Few-Shot Learning with Collateral Location Coding and Single-Key Global Spatial Attention for Medical Image Classification
Abstract
:1. Introduction
- (1)
- A complete classification framework is presented for few-shot learning of medical images, which achieves excellent performance compared with the well-known few-shot learning algorithms.
- (2)
- A collateral location coding is proposed to help the network explicitly utilize the location information.
- (3)
- A single-key global spatial attention is designed to make the pixels at each location perceive the global spatial information in a low-cost way.
- (4)
- Experimental results on three medical image datasets demonstrate the compelling performance of our algorithms in the few-shot task.
2. Related Work
2.1. Medical Image Classification
2.2. Few-Shot Learning
3. Method
3.1. Overview
3.2. Collateral Location Coding
3.3. Single-Key Global Spatial Attention
3.4. Classification
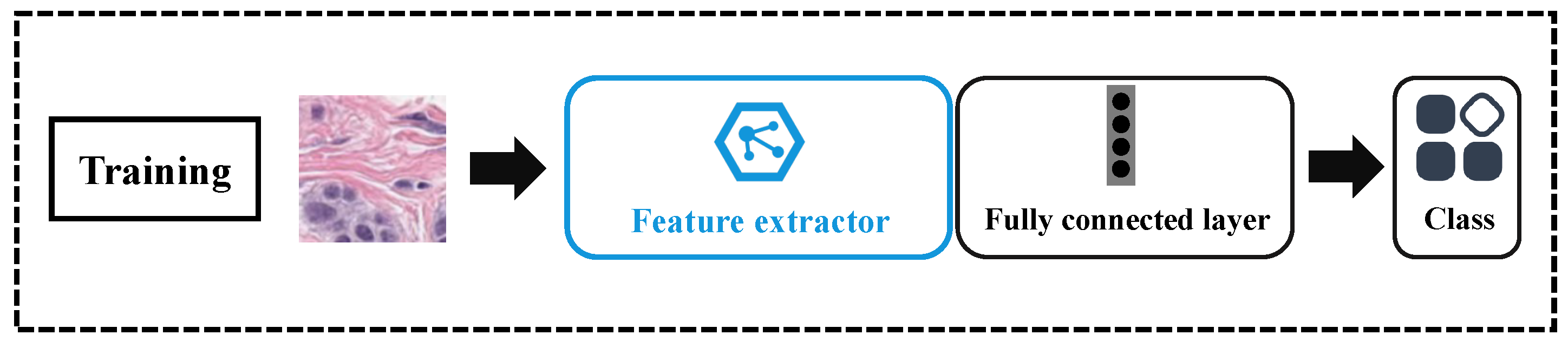
3.4.1. Training
3.4.2. Testing
4. Experimental Results and Analysis
4.1. Dataset Description
4.2. Experimental Setup
4.3. Comparing with State-of-the-Art Algorithms
4.4. Ablation Experiments
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Wu, Y.; Ma, W.; Gong, M.; Su, L.; Jiao, L. A novel point-matching algorithm based on fast sample consensus for image registration. IEEE Geosci. Remote Sens. Lett. 2014, 12, 43–47. [Google Scholar] [CrossRef]
- Wu, Y.; Li, J.; Yuan, Y.; Qin, A.; Miao, Q.G.; Gong, M.G. Commonality autoencoder: Learning common features for change detection from heterogeneous images. IEEE Trans. Neural Netw. Learn. Syst. 2021, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, H.; Liu, Y.; Gong, M. Multi-fidelity evolutionary multitasking optimization for hyperspectral endmember extraction. Appl. Soft Comput. 2021, 111, 107713. [Google Scholar] [CrossRef]
- Gong, M.; Liang, Y.; Shi, J.; Ma, W.; Ma, J. Fuzzy c-means clustering with local information and kernel metric for image segmentation. IEEE Trans. Image Process. 2012, 22, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Gong, M.; Zhou, Z.; Ma, J. Change detection in synthetic aperture radar images based on image fusion and fuzzy clustering. IEEE Trans. Image Process. 2011, 21, 2141–2151. [Google Scholar] [CrossRef]
- Gong, M.; Feng, K.y.; Fei, X.; Qin, A.K.; Li, H.; Wu, Y. An Automatically Layer-wise Searching Strategy for Channel Pruning Based on Task-driven Sparsity Optimization. IEEE Trans. Circ. Syst. Video Technol. 2022, 1. [Google Scholar] [CrossRef]
- Wu, Y.; Liu, J.W.; Zhu, C.Z.; Bai, Z.F.; Miao, Q.G.; Ma, W.P.; Gong, M.G. Computational intelligence in remote sensing image registration: A survey. Int. J. Autom. Comput. 2021, 18, 1–17. [Google Scholar] [CrossRef]
- Wu, Y.; Mu, G.; Qin, C.; Miao, Q.; Ma, W.; Zhang, X. Semi-supervised hyperspectral image classification via spatial-regulated self-training. Remote Sens. 2020, 12, 159. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Xiao, Z.; Liu, S.; Miao, Q.; Ma, W.; Gong, M.; Xie, F.; Zhang, Y. A Two-Step Method for Remote Sensing Images Registration Based on Local and Global Constraints. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2021, 14, 5194–5206. [Google Scholar] [CrossRef]
- Li, H.; Li, J.; Zhao, Y.; Gong, M.; Zhang, Y.; Liu, T. Cost-Sensitive Self-Paced Learning With Adaptive Regularization for Classification of Image Time Series. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2021, 14, 11713–11727. [Google Scholar] [CrossRef]
- Wang, Z.; Li, J.; Liu, Y.; Xie, F.; Li, P. An Adaptive Surrogate-Assisted Endmember Extraction Framework Based on Intelligent Optimization Algorithms for Hyperspectral Remote Sensing Images. Remote Sens. 2022, 14, 892. [Google Scholar] [CrossRef]
- García Seco de Herrera, A.; Markonis, D.; Joyseeree, R.; Schaer, R.; Foncubierta-Rodríguez, A.; Müller, H. Semi–supervised learning for image modality classification. In International Workshop on Multimodal Retrieval in the Medical Domain; Springer: Berlin, Germany, 2015. [Google Scholar]
- Peikari, M.; Salama, S.; Nofech-Mozes, S.; Martel, A.L. A cluster-then-label semi-supervised learning approach for pathology image classification. Sci. Rep. 2018, 8, 7193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lecouat, B.; Chang, K.; Foo, C.S.; Unnikrishnan, B.; Brown, J.M.; Zenati, H.; Beers, A.; Chandrasekhar, V.; Kalpathy-Cramer, J.; Krishnaswamy, P. Semi-supervised deep learning for abnormality classification in retinal images. arXiv 2018, arXiv:1812.07832. [Google Scholar]
- Springenberg, J.T. Unsupervised and semi-supervised learning with categorical generative adversarial networks. arXiv 2015, arXiv:1511.06390. [Google Scholar]
- Madani, A.; Moradi, M.; Karargyris, A.; Syeda-Mahmood, T. Semi-supervised learning with generative adversarial networks for chest X-ray classification with ability of data domain adaptation. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 1038–1042. [Google Scholar]
- Armato, S.G.; Li, F.; Giger, M.L.; MacMahon, H.; Sone, S.; Doi, K. Lung cancer: Performance of automated lung nodule detection applied to cancers missed in a CT screening program. Radiology 2002, 225, 685–692. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Yu, L.; Luo, L.; Dou, Q.; Heng, P.A. Semi-Supervised Medical Image Classification With Relation-Driven Self-Ensembling Model. IEEE Trans. Med. Imaging 2020, 39, 3429–3440. [Google Scholar] [CrossRef]
- Gyawali, P.K.; Ghimire, S.; Bajracharya, P.; Li, Z.; Wang, L. Semi-supervised medical image classification with global latent mixing. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2020; pp. 604–613. [Google Scholar]
- Phan, H.; Krawczyk-Becker, M.; Gerkmann, T.; Mertins, A. DNN and CNN with weighted and multi-task loss functions for audio event detection. arXiv 2017, arXiv:1708.03211. [Google Scholar]
- Khan, S.H.; Hayat, M.; Bennamoun, M.; Sohel, F.A.; Togneri, R. Cost-sensitive learning of deep feature representations from imbalanced data. IEEE Trans. Neural Netw. Learn. Syst. 2017, 29, 3573–3587. [Google Scholar]
- Han, W.; Huang, Z.; Li, S.; Jia, Y. Distribution-sensitive unbalanced data oversampling method for medical diagnosis. J. Med Syst. 2019, 43, 1–10. [Google Scholar] [CrossRef]
- Yu, H.; Sun, C.; Yang, X.; Yang, W.; Shen, J.; Qi, Y. ODOC-ELM: Optimal decision outputs compensation-based extreme learning machine for classifying imbalanced data. Knowl. Based Syst. 2016, 92, 55–70. [Google Scholar] [CrossRef]
- Li, C.H.; Yuen, P.C. Semi-supervised learning in medical image database. In Pacific-Asia Conference on Knowledge Discovery and Data Mining; Springer: Berlin, Germany, 2001; pp. 154–160. [Google Scholar]
- Cubuk, E.D.; Zoph, B.; Shlens, J.; Le, Q.V. Randaugment: Practical automated data augmentation with a reduced search space. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, Seattle, WA, USA, 14–19 June 2020; pp. 702–703. [Google Scholar]
- Su, H.; Shi, X.; Cai, J.; Yang, L. Local and global consistency regularized mean teacher for semi-supervised nuclei classification. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2019; pp. 559–567. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. In Proceedings of the 28th Conference on Neural Information Processing Systems (NIPS 2014), Montreal, QC, Canada, 8–13 December 2014; p. 27. [Google Scholar]
- Antoniou, A.; Storkey, A.; Edwards, H. Data augmentation generative adversarial networks. arXiv 2017, arXiv:1711.04340. [Google Scholar]
- Chen, Z.; Fu, Y.; Chen, K.; Jiang, Y.G. Image block augmentation for one-shot learning. Proc. AAAI Conf. Artif. Intell. 2019, 33, 3379–3386. [Google Scholar] [CrossRef] [Green Version]
- Vinyals, O.; Blundell, C.; Lillicrap, T.; Wierstra, D. Matching networks for one shot learning. In Proceedings of the 30th Conference on Neural Information Processing Systems (NIPS 2016), Barcelona, Spain, 5–10 December 2016; p. 29. [Google Scholar]
- Snell, J.; Swersky, K.; Zemel, R. Prototypical networks for few-shot learning. In Proceedings of the 31st Conference on Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017; p. 30. [Google Scholar]
- Li, W.; Xu, J.; Huo, J.; Wang, L.; Gao, Y.; Luo, J. Distribution consistency based covariance metric networks for few-shot learning. Proc. AAAI Conf. Artif. Intell. 2019, 33, 8642–8649. [Google Scholar] [CrossRef]
- Finn, C.; Abbeel, P.; Levine, S. Model-agnostic meta-learning for fast adaptation of deep networks. In Proceedings of the 34th International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017; pp. 1126–1135. [Google Scholar]
- Ravi, S.; Larochelle, H. Optimization as a model for few-shot learning. In Proceedings of the 34th International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017; pp. 1–11. [Google Scholar]
- Cheng, Y.; Yu, M.; Guo, X.; Zhou, B. Few-shot learning with meta metric learners. arXiv 2019, arXiv:1901.09890. [Google Scholar]
- Islam, M.A.; Jia, S.; Bruce, N.D. How much position information do convolutional neural networks encode? arXiv 2020, arXiv:2001.08248. [Google Scholar]
- Wang, X.; Kong, T.; Shen, C.; Jiang, Y.; Li, L. Solo: Segmenting objects by locations. In European Conference on Computer Vision; Springer: Berlin, Germany, 2020; pp. 649–665. [Google Scholar]
- Gonzalez, J.L.; Kim, M. PLADE-Net: Towards Pixel-Level Accuracy for Self-Supervised Single-View Depth Estimation with Neural Positional Encoding and Distilled Matting Loss. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 6851–6860. [Google Scholar]
- Hendrycks, D.; Gimpel, K. Gaussian error linear units (gelus). arXiv 2016, arXiv:1606.08415. [Google Scholar]
- Liu, H.; Liu, F.; Fan, X.; Huang, D. Polarized self-attention: Towards high-quality pixel-wise regression. arXiv 2021, arXiv:2107.00782. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Yang, J.; Shi, R.; Ni, B. MedMNIST Classification Decathlon: A Lightweight AutoML Benchmark for Medical Image Analysis. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021; pp. 191–195. [Google Scholar]
- Yang, J.; Shi, R.; Wei, D.; Liu, Z.; Zhao, L.; Ke, B.; Pfister, H.; Ni, B. MedMNIST v2: A Large-Scale Lightweight Benchmark for 2D and 3D Biomedical Image Classification. arXiv 2021, arXiv:2110.14795. [Google Scholar]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 180161. [Google Scholar] [CrossRef]
- Codella, N.; Rotemberg, V.; Tschandl, P.; Celebi, M.E.; Dusza, S.; Gutman, D.; Helba, B.; Kalloo, A.; Liopyris, K.; Marchetti, M.; et al. Skin lesion analysis toward melanoma detection 2018: A challenge hosted by the international skin imaging collaboration (isic). arXiv 2019, arXiv:1902.03368. [Google Scholar]
- Kather, J.N.; Krisam, J.; Charoentong, P.; Luedde, T.; Herpel, E.; Weis, C.A.; Gaiser, T.; Marx, A.; Valous, N.A.; Ferber, D.; et al. Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study. PLoS Med. 2019, 16, e1002730. [Google Scholar] [CrossRef] [PubMed]
- Bilic, P.; Christ, P.F.; Vorontsov, E.; Chlebus, G.; Chen, H.; Dou, Q.; Fu, C.W.; Han, X.; Heng, P.A.; Hesser, J.; et al. The liver tumor segmentation benchmark (lits). arXiv 2019, arXiv:1901.04056. [Google Scholar]
- Xu, X.; Zhou, F.; Liu, B.; Fu, D.; Bai, X. Efficient Multiple Organ Localization in CT Image Using 3D Region Proposal Network. IEEE Trans. Med. Imaging 2019, 38, 1885–1898. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Sung, F.; Yang, Y.; Zhang, L.; Xiang, T.; Torr, P.H.; Hospedales, T.M. Learning to compare: Relation network for few-shot learning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 1199–1208. [Google Scholar]
- Liu, Y.; Lee, J.; Park, M.; Kim, S.; Yang, E.; Hwang, S.J.; Yang, Y. Learning to propagate labels: Transductive propagation network for few-shot learning. In Proceedings of the International Conference on Learning Representations, New Orleans, LA, USA, 6–9 May 2019; pp. 1–14. [Google Scholar]


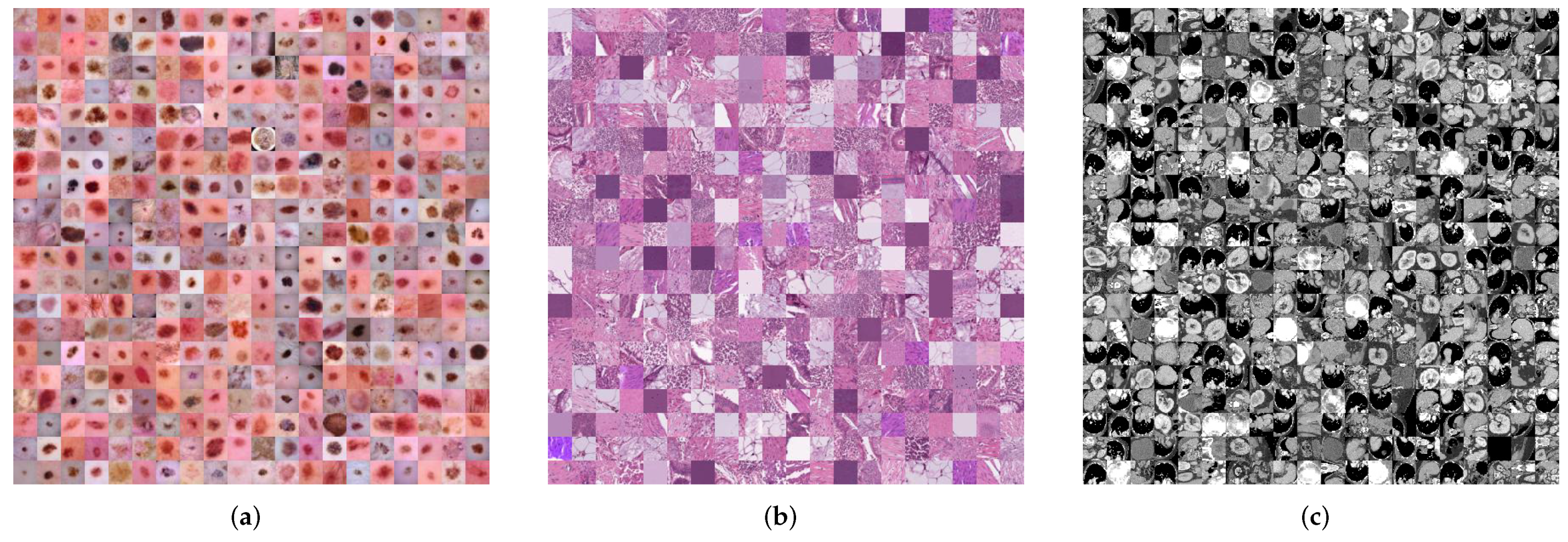

| Datasets | Classes | Training | Validation | Test | Image Modality |
|---|---|---|---|---|---|
| DermaMNIST [44,45] | 7 | 7007 | 1003 | 2005 | Dermatoscope |
| PathMNIST [46] | 9 | 89,996 | 10,004 | 7180 | Pathology |
| OrganMNIST (Axial) [47,48] | 11 | 34,581 | 6491 | 17,778 | Abdominal CT |
| Method | 2-Way | |
|---|---|---|
| 1-Shot | 5-Shot | |
| MatchingNet | ||
| MAML | ||
| PrototypeNet | ||
| Relation Net | ||
| TPN | ||
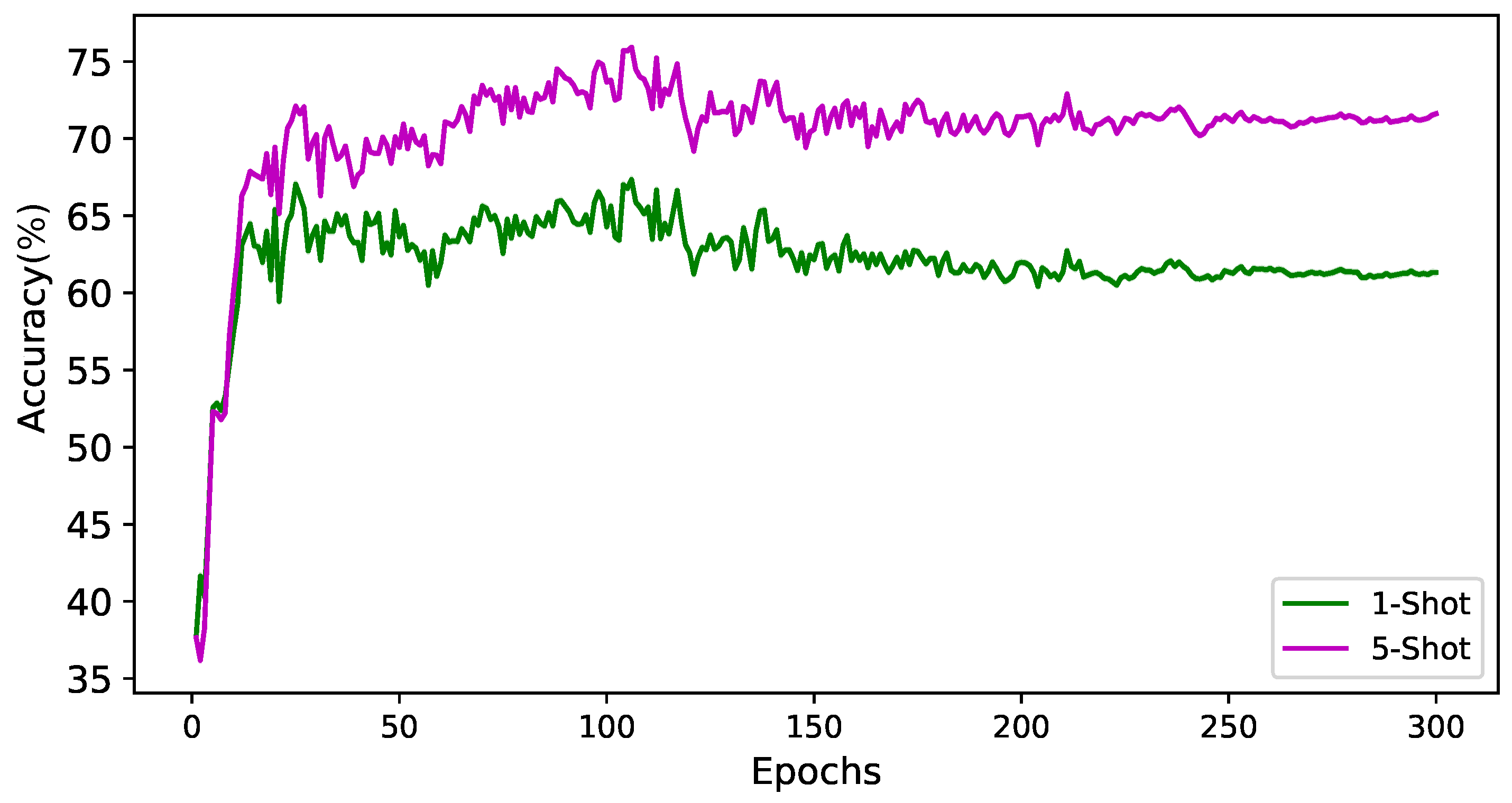
| Ours | 63.37 ± 0.80% | 69.38 ± 1.03% |
| Method | 3-Way | |
|---|---|---|
| 1-Shot | 5-Shot | |
| MatchingNet | ||
| MAML | ||
| PrototypeNet | ||
| elation Net | ||
| TPN | ||
| Ours | 54.82 ± 0.78% | 61.92 ± 0.81% |
| Method | 3-Way | |
|---|---|---|
| 1-Shot | 5-Shot | |
| MatchingNet | ||
| MAML | ||
| PrototypeNet | ||
| Relation Net | ||
| TPN | % | |
| Ours | 53.48 ± 0.81% | 59.38 ± 0.84% |
| Method | 2-Way | |
|---|---|---|
| 1-Shot | 5-Shot | |
| Baseline | ||
| + Collateral Location Coding | ||
| + Single-Key Global Spatial Attention | ||
| Full | 63.37 ± 0.80% | 69.38 ± 1.03% |
| Method | 3-Way | |
|---|---|---|
| 1-Shot | 5-Shot | |
| Baseline | ||
| + Collateral Location Coding | ||
| + Single-Key Global Spatial Attention | ||
| Full | 54.82 ± 0.78% | 61.92 ± 0.81% |
| Method | 3-Way | |
|---|---|---|
| 1-Shot | 5-Shot | |
| Baseline | ||
| + Collateral Location Coding | ||
| + Single-Key Global Spatial Attention | ||
| Full | 53.48 ± 0.81% | 59.38 ± 0.84% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shuai, W.; Li, J. Few-Shot Learning with Collateral Location Coding and Single-Key Global Spatial Attention for Medical Image Classification. Electronics 2022, 11, 1510. https://doi.org/10.3390/electronics11091510
Shuai W, Li J. Few-Shot Learning with Collateral Location Coding and Single-Key Global Spatial Attention for Medical Image Classification. Electronics. 2022; 11(9):1510. https://doi.org/10.3390/electronics11091510
Chicago/Turabian StyleShuai, Wenjing, and Jianzhao Li. 2022. "Few-Shot Learning with Collateral Location Coding and Single-Key Global Spatial Attention for Medical Image Classification" Electronics 11, no. 9: 1510. https://doi.org/10.3390/electronics11091510
APA StyleShuai, W., & Li, J. (2022). Few-Shot Learning with Collateral Location Coding and Single-Key Global Spatial Attention for Medical Image Classification. Electronics, 11(9), 1510. https://doi.org/10.3390/electronics11091510






