rapidly worldwide, seriously threatening the lives and health of people [
2]. According to the Global New Coronavirus Pneumonia Epidemic Real-Time Big Data Report, as of October 2022, more than 200 countries and regions worldwide have been infected with the new coronavirus pneumonia, and the global number of confirmed cases cumulatively exceeds 600 million and the number of deaths exceeds 6 million.
COVID-19 is a novel infectious disease caused by severe acute respiratory syndrome coronavirus-2 infection, whose early clinical features are mainly fever, dry cough, and malaise, with a few accompanying symptoms such as runny nose and diarrhea. Severe cases can cause dyspnea and organ failure, which even can lead to death [
3,
4]. For more than two years, due to the instability of the COVID-19 gene sequence, several variants of COVID-19 have been generated. These variants are characterized by greater concealment, which makes it extremely difficult to diagnose them accurately.
Nucleic acid testing is the most common method to diagnose COVID-19. This method detects viral fragments by using a reverse transcription polymerase chain reaction (RT-PCR) [
4] technique. However, the nucleic acid test has disadvantages such as being time consuming, low sensitivity, high false negative rate, and the need for special test kits [
5,
6], which limits its development. In recent years, medical imaging techniques have been widely used for the diagnosis of various diseases. Chest X-ray (CXR) and computed tomography (CT) [
7] are used to diagnose COVID-19. Compared with the nucleic acid test, medical-imaging-based diagnostics are faster and more effective. CT generates a lot of radiation and is not suitable for pregnant women and infants, while CXR contains little radiation and can reduce the risk of cross-infection to some extent. Furthermore, CXR is less costly and more widely used than CT. However, the manual analysis and diagnostic process based on CXR images depends heavily on the expertise of healthcare professionals and the analysis of image characteristics is time consuming, which makes it difficult to observe occult lesions at an early stage and distinguish between other viral and bacterial pneumonias [
8]. Due to the urgent need, experts recommend the use of computer-aided diagnosis to replace manual diagnosis to improve the efficiency of detection and help doctors diagnose more accurately.
With the development of artificial intelligence, deep learning methods [
9,
10] have achieved good success in the field of computer vision. Several studies [
11,
12,
13] have shown that convolutional neural networks (CNNs) have excellent feature extraction capabilities and can accurately extract image features of different scale sizes. Medical image classification using CNN requires the fusion of feature maps from different scale sizes, while taking into account both local and global information. The representative models used for COVID-19 classification include VGG networks, ResNet networks, and high-resolution networks [
14]. These experimental results suggest that local feature extraction of medical images using CNN is feasible. CNN has a fixed sampling location and its limited perceptual field leads to poor global modeling capabilities, which cannot learn image features effectively according to the complex changes of lesion regions. COVID-19 CXR images showed consolidation in the lung and ground-glass clouding with commonly irregular shapes, such as hazy, patchy, diffuse, and reticular nodular patterns, which greatly increased the difficulty of COVID-19 detection [
15,
16]. Consequently, improving feature extraction from infected regions with complex shapes and establishing long-distance dependencies between features is the key to recognize COVID-19 accurately. Transformer is the most advanced sequence encoder, whose core idea is self-attention [
17], which can establish long-range dependencies between feature vectors and improve feature extraction and representation. Vision Transformer (ViT) [
18,
19] is the representative model. The experimental results indicate that the extraction of global information from medical images using pure Transformer is practicable, but it tends to lead to excessive memory and computational costs. As a result, some studies [
20,
21] have shown that combining convolution and Transformer as a hybrid network model can help improve classification performance while reducing computational cost.
In this paper, we design a new deep learning network model (BoT-ViTNet) based on ResNet50 for automatic image classification to help doctors more accurately identify COVID-19 and other viral pneumonias. The network model first combines the advantages of CNN for extracting local feature representations and multi-headed self-attention for global information modeling. Then, the TRT-ViT blocks are used in the final stage to fuse global and local feature information. This well solves the problem of learning feature representations from different complexly infected regions of CXR images, thus significantly improving the classification performance of the model. The main work includes:
A novel model called BoT-ViTNet is constructed for COVID-19 image classification and it can simultaneously extract both local feature information and global semantic information from infected regions with complex shapes of CXR images, achieving good classification performance.
Introducing the multi-headed self-attention (MSA) to the last Bottleneck block of the first three stages in the ResNet50 to enhance the ability to model global information.
In the final stage of ResNet50, the TRT-ViT block [
22] is used to replace the original bottleneck block, which can extract both global and local feature information to enhance the feature representation and correlation between feature locations, so as to help identify complex lesion areas in CXR images.
The remainder of this paper is described as follows. The related work of this paper, which includes convolutional neural network, vision Transformer, and hybrid network models, is briefly mentioned in
Section 2.
Section 3 presents the proposed BoT-ViTNet in detail and describes it in part. In
Section 4, extensive experiments are conducted to prove the effectiveness of BoT-ViTNet and the experimental results are discussed and analyzed. Finally, the conclusion of the whole paper is given in
Section 5.
3. Method
To identify COVID-19 CXR images accurately, we propose a novel model called BoT-ViTNet in this paper, whose architecture is shown in
Figure 1. The BoT-ViTNet model contains three parts. For the first part, we use the bottleneck block for local feature extraction of the lesion region of CXR images. For the second part, multi-headed self-attention block (MSA) is introduced to learn the contextual information of the extracted features, which can enhance the global modeling capability of the feature information. For the last part, the TRT-ViT block is used to extract both local and global feature information to further enhance the feature representation and correlation between feature locations.
The general structure of BoT-ViTNet is similar to ResNet50, which also goes through four stages and each stage consists of 3, 4, 6, and 3 blocks, respectively, as shown in
Figure 1. The CXR image is first passed through a 7 × 7 convolution layer with a step size of 2 and a 3 × 3 pooling layer with a step size of 2 to obtain a feature map with a resolution of 56 × 56 × 64. Then, the feature map is input into the Bottleneck block, which consists of two types of residual convolution structure, as shown in
Figure 1a,b. The Bottleneck block conducts a channel expansion of the feature map when the step size is 1 and performs a downsampling operation to increase the perceptual field on the feature map when the step size is 2. After passing 2 Bottleneck blocks sequentially, the feature map is input to the MSA block, whose structure is shown in
Figure 1c. The MSA block learns the global information of image features and establishes long-range dependencies of features to enhance the expression of features. To further fuse the global and local information, the feature maps are input to the TRT-ViT block after being processed by multiple Bottleneck blocks and MSA blocks. The structure of the TRT-ViT block is shown in
Figure 1d. The TRT-ViT block extracts global features and local information by Transformer and Bottleneck, respectively, and then fuses the global features and local information, which improves the expression of features and the correlation of positions between features. The global average pooling layer is used to integrate the global spatial information for the fused features. The output features are mapped to the softmax layer for probability prediction. BoT-ViTNet can not only capture deep global semantic information of CXR images but also extract shallow local texture information of CXR images. It inherits the advantages of Transformer and CNN, improving the recognition performance.
Table 1 shows the structural details of the BoT-ViTNet model.
3.1. Bottleneck Block
Unlike the extraction of features using standard convolution, the Bottleneck block can reduce the computational complexity of the model while extracting features. Therefore, we used the Bottleneck block for local feature extraction of complex lesion regions in CXR images. The Bottleneck block consists of two 1 × 1 convolutions and a 3 × 3 depth-wise convolution. The first 1 × 1 convolution is used to reduce the number of channels of the feature map so that feature extraction can be performed more efficiently and intuitively. The 3 × 3 depth-wise convolution is used to extract the local feature information of the image. The second 1 × 1 convolution is used to expand the number of channels of the feature map so that the number of channels of the output feature map is equal to the number of channels of the input feature map, and to perform summation. The use of the Bottleneck structure greatly reduces the number of parameters and computation, thus improving computational efficiency. In addition, a residual structure is added to each output to avoid causing network degradation and over-fitting problems. The residual block is computed as follows:
where
denotes the input feature map,
indicates the output feature map, and
represents the convolution operation. The residual network can span the previous layers of the network and act on the later layers, which can improve the gradient disappearance problem when the network is trained for back propagation.
3.2. MSA Block
To enhance the long-term dependencies of the features, we introduce the MSA block in ResNet50, as shown in
Figure 1c. MSA [
37] is an essential component of Transformer, which can unite feature information from different locations representing different subspaces. It is an extension of Self-attention (SA), which runs k SA operations in parallel at the same time and projects their concatenated outputs. We first review the basic SA modules that are widely used in neural network architectures. SA is the core idea of Transformer, which has the feature of a weak inductive bias. By performing similarity calculation, it can establish long-distance dependency between feature vectors and improve feature extraction and expression ability. The input of each SA consists of query
Q, key
K, and value
V, which are linear transformations of the input sequence. The new vectors
Q,
K, and
V are obtained by multiplying the original
Q,
K, and
V with the weight matrices
WQ,
WK, and
WV, which are learned during the training process, respectively. In this section, we use Scaled Dot-Product Attention for the similarity calculation among vectors with the following equation:
where
denotes the input sequence,
represents the SA operation, and
means the dimension of the head.
MSA concatenates k single-head self-attentions and performs a linear projection operation on them with the following equation:
In Equation (3), is the output of MSA, means the MSA operation, denotes the connection of feature maps with the same dimension, and is the learnable linear transformation.
CNN has strong inductive bias and can effectively extract local texture information of feature maps in shallow networks, while MSA has weak inductive bias and can establish long-range dependencies of features in deep networks. Consequently, combining CNN with MSA can obtain powerful feature representation capability and high accuracy.
3.3. TRT-ViT Block
To further improve the feature representation, the TRT-ViT block [
22], consisting of Transformer and Bottleneck is introduced in the last stage of ResNet50, which takes a global-then-local hybrid block pattern for feature extraction. As described in the paper [
38], usually the Transformer with a larger receptive field can extract global information from the feature map and enable information exchange. In contrast, a convolution with a small receptive field can only extract local information from the feature map. TRT-ViT block fully combines the advantages of the Transformer and Bottleneck, which enhance the expression of features and the correlation of positions between features, thus helping to identify complex lesion regions in CXR images.
The network structure of the TRT-ViT block is shown in
Figure 1d, which first uses Transformer to model the global information and then uses Bottleneck to extract the local information. Transformer is calculated as follows:
where
is the input feature map and
is the output feature map. We firstly perform an operation of channel reduction using a 1 × 1 convolution with a step size of 1, reducing the number of channels of the feature map to half of the original feature map. Then, to capture the long-range dependencies of features in complex lesion regions of the image, we use MSA in Transformer to extract global information of the feature map and implement information exchange within each channel. Finally, the global features after information exchange are delivered to the multilayer perceptron (MLP) layer to improve the ability of the network to acquire image background information, which helps to identify complex lesions in CXR images. After Transformer operation, the feature map containing global information is input into Bottleneck blocks to learn the local space information. We connect the extracted global features with local features to enhance the expression of the features and the correlation between the positions of the features, improving the recognition accuracy.
Transformer aims to establish global connections between features, whereas convolution captures only local information. The computational effort of the Transformer and Bottleneck is almost equal when the resolution of the input image is low, indicating that the placement of Transformer at a later stage of the network helps to balance performance and efficiency [
39]. It was further demonstrated that using a hybrid block pattern of global-then-local can be helpful to identify complex lesion regions in CXR images. Consequently, in this section, the Bottleneck block is replaced by the TRT-ViT block in the last stage of the ResNet50 network and cross-stacked, which can effectively extract local texture information and global semantic information from infected regions with complex shapes and performs feature fusion, while achieving high performance and high accuracy.
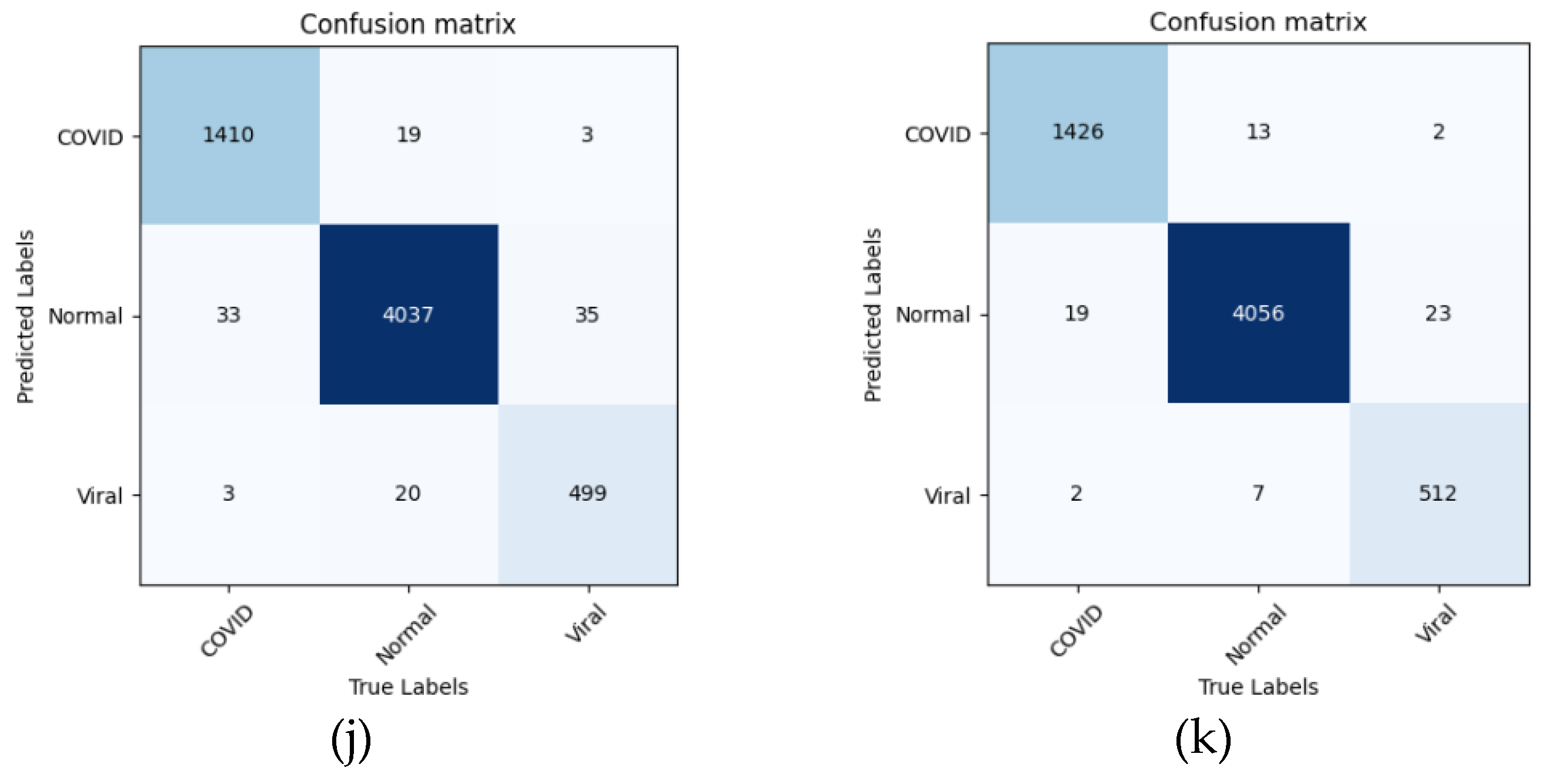
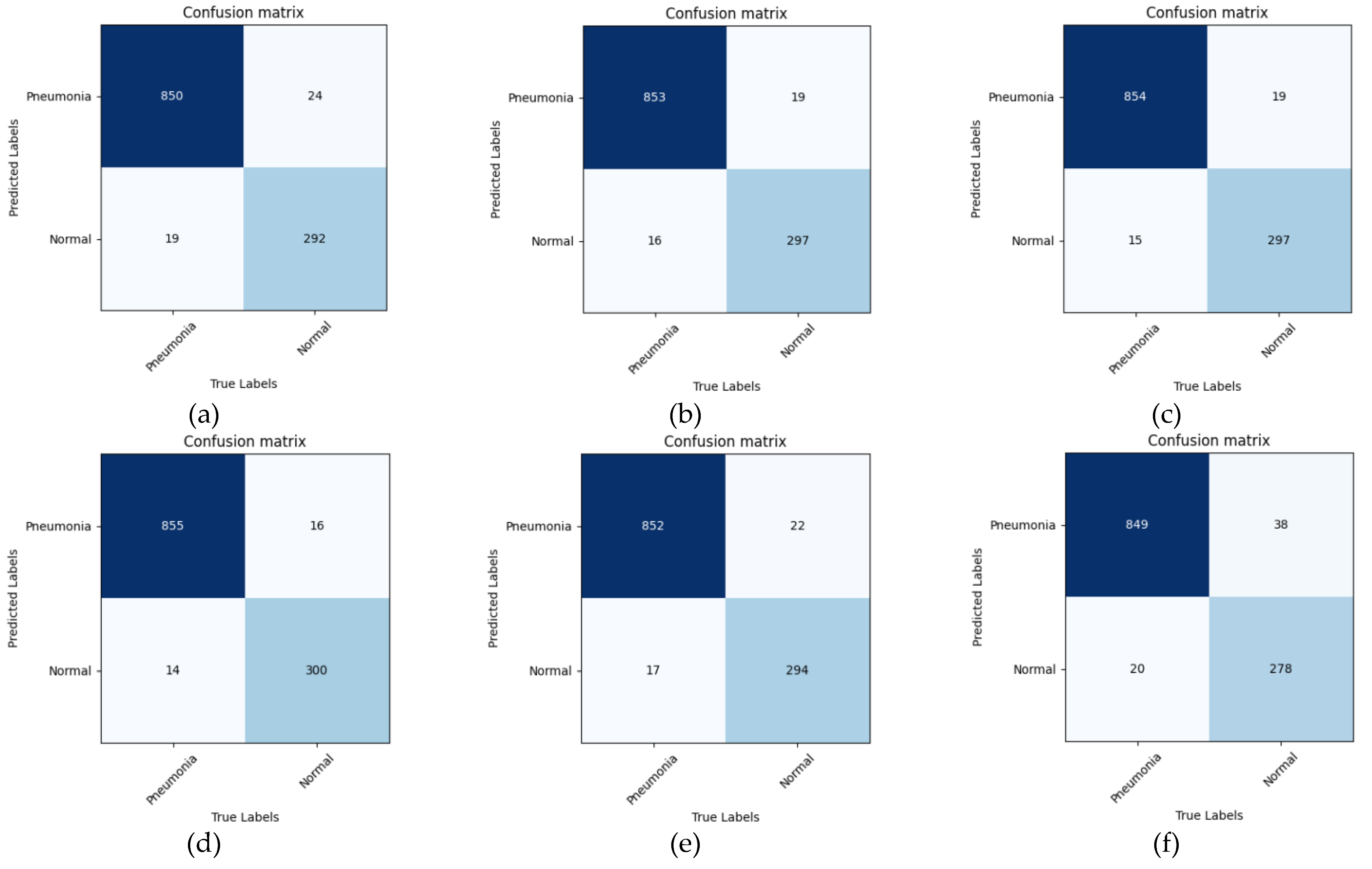
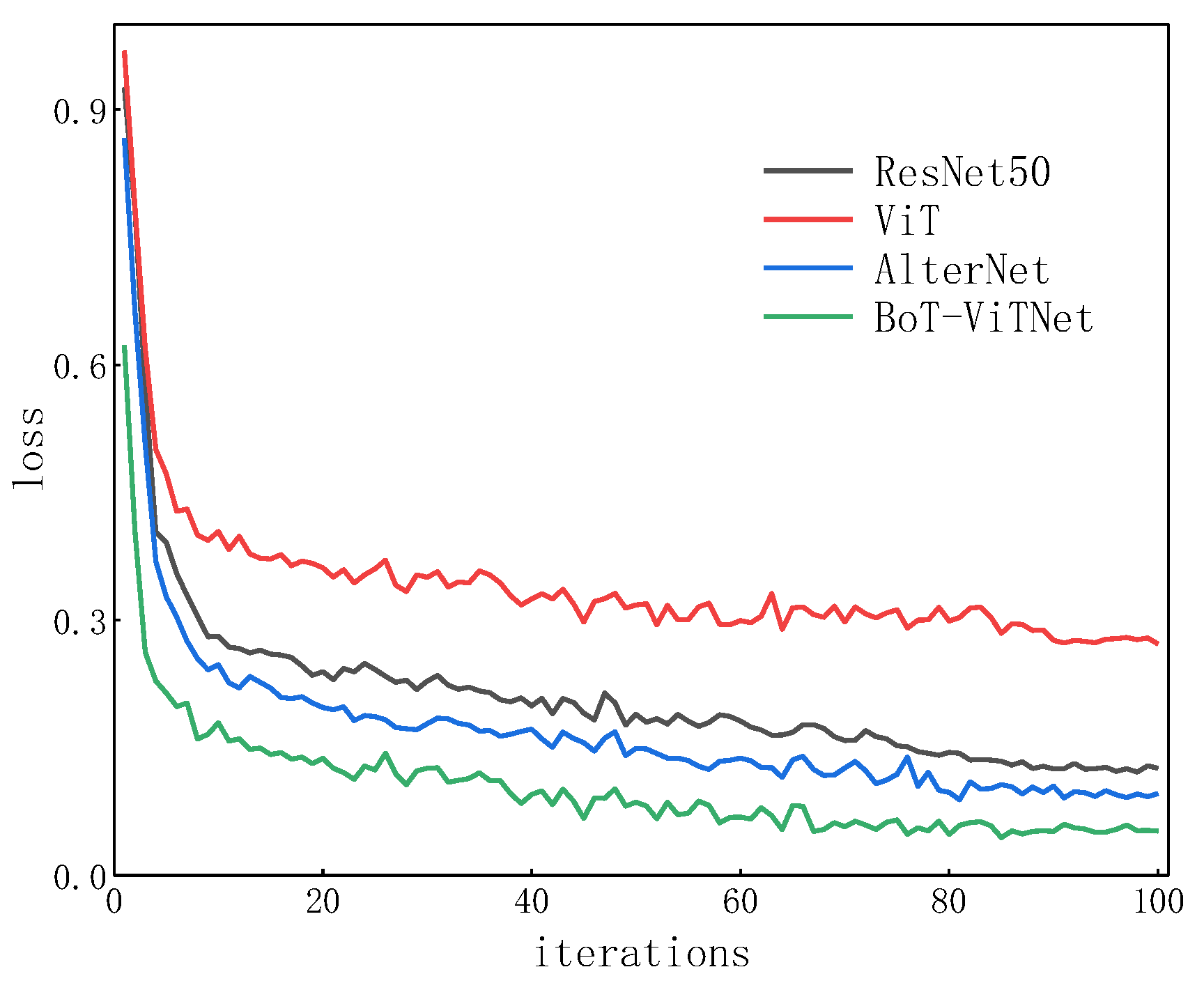
5. Conclusions
In this paper, we designed a BoT-ViTNet model for COVID-19 image classification based on the ResNet50. Firstly, the MSA block is introduced in the last Bottleneck block of the first three stages of ResNet50 to enhance the ability to model global information. Then, to further enhance the correlation between features and the representation of features, the TRT-ViT block, which consists of Transformer and Bottleneck, is used in the final stage of ResNet50 to fuse global and local information for improving the recognition of complex lesion regions in CXR images. Finally, the extracted features are delivered to the global average pooling layer for global spatial information integration in a concatenated way and used for classification. The experimental results of image classification on the publicly accessible COVID-19 Radiography database and Coronahack dataset show that BoT-ViTNet model achieves the better results. The overall accuracy, precision, sensitivity, specificity, and F1-score of the BoT-ViTNet model on the COVID-19 Radiography database are 98.91%, 97.80%, 98.76%, 99.13%, and 98.27%, respectively. The BoT-ViTNet model has better recognition effect on COVID-19 with 98.55%, 99.00%, 99.67%, and 98.77% in precision, sensitivity, specificity, and F1-score, respectively. Compared with other classification models, the BoT-ViTNet model has better performance in recognizing and classifying COVID-19 images. Although the BoT-ViTNet model can achieve good results for the classification of COVID-19 images, further clinical studies and tests are still required.