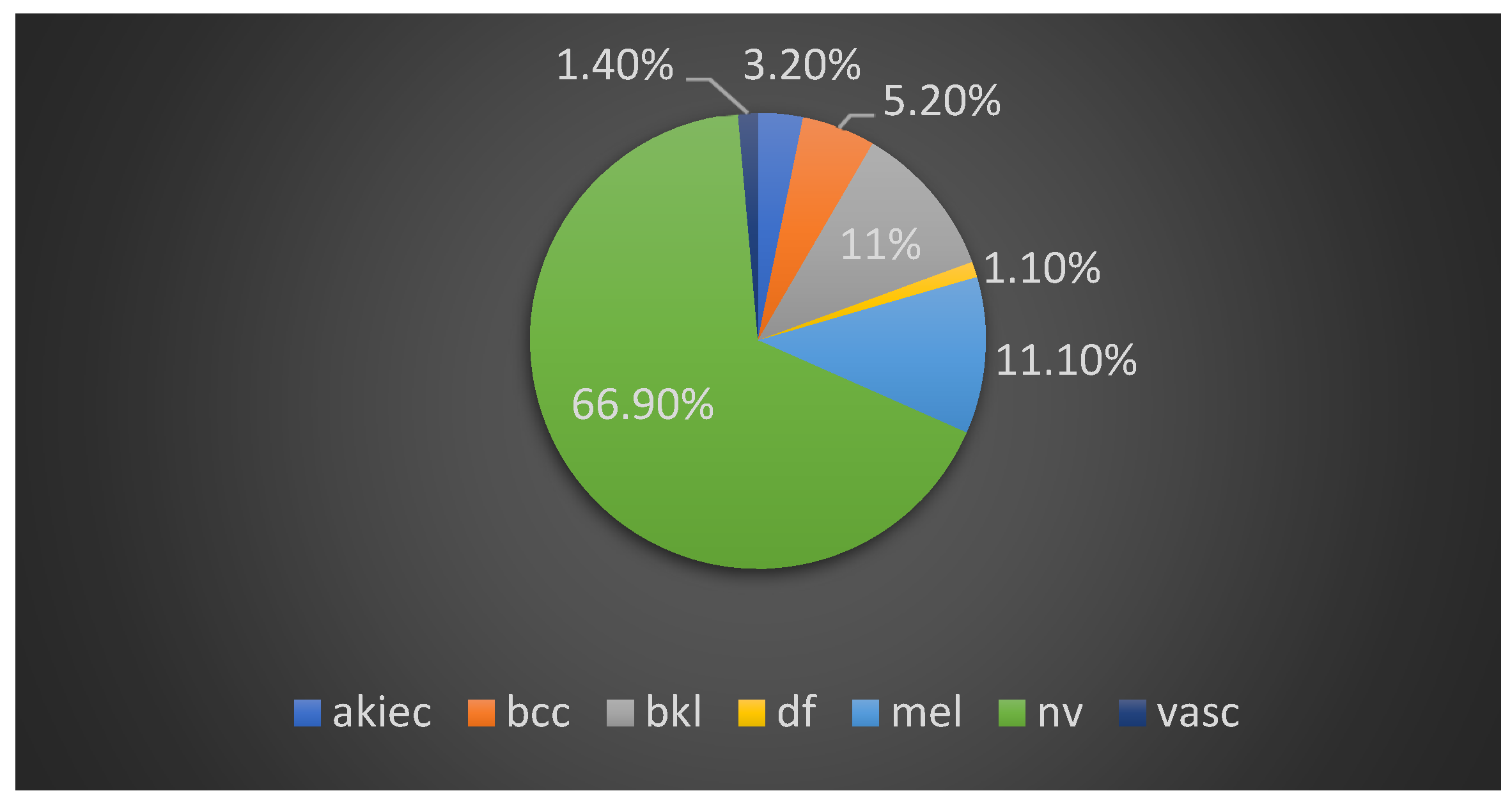
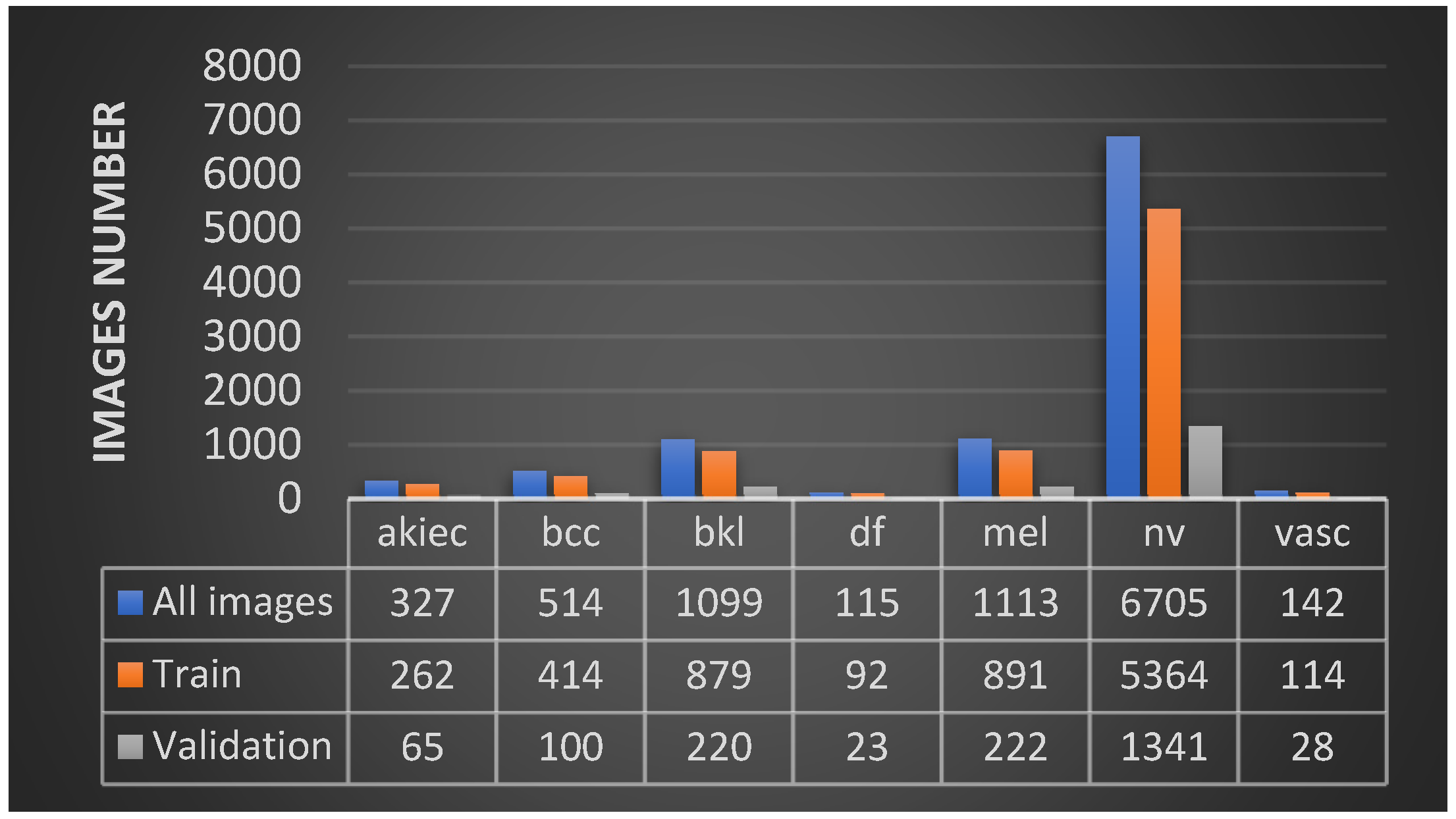
1. Introduction
Skin cancer constitutes one of the most common types of cancer, and according to the World Health Organization, it accounts for one in three diagnosed cancer types [
1]. Skin cancer reports have constantly increasing incidence rates, thus making the development of more efficient and accurate diagnostic methods a true necessity. One in five people will develop skin cancer before the age of 70 [
2]. More than 3.5 million new cases occur annually in the USA, and that number continues to rise [
3]. The most dangerous type of skin cancer is melanoma, a type that causes 75% of skin cancer deaths [
4]. It is more common in people with a sunburn history, fair skin and unnecessary exposure to UV light, and people who use tanning beds [
5]. The rates of melanoma occurrence and corresponding mortality are expected to rise over the next decades [
6].
The crucial point for treating skin cancer is the early and accurate detection of it. For example, if melanoma is not diagnosed in the early stages, it starts to grow and continues to spread throughout the outer skin layer, finally penetrating the deep layers, where it connects with the blood and the lymph vessels. That is why it is very important to diagnose it in the early stages, when the mortality rate is very low and a successful treatment is possible. The estimated 5 year survival rate of diagnosed patients ranges from 15% if detected at its latest stage, to over 97% if detected at its earliest stages [
7].
Skin cancer diagnosis is a difficult process, and even experienced specialist dermatologists had a success rate of only 60% until the invention of dermοscopic images, which increased success to between 75% and 84% [
8]. The difficulty lies in the fact that malignant lesions are often very similar to the benign moles, and both have small diameter which does not allow for clear images with normal cameras. For example, melanoma and nevus are both melanotic types, and for that reason, the classification difficulty between them is even bigger [
9]. Furthermore, most people do not regularly visit their dermatologist, and so they end up with a fatal, late diagnosis.
Thus, there is need, for those cases, to provide an easy alternative solution. The most ubiquitous digital technology typically available for individuals is that of smartphones. The imaging capability of a smartphone could be a natural way for dermatologists, general practitioners and patients to exchange information about skin lesion changes that might be worrisome [
10].
Most of the existing state-of-the-art efforts use either hybrid models [
11,
12] or ensembles of deep learning classifiers [
13,
14,
15], which are quite heavy to be used in a mobile application. To be able to build an effective mobile application, it is required to find a deep learning model that achieves state-of-the-art performance and is relatively light. Thus, the main objective of this paper is to find a single, relatively light deep learning model, which, combined with appropriate image processing methods, achieves a state-of-the-art performance. To that end, we investigate 11 CNN (convolutional neural networks) single architectures configurations to compare their ability to correctly classify skin lesions. The best model in terms of performance, required memory size and number of parameters to be specified is chosen for our mobile application to be able to classify skin lesions using a common smartphone.
The reminder of the article is structured as follows:
Section 2 presents background knowledge on skin cancer types.
Section 3 presents an extensive number of the most recent, related works on methods and approaches to skin cancer detection.
Section 4 presents the CNN architectures and the datasets, while
Section 5 presents the conducted experiments, the collected results and a discussion on them, including a comparison with existing best efforts. The development of the android application for the recognition of skin cancer is presented in
Section 6. Finally,
Section 7 concludes the paper and provides directions for future work.
2. Medical Knowledge
There are various types of skin cancer. In this section, we present the most significant of them, giving some medical information.
Melanoma (MEL): The most dangerous form of skin cancer, it develops when non-amendable DNA damage occurs to skin cells and causes mutations that lead the skin cells to proliferate rapidly and form malignant tumors. The cause of this phenomenon is usually exposure to ultraviolet (UV) radiation or artificial tanning devices. If it spreads (gives metastases) to the lymphatic system or internal organs, it is fatal at percentages of 38% and 86%, respectively. However, if it is diagnosed and treated at an early stage, the mortality rate is only 0.2% over the next 5 years. The difficulty of diagnosis lies in the fact that melanoma, at its early stages, is similar to other benign skin lesions, and yet, it often develops through them.
Basal Cell Carcinoma (BCC): It is the most common form of skin cancer, and at the same time, the most common form of cancer of all cancers. This type of cancer occurs in the basal cells, which are found in the deeper layers of the epidermis (the surface layer of skin). Almost all basal cell carcinomas occur οn parts of the body that have been extensively exposed to sun—especially on the face, ears, neck, head, shoulders and back. In rare cases, however, tumors also develop in non-exposed areas. Further, contact with arsenic, exposure to radioactivity, open wounds that do not heal, chronic inflammatory skin conditions and complications from burns, scars, infections, vaccines or even from tattoos are compounding factors.
Actinic Keratosis and Intraepithelial Carcinoma (AKIEC): Approximately 450,000 new cases of squamous cell carcinoma (SCC), the main type of AKIEC, are diagnosed annually. This makes this form of cancer the second most common skin cancer (after BCC). This form of cancer is found in the acanthocytes, which constitute the epidermis. SCC can occur at any area of the skin, including the mucous membranes of the mouth and genitals. Of course, it is more often observed in places that are exposed to the sun, like the ears, lower lip, face, bald part of a skull, neck, hands, arms and legs. Often, the skin at those points appears visually as if a sun damage has occurred, displaying wrinkles, changes in color and loss of elasticity.
Melanocytic nevus (NV): A nevus is the reference point (colored) characterized by the accumulation of melanocytes in different layers of the skin. They occur in the embryonic period of life, childhood and adolescence and less often in adults and the elderly. The epidermal nevus is a brown or black smooth spot and is rarely prone to enlargement and malignancy.
Benign Keratosis Lesions (BKL): This kind of benign tumor is the most common and mainly appears in middle-aged and elderly patients as seborrheic hyperkeratosis. Seborrheic hyperkeratosis is caused by the addition of keratin to the stratum corneum. In some cases, it can develop rapidly and sometimes resembles skin cancer.
Vascular lesion (VASC): Skin vascular lesions are due to the expansion of a small group of blood vessels located just below the skin’s surface. They are often created on the face and feet. They are not lethal, but they can cause severe leg pain after prolonged standing, and sometimes indicate more serious venous conditions.
Dermatofibroma (DF): A common benign fibrotic skin lesion, it is caused by the non-cancerous growth of the tissue cells of the skin. It is generally a single round or oval, brownish, or sometimes yellowish, nodule of 0.5 to 1 cm in diameter.
In
Table 1, the characteristics of various lesions are presented.
3. Related Work
There many works related to the diagnosis of skin cancer. We refer here to more recent and important works.
Afonso Menegolay et al. [
16] tried to classify lesions as melanoma vs. benign, malignant vs. benign and carcinoma vs. melanoma vs. benign, and they showed that the last one gives the best results. They also proved that fine-tuning improves classification, but on the other hand, transfer learning should be implemented with care concerning the dataset they used. They used the Atlas and ISBI Challenge 2016 datasets for the lesions and ImageNet and Retinopathy datasets for fine-tuning and transfer learning. They also argued that DNNs (deep neural networks) perform better when based on VGG-M and VGG-16 architectures.
Esteva et al. [
17] use a GoogleNet Inception v3 CNN architecture that was pretrained on approximately 1.28 million images from the 2014 ImageNet Large Scale Visual Recognition Challenge. They trained it end-to-end on their dataset using transfer learning. Their dataset consisted of 129,450 dermatologist-labelled clinical images, including 3374 dermoscopic images. They split it into 127,463 training and validation images and 1942 biopsy-labelled test images. Nine-fold cross-validation was used. Diseases were organized in a tree structure. For the three-class case (benign lesions, malignant lesions and non-neoplastic lesions), which concerned the first-level nodes of the tree, CNN achieved 72.1 ± 0.9% (mean ± sd) overall accuracy, which was better compared to that of two dermatologists (65.56% and 66.0%, respectively). For the nine-class case (dermal, epidermal, melanocytic–melanoma, epidermal, dermal, cutaneous lymphoma–genodermatosis and inflammatory), which are the second-level nodes, it achieved 55.4 ± 1.7% accuracy, again better than that of the same two dermatologists (53.3% and 55.9%, respectively).
Li and Shen [
18] developed a lesion index calculation unit (LICU) for the lesion segmentation and classification task of ISIC 2017 Challenge, which contains 2000 skin lesion images, 374 melanoma cases, 254 seborrheic keratosis and 1372 nevus. They used the ISIC 2017 dataset, and they achieved an accuracy of 0.912. They pre-processed the data using rescaling in the high-resolution images after cropping the image in the center of the lesion, and they applied data augmentation to increase the number of the images by flipping them on the x or y axis. They additionally proposed the use of weighted softmax loss function and batch normalization.
The Recod Titans Group [
11] created ensemble deep learning models to classify the skin lesion images. They used open datasets from the ISIC 2018 Challenge, the official challenge dataset with 10,015 dermoscopic images; ISIC Archive1, with over 13,000 dermoscopic images; Interactive Atlas of Extreme Learning, with 1000+ clinical cases, each with dermoscopic and close-up clinical images; Dermofit Image Library, with 1300 images; and PH2 Dataset, with 200 dermoscopic images. In addition, they used Dermatology Atlas, Derm101 and DermIS to gain an additional 631 images and reach the number of 30,726 images for their final dataset. Further, they increased that number with the use of synthetic skin lesions using GANs (generative adversarial networks) [
19] and other data augmentation techniques, such as traditional color and geometric transforms, and more unusual augmentations, such as elastic transforms, random erasing and a novel augmentation, that mix two different lesions. They trained the hybrid models of Inceptionv4, ResNet-152, and DenseNet-161, which were all pretrained on the ImageNet dataset. Finally, they proposed three hybrid models: (1) the XGBoost ensemble of 43 deep learning models, (2) an average of eight best deep learning models (on the holdout set), augmented with synthetic images, and (3) an average of 15 deep learning models. Their final results were 0.732, 0.725 and 0.803, respectively, for the normalized multi-class accuracy.
The authors in [
20] used a CNN (Microsoft ResNet-152 model) for classifying clinical skin images in 1 of 12 skin diseases: basal cell carcinoma, squamous cell carcinoma, intraepithelial carcinoma, actinic keratosis, seborrheic keratosis, malignant melanoma, melanocytic nevus, lentigo, pyogenic granuloma, hemangioma, dermatofibroma and warts. The CNN was trained using the training sets of the Asan and MED-NODE datasets, as well as the Atlas site images (19,398 images in total). It was tested via test sets of the Asan and Edinburgh datasets. The average values for AUC (area under curve), sensitivity and specificity were: 0.91 ± 0.01, 86.4 ± 3.5 and 85.5 ± 3.2 (Asian test set), and 0.89 ± 0.01, 85.1 ± 2.2 and 81.3 ± 2.9 (Edinburgh test set). Especially for malignant melanoma, they were: 0.96 ± 0.00, 91.0 ± 4.3 and 90.4 ± 4.5 (Asian test set), and 0.88 ± 0.01, 85.5 ± 2.3 and 80.7 ± 1.1 (Edinburgh test set). A test with 16 dermatologists showed that the results of the system were comparable to those of the dermatologists. The creation of a user-friendly, PC- and smartphone-based automatic skin disease recognition platform is also reported.
Harangi [
13] investigated whether an ensemble of CNNs could increase the individual deep classifiers’ performance to compensate for the shortage of large numbers of labelled melanoma images. Four CNNs of different type were ensembled: GoogLeNet, AlexNet, ResNet and VGGNet. Only 2000 images were used for training, 150 for validation and 600 for testing, as related to three types of skin cancer (nevus, melanoma and seborrheic keratosis). Using image transformations, the images increased to 14,300. A weighted voting scheme was used for aggregating the results of individual CNNs. The ensemble achieved an average accuracy of 0.866, an average sensitivity of 0.556 and an average specificity of 0.785.
After RGB skin cancer images pre-processing, pre-trained AlexNet was used for feature extraction, and afterwards, a combined ECOC SVM algorithm was used for cancer classification in [
21]. A total of 3753 images collected from the internet were used (2985 as the training set and 768 as the test set) concerning the four classes of carcinomas (actinic keratoses, basal cell carcinoma, squamous cell carcinoma and melanoma). The system achieved accuracy equal to 94.2%, sensitivity equal to 97.83 and specificity equal to 90.74 for melanoma and almost the same for the rest of the classes of carcinomas.
The authors in [
22] used an attention residual learning convolutional neural network (ARL-CNN) model. It was composed of multiple ARL (attention residual learning) blocks, a global average pooling (GAP) layer and a classification layer. The ISIC-skin 2017 dataset, consisting of 2000 training, 150 validation, and 600 test images, was used for training and evaluation, enhanced with 1320 additional dermoscopic images from the ISIC archive related to melanoma, nevus and seborrheic keratosis. The model achieved an accuracy of 0.850, a sensitivity of 0.658 and specificity of 0.896 for melanoma classification, and 0.868, 0.878 and 0.867, respectively, for seborrheic keratosis classification.
Khan et al. [
9] proposed an intelligent system to detect and distinguish between melanoma and nevus cases. A Gaussian filter was used for removing noise from the skin lesion and an improved k-means clustering was utilized to segment out the lesion. Textual and color features were afterwards extracted. A support vector machine (SVM) was used for the classification of skin cancer into melanoma and nevus. The DERMIS dataset, having a total number of 397 skin cancer images (146 of melanoma and 251 of nevus type), was used. The system achieved 96% accuracy, 97% sensitivity, 96% specificity and 97% precision.
Sarkar et al. [
23] used what they called a deep, depthwise separable residual convolutional algorithm for binary melanoma classification. Dermoscopic skin lesion images were pre-processed by noise removal using a non-local means filter, followed by enhancement using contrast-limited adaptive histogram equalization over a discrete wavelet transform algorithm. Images were fed to the model as multi-channel image matrices, with channels chosen across multiple color spaces based on their ability to optimize the performance of the model. The proposed model achieved an accuracy of 99.50% on the international skin imaging collaboration (ISIC), 96.77% on PH2, 94.44% on DermIS and 95.23% on MED-NODE datasets.
The approach in [
14] dealt with the problem of skin lesion classification in eight classes: melanoma (MEL), melanocytic nevus (NV), basal cell carcinoma (BCC), actinic keratosis (AK), benign keratosis (BKL), dermatofibroma (DF), vascular lesion (VASC) and squamous cell carcinoma (SCC). The data set used was the ISIC 2019. Data augmentation was also used. They set up an ensemble of CNN architectures. The optimal ensemble comprised six EfficientNet configurations, two ResNext variants and an SENet. They achieved accuracy of 75.2% using five-fold cross-validation. The system ranked first in the ISIC 2019 Skin Lesion Classification Challenge (with a balanced accuracy of 63.6% for task 1 and 63.4% for task on official test set).
The approach in [
24] dealt with the problem of skin disease classification in six classes: seborrheic keratosis (SK), actinic keratosis (AK), rosacea (ROS), lupus erythematosus (LE), basal cell carcinoma (BCC) and squamous cell carcinoma (SCC), based on clinical images. They compared five CNN architectures: ResNet-50, Inception-v3, DenseNet121, Xception and Inception-ResNet-v2, all in two modes. In the first, the five models were trained using only clinical facial images, whereas in the second, they used transfer learning: they pretrained the models with the data of other body parts and used the parameters from the pretrained model as the initial parameters for the new model. They constructed a data set containing 4394 images (2656 of which were facial images) from a large Chinese database of clinical images. The best result (an average precision of 70.8% and average recall of 77%) was achieved by the Inception-ResNet-v2 model.
Ameri [
25] built a CNN based on a pre-trained AlexNet model that was modified for binary classification using transfer learning. All images were resized to 227 × 227 to match the input size of the AlexNet model. The model was trained using the SGDM algorithm, with an initial learning rate of 0.0001 and a mini-batch size of 30 and 40 epochs. A total of 3400 images (1700 benign and 1700 malignant) were used from the HAM10000 dataset, and 90% were randomly selected as a train set and the rest as a test set. A classification accuracy of 84%, a sensitivity of 81% and a specificity of 88% were achieved.
The authors in [
26] investigated the use of Faster R-CNN and MobileNet v2 for skin cancer detection on mobile devices, such as smartphones. They trained and tested them using a perfectly balanced dataset of 600 images concerning two skin diseases: actinic keratosis and melanoma. The images were obtained from the ISIC archive. For training, 90% were used, while 10% were used for testing, and they were enhanced with 40 extra images taken via a smartphone. Experiments showed the superiority of Faster R-CNN both in accuracy and training time. The best accuracy result for Faster R-CNN was 87.2% (on notebook) and 86.3% (on smartphone), but for different learning rates.
Mporas et al. [
27] extracted lesion features after image preprocessing and segmentation based on different color models (RGB, HSV, YIQ, etc.). They use the HAM10000 dataset to train several classifiers (SVM, NN, Bagging, AdaBoost, REP, RF, J48 and IBk). Their experiments showed better results for classifiers trained with the RGB-model-produced dataset. The winner was AdaBoost (RF), with an accuracy of 73.08%. They improved this result by combining the features of different color models. The combination of RGB plus HSV plus YIQ features and the AdaBoost (RF) achieved an accuracy of 74.26%.
In [
28], the authors built a CNN for skin cancer diagnosis. The CNN consists, excepting convolution and pooling layers, of three hidden layers, using 3 × 3 filter sizes with 16, 32, and 64 channel outputs in sequence; a fully connected layer; and softmax activation. A total of 3000 training images and 1000 validation images concerning four carcinoma classes (dermatofibroma, nevus pigmentosus, squamous cell carcinoma and melanoma) were used from the ISIC database. Using the Adam optimizer, they achieved a 99% accuracy and an F1-score of almost 1.
The authors in [
29] proposed a combination of CNN and a one-versus-all (OVA) approach to classify skin diseases. The HAM10000 dataset was used, enhanced by data augmentation techniques (e.g., rotating, scaling, translation, adding noise, etc.). No feature extraction was used. The model was trained via raw images following an end-to-end approach. The model achieved an accuracy of 92.90%, which was much better than that achieved by the CNN alone (77%).
Huang et al. [
30] built two deep learning models and used two datasets for skin disease classification. First, they built a DenseNet network for the binary (malignant or benign) classification problem based on the Kaohsiung Chang Gung Memorial Hospital (KCGMH) dataset (1287 clinical images certified by biopsy) concerning five classes: BCC, SCC, MEL, benign keratosis and NV. It achieved an accuracy of 89.5%, a sensitivity of 90.8% and a specificity of 88.1%. The second model was an EfficientNet model used for multi-class skin disease classification on two datasets, the KCGMH and the HAM10000 (10,015 dermatoscopic images and seven classes). The EfficientNet B4 version achieved the best overall accuracy results: 72.1% and 85.8%, respectively.
The authors in [
31] used a mixture of features: handcrafted features based on shape, color and texture (after image preprocessing), and deep learning features obtained using a selected CNN architecture (VGG, Mobilenet, ResNet, DenseNet, Inception V3 and Xception), which was pre-trained in an Imagenet classification task. Then, a selected classifier (one of LR, SVM or RVM) is trained as a binary classifier (malignant or benign) on the ISIC 2018 dataset (comprised of both malignant and other benign skin lesion images), augmented by the SMOTE method, to eliminate data imbalance. The system achieved an accuracy of 89.71% (on the test set), sensitivity of 86.41%, specificity of 90% and precision of 92.08%.
The proposed system in [
12] worked in two stages. In the first stage, an end-to-end resolution-preserving deep model for segmentation, namely FrCN, was used. In the second stage, the segmented lesions were passed into a deep learning classifier (Inception-v3, ResNet-50, Inception-ResNet-v2 and DenseNet-201). Classifiers were trained and evaluated on three dermoscopic images datasets: ISIC 2016 (two output classes), ISIC 2017 (three output classes) and ISIC 2018 (seven output classes), all augmented (using rotation and flipping). The best results for the first dataset were achieved by the Inception-ResNet-v2 deep classifier (accuracy of 81.79%, sensitivity of 81.80%, specificity of 71.40% and F1-score of 82.59%). ResNet-50 was the winner of the second (accuracy of 81.57%, sensitivity of 75.33%, specificity of 80.62% and F1-score of 75.75%) and third datasets (accuracy of 89.28%, sensitivity of 81.00%, specificity of 87.16% and F1-score of 81.28%).
The authors in [
32] used Deep Learning Studio (DLS) to build four different pre-trained CNN models that were based on ResNet, SqueezNet, DenseNet and Inception v3, respectively. They trained and tested them using the HAM10000 dataset divided into training (80%), validation (10%) and test (10%) sets. The best model (Inception v3) achieved a 95.74% F1-score for the two-class prediction case.
Salian et al. [
33] configured their own CNN and trained it end-to-end using two datasets of dermoscopic images, PH2 and HAM10000. They used two versions of the datasets, the original and another one produced after augmentation. Results showed that augmentation was not beneficial in all cases. Their system achieved the best accuracy values on the version with augmentation for the PH2 dataset (97.25%) and on the version without augmentation for the HAM10000 dataset (83.152%). Compared to other state-of-the-art systems (e.g., Mobilenet and VGG-16), it was much better than VGG-16 and a bit better, on average, than Mobilenet.
The authors in [
15] investigated the effect of image size on skin lesion classification based on pre-trained CNNs and transfer learning. Images from the ISIC datasets were either resized to or cropped to six different sizes. Three pre-trained CNNs, namely EfficientNetB0, EfficientNetB1 and SeReNeXt-50, were used in the experiments in single and ensemble mode (called a MSM-CNN: multi-scale multi-CNN). The results showed that image cropping was a better strategy than image resizing. The best accuracy (86.2%) was achieved on the ISIC 2018 dataset by the MSM-CNN configuration.
A hair detection methodology was followed in [
34] for image pre-processing using color enhancement, hair lines detection and thresholding for lesion segmentation. Afterwards, features were extracted (texture descriptors, color, convexity, circularity and irregularity index). They chose 640 images from the ISIC database and used 512 of them for training and the rest for testing. They used an ensemble voting classifier consisting of a KNN (k-nearest neighbor), an SVM (support vector machine) and a CNN classifier. The CNN took as inputs whole images, sized 124 × 124 pixels, and was composed of nine layers (three convolution layers with a filter size of 3 × 3 with a ReLU activation function, proceeded by three spatial max pooling layers, which down-sampled the feature maps). It achieved an accuracy of 88.4%, which was better than the accuracy of each individual classifier.
The authors in [
35] built an architecture combining the MobileNet and LSTM architectures for skin disease classification. They used the HAM10000 dataset (seven classes) for training and evaluation. Compared to other state-of-the-art approaches, it achieved better results (accuracy of 85.34%, sensitivity of 88.24% and specificity of 92.00%). The proposed model also exhibited reduced execution time compared to other models. The authors implemented a mobile application, based on the MobileNet-LSTM model, called esmart-Health.
Acosta et al. [
36] proposed a two-stage classification process: (a) create a cropped bounding box around a skin lesion, and (b) classify the cropped area using a CNN. In the first stage, Mask R_CNN was used, whereas ResNet152 was used in the second. They used 1995 dermatoscopic images for training, 149 for validation and 598 for testing, from the ISIC 21i challenge database. There were two system output classes: benign and malignant (meaning “melanoma”). Due to the small number of training examples, they used transfer learning. They produced five different models after training. The best model, called eVida, outperformed all participants in the 2017 ISBI Challenge, achieving an average accuracy of 0.904, sensitivity of 0.820 and specificity of 0.925.
Wang and Hamian [
37] used an extreme learning model (ELM) neural net model optimized by the thermal exchange optimization (TEO) algorithm. It was used to classify skin images represented by their features. Features were extracted after dermoscopy image pre-processing for noise reduction, contrast enhancement and segmentation (Otsu algorithm). The extracted features were geometric, texture and statistical. They used the SIIM-ISIC melanoma dataset with image size of 1024 × 1024 pixels. Compared with other methods (i.e., CNN, Genetic Algorithm, SVM, etc.), their approach achieved the best results: an accuracy of 92.65%, sensitivity of 91.18% and specificity of 89.70%.
In our approach, the objective was to find a single CNN-based model and suitable image processing methods to achieve state-of-the-art results. We chose the HAM10000 dataset, which includes seven unbalanced classes of skin cancer. We employed classical data augmentation, transfer learning and fine-tuning to improve the performance of our models. We compared 11 deep learning network models. DenseNet169 was the winner model, achieving an accuracy of 92.25% and a recall (sensitivity) of 93.59%, which outperformed deeper models. We have achieved with a single standard, deep learning, light model better results than other attempts that use hybrid or ensemble deep learning models. We also implemented a mobile application for skin cancer prevention and two-class (benign–malignant) detection.
5. Results and Discussion
We used the TensorFlow 1.4 framework in combination with Keras API version 2.0.8 and python 3.5 to implement and run our models on a Linux system with a GTX 1060 6GB graphics card. To increase the performance of our models, we used the image techniques mentioned above. First, we used data augmentation with image random crops, rotations, zoom and horizontal and vertical flips. In addition, we employed transfer learning from the ImageNet [
51] dataset while retraining the last layers, and after that, fine-tuning of the model, retraining the whole model with a smaller training rate. We also tried to train the models from the start without transfer learning, but the results were poorer, especially in the deeper models. Furthermore, we changed the color space from RGB to HSV and grayscale, but even then, the results were worse, especially for the tests with the grayscale images.
In
Table 3, we present the final results based on the average (weighted) accuracy [
51] of the 11 models that were trained with data augmentation, transfer learning, fine-tuning and the SGD optimizer on the original RGB images. Because of the highly unbalanced data, we used the appropriate class weights during the training process to treat the classes equally.
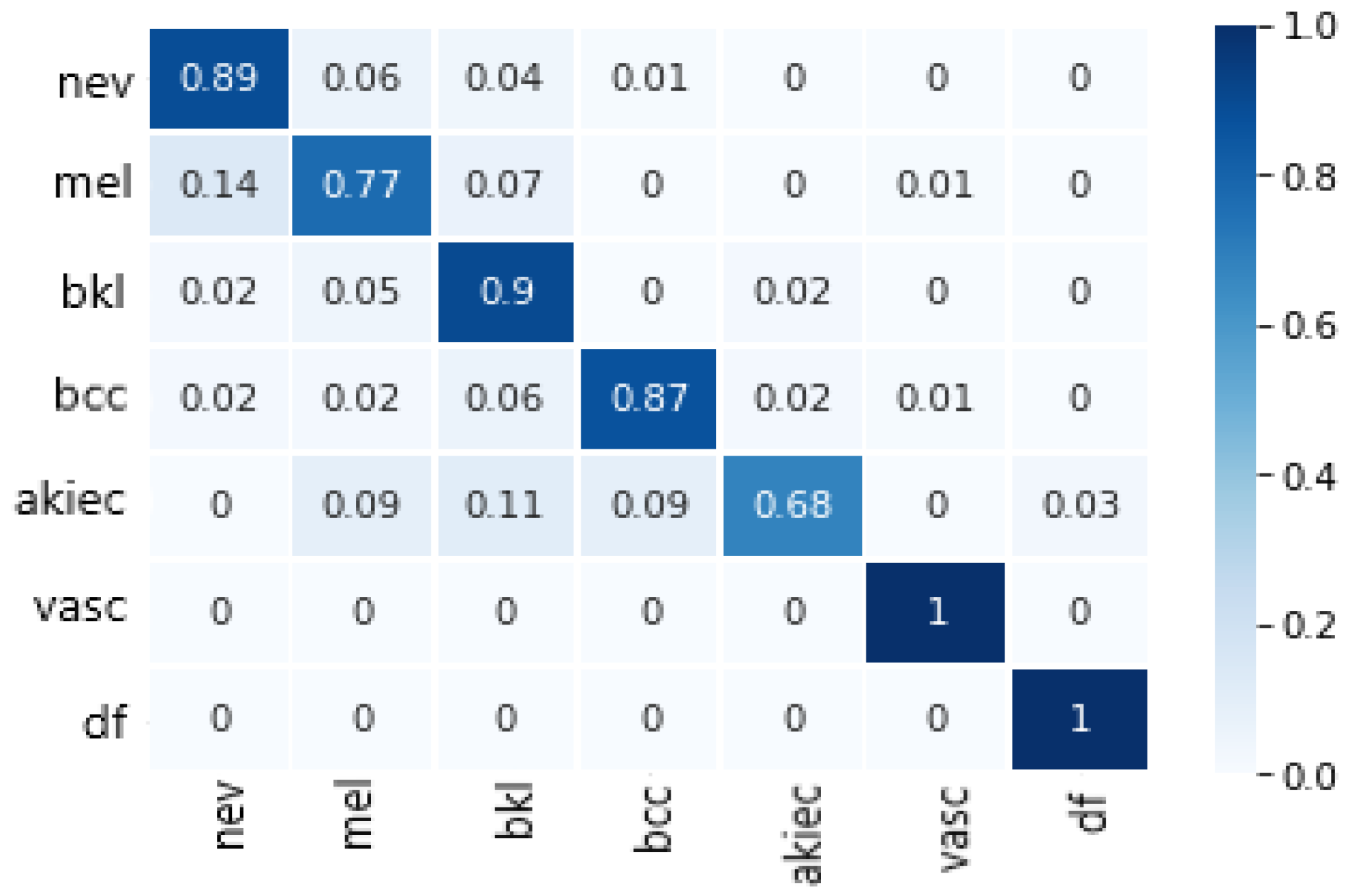
Finally, the model we chose was DenseNet169, which achieved the best values for all metrics and requires relatively less memory size and specification of relatively less parameters than other non-DenseNet candidates, e.g., InceptionV3, ResNet50 and VGG16 (see
Table 2). The confusion matrix of DenseNet169 is depicted in
Figure 3.
In order to face up with the image quality problem, we tried to insert each image during the validation phase more than one time and make a diagnosis based on their median average. More specifically, we inserted each validation image four times into the model by making a different flip each time, and then we calculated the median average of the results in order to classify the image. This technique was used in five randomly selected models during the last 50 training epochs of DenseNet169. The results showed that there was an improvement of +1.004% on average compared to the original image case (see
Table 4). From
Table 4, it is also obvious that vertical flip was the main contributor to the improvement.
For the application, given its qualitative orientation and its use as a first diagnostic hint, we decided to use a two-class mapping of the DenseNet169 model to distinguish between benign (nv, bkl, vasc or df) and malignant (mel, bcc or akiec) cases. So, the seven-class model was used, but it gives as output to the user “benign” or “malignant”, depending on the predicted class. The metrics of the two-class mapping model are presented in
Table 5.
From the tested model architectures, DenseNet169 achieved the highest values for all metrics, while DenseNet121 the second highest. In contrast to our expectations, DenseNet201 did not do better than the other, less deep DenseNet models. It seems that the size of the training set was no more adequate for deeper training than that of DenseNet169. From the rest of the models, InceptionResNetV2 and VGG16 did better than the others, being very close between each other. Again, the deeper VGG19 model did not outperform the less deep VGG16 model.
The experimental results show that our model did very well in distinguishing between melanoma and nevus, which is considered a hard case given that both are melanocyte lesions and they visually look alike. Furthermore, we managed to reach a higher accuracy than specialist dermatologists.
In
Table 6, a comparison with state-of-the-art approaches is attempted. In the table, we have included models that deal with comparable datasets. We also distinguish them in two groups, the two-class and seven-class groups. As is clear from
Table 6, our model is the best in the seven-class category, whereas it is the second best, in terms of achieved accuracy, in the two-class category. However, compared to the first one, it has the advantage of being a single model.
6. Application Development
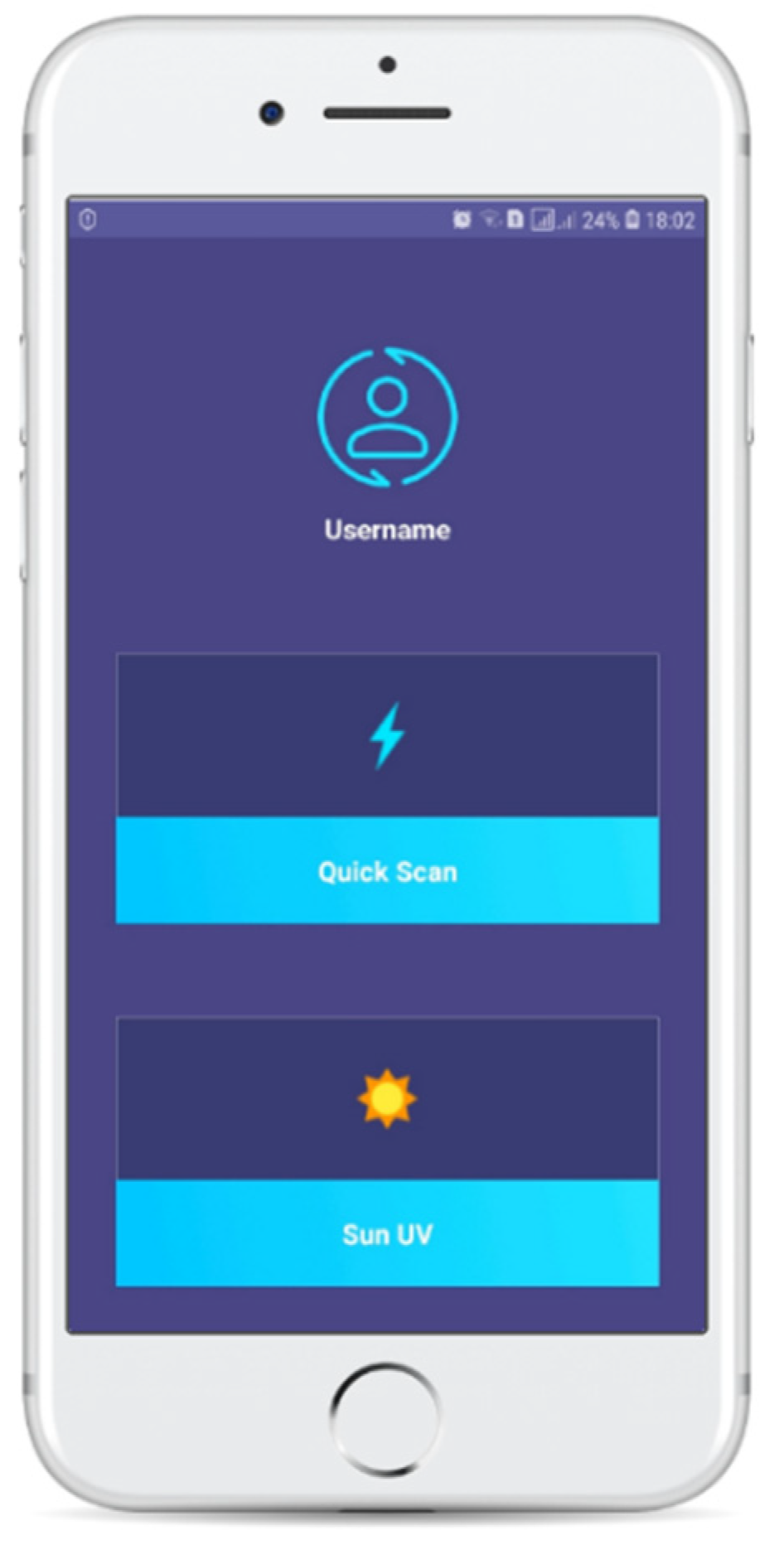
We developed a mobile application to constitute an integrated system for the prevention and diagnosis of skin lesions (
Figure 4). We used Android-Studio έκδοση-3.1.3 and Android 8.0. It was designed to be a useful and easy to use tool for every smartphone user, minimizing the complex processes and making the application environment as simple and easy to understand as possible. We placed the DenseNet169 two-class mapping model inside it, after we transformed it to a TensorFlowLite model, in order to be optimized for smartphones. The application enables us to take a photo of the skin lesion that we want to examine, manually crop the photo and keep the area that is of interest (ROI) and insert it into the classification model. Other features of the application concern the creation of a user account to provide authentication, as well as the ability to calculate the amount of time that the user can remain exposed to the sun without any burns. Studies show that the possibility of melanoma is doubled every five sunburns. For this calculation, we use the user’s skin phototype, the sun UV indicator present in the user’s area at the given time and the user’s sunscreen index.
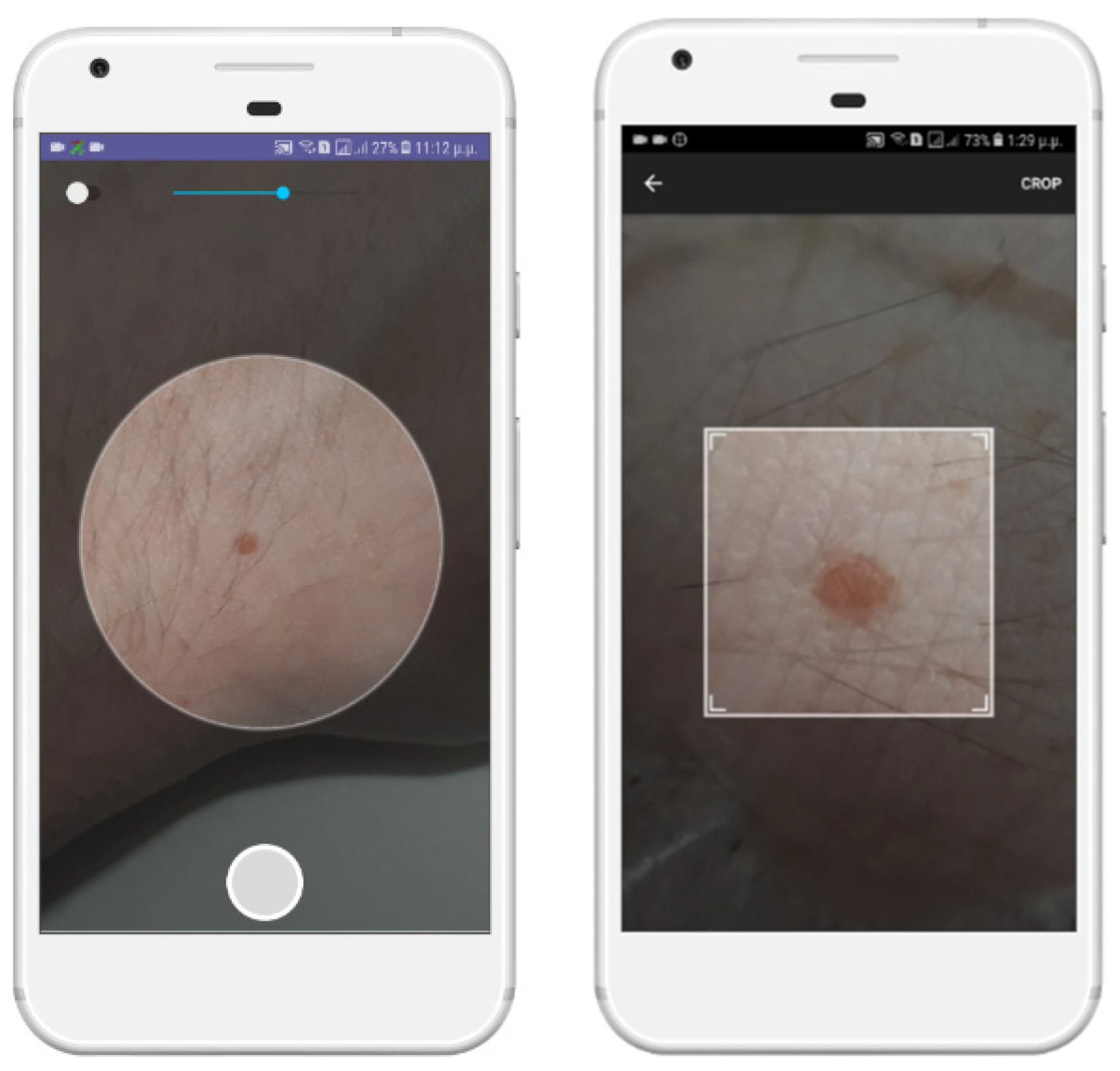
To make the photo quality as high as possible, we provided the option of zooming in on the area of skin damage or to open the flashlight of the mobile, if available. Then, we ask the user to manually crop the photo into a square area near the outer perimeter of the skin lesion (
Figure 5).
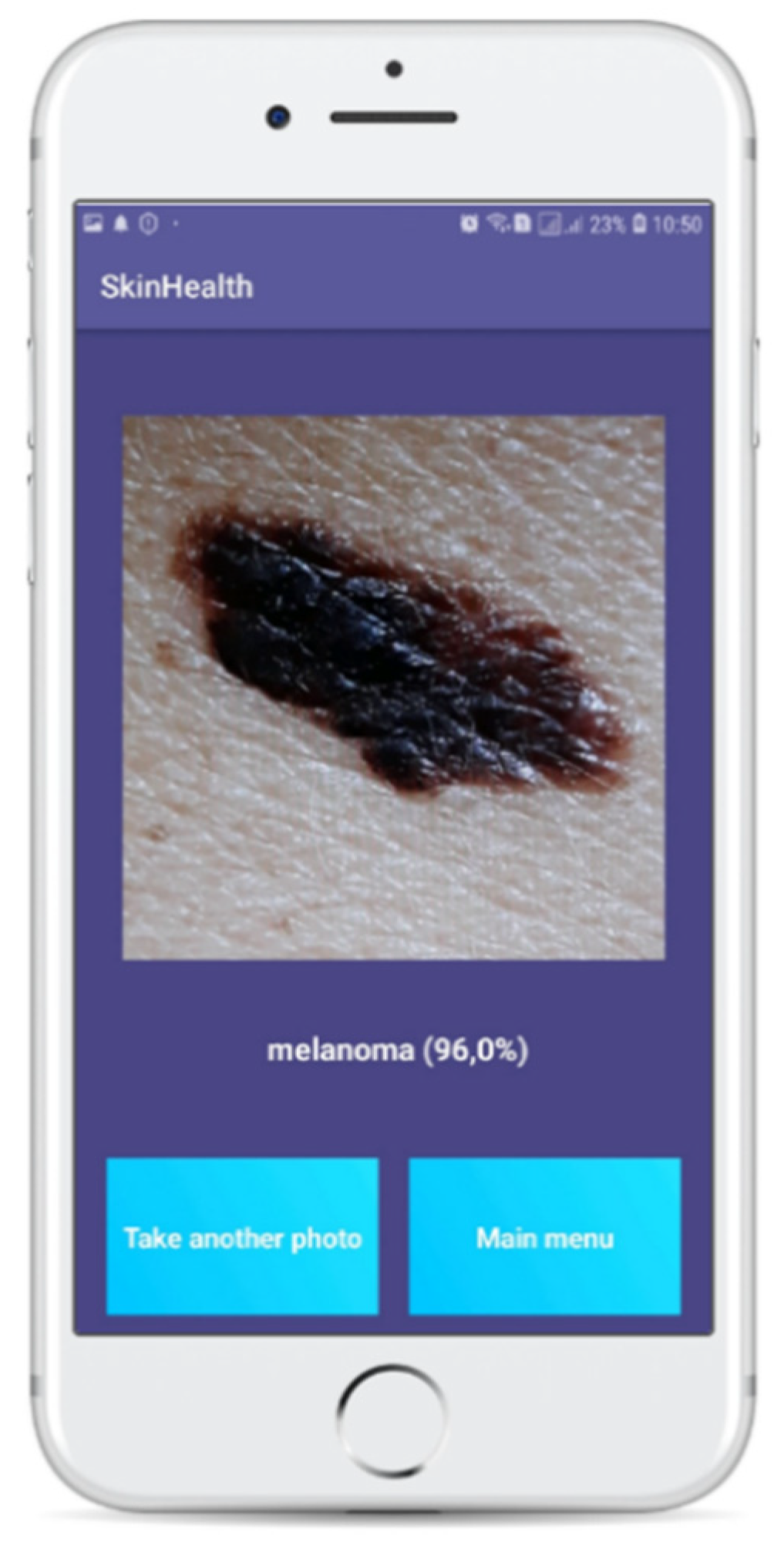
Figure 6 presents a classification result after the use of the application for a specific case.
In addition, to reduce the likelihood of malignant skin damage of users, we developed another function in our application. Since the main factor for melanoma is the irradiated sunlight, and, more specifically, the hours exposed to the sun, according to the degree of ultraviolet radiation that we receive at that time, we inform the user of the time that he/she can safely be exposed to the sun without resulting in sunburns. We do this with the following steps:
Collect the user’s location via the mobile phone’s GPS system.
Send the location and the current time to OPENUV API (an open forecast and update platform for solar radiation). This returns the data we need to inform the user about sun UV index in his/her location and the time he/she can stay in the sun for every skin phototype.
The user selects his/her skin phototype and the degree of sunscreen (if he/she uses one) to be informed of the time that he/she can be exposed to the sun without having unpleasant side effects for his/her health.
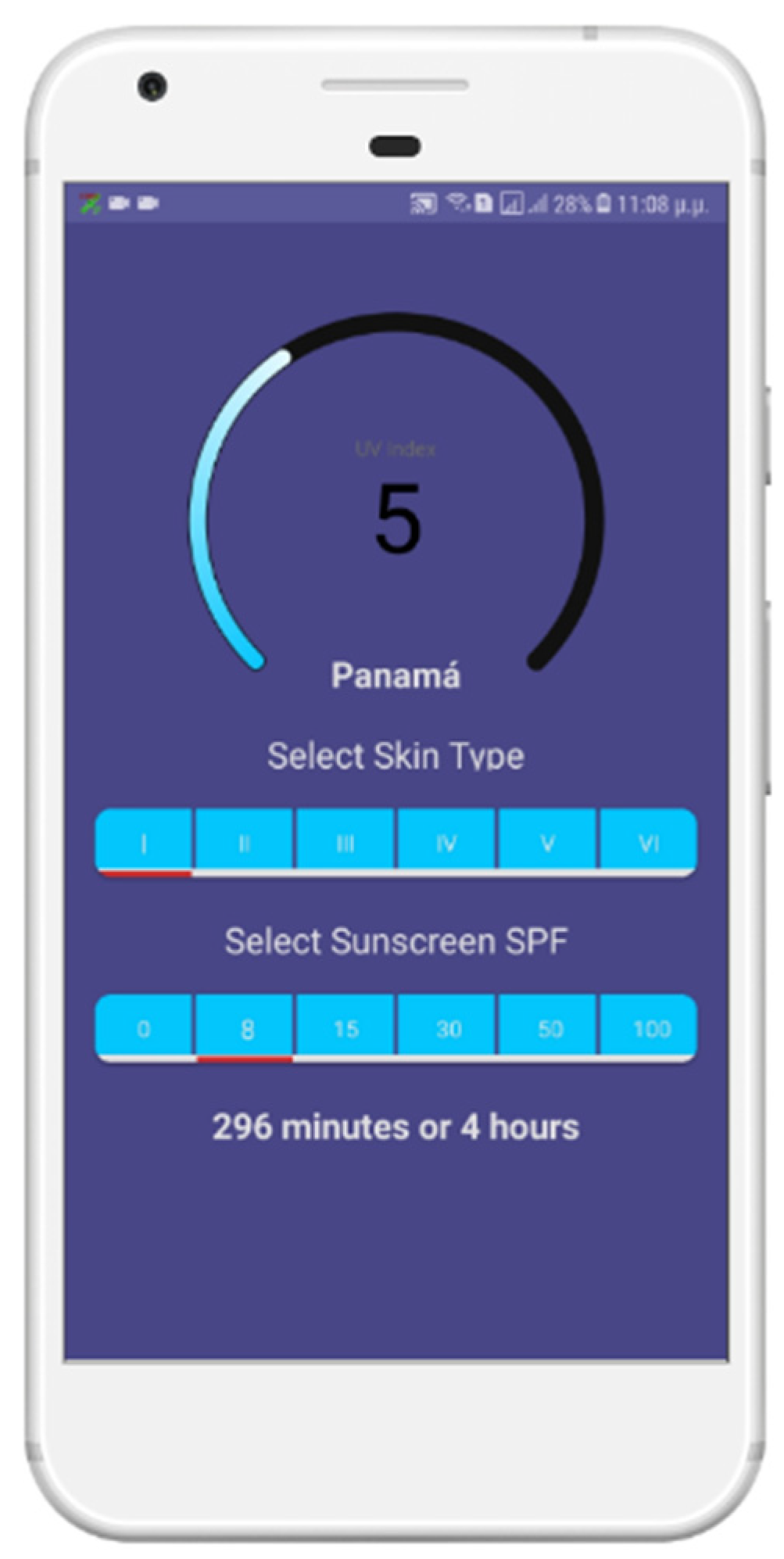
In
Figure 7, we show the corresponding snapshot of the application in which we see that in Panama City, the sun UV index is currently 5, and the user with a phototype 1 can stay in the sun for 296 min using a sunscreen with an SPF of 8.
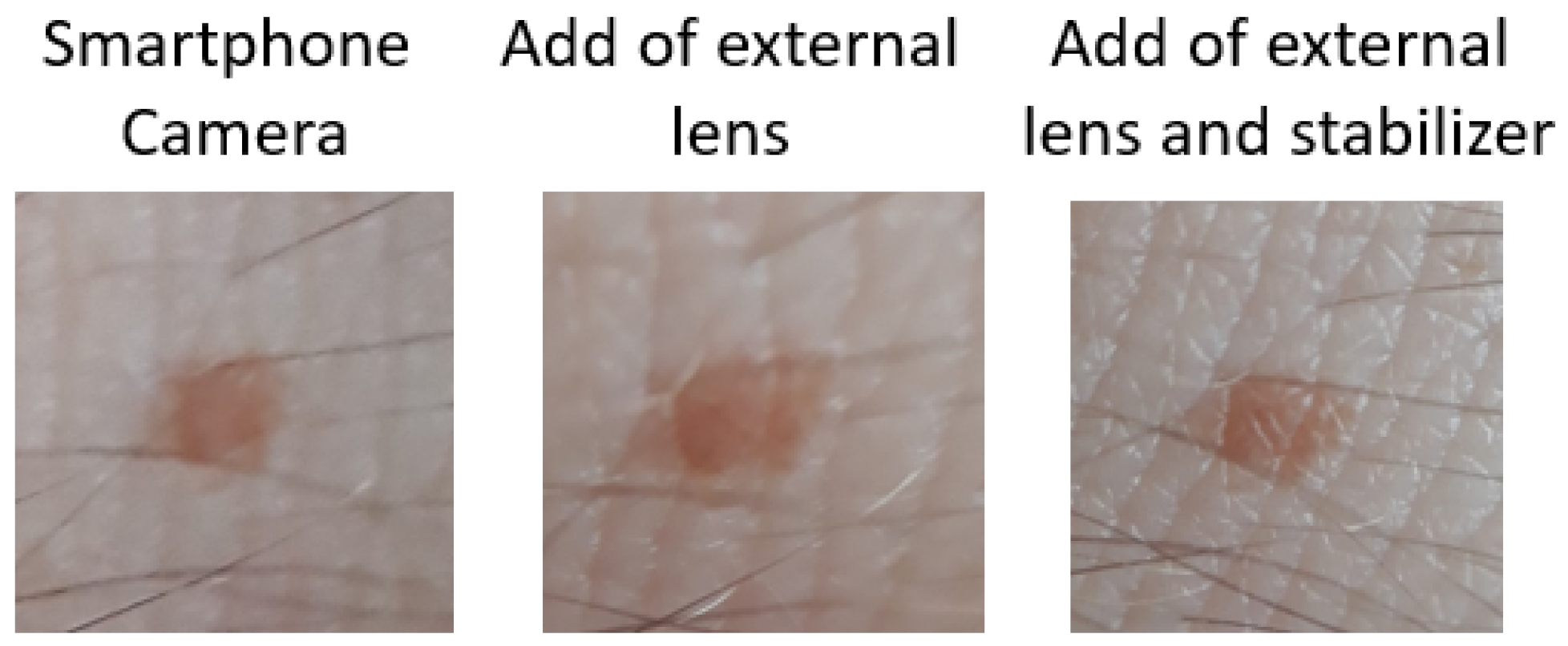
Finally, we tested the app in a real environment, using at first only the phone camera lens. After that, we tried it again with an external 10× macro lens, and the result was a bit better. We also tried it with the external macro lens and a handmade stabilizer, which gave us the best result. The quality of the image is very close to the dataset images. We strongly believe that this sector of the app is critical for the average accuracy of the model. We can see the results of the images in
Figure 8.
After that, the image is automatically passed to the model to be categorized. Within a few seconds (about 2–4), depending on the capabilities of the smartphone, the classification result is displayed to the user. The user then has the option to return to the original menu or restart the process from scratch (see
Figure 6).
Given the shortage of smartphone-taken skin lesion image databases, we could not test our application in a systematic way; however, small scale tests were very successful.
To use such a system may require the transfer of the image to a server, where the system could be implemented and run more efficiently. Further, to achieve better performance for the data transmition, new technologies for wireless sensor networks [
52,
53] could be used.