Assessment of Dual-Tree Complex Wavelet Transform to Improve SNR in Collaboration with Neuro-Fuzzy System for Heart-Sound Identification
Abstract
:1. Introduction
2. Materials and Methods
- (1)
- MIT heart sounds database (MITHSDB) with 409 PCG recordings made at nine different recording positions and orientations from 121 subjects (age and gender unknown). Each subject contributed several recordings. The subjects were divided into:
- (i)
- normal control group: 117 recordings from 38 subjects;
- (ii)
- murmurs and mitral-valve prolapse (MVP): 134 recordings from 37 patients;
- (iii)
- 118 benign murmurs recorded from 34 patients;
- (iv)
- aortic disease (AD): 17 recordings from 5 patients;
- (v)
- other miscellaneous pathological conditions (MPC).
- (2)
- Aalborg University heart sounds database (AADHSDB). The signals were recorded from 151 subjects (coronary angiography disease). Age and gender are unknown.
- (3)
- Aristotle University of Thessaloniki heart sounds database (AUTHHSDB). Forty-five subjects were enrolled with ages between 18 and 90 years.
- (4)
- Toosi Technology University heart sounds database (TUTHSDB): includes 16 patients with different types of cardiac-valve diseases (age and gender are unknown).
- (5)
- University of Haute Alsace heart sounds database (UHAHSDB). Nineteen normal subjects were recorded, with ages between 18 and 40 years; instead, the abnormal recordings were from 30 patients (10 females and 20 males) aged from 44 to 90 years.
- (6)
- Dalian University of Technology heart sounds database (DLUTHSDB). Subjects included 174 healthy volunteers (2 females and 172 males, aged from 4 to 35 years), and 335 patients (227 females and 108 males, aged from 10 to 88 years).
- (7)
- Shiraz University adult heart sounds database (SUAHSDB). The subjects included are 69 females and 43 males, aged from 16 to 88 years.
- (8)
- Skejby Sygehus Hospital heart sounds database (SSHHSDB): comprises 35 recordings from 12 normal subjects and 23 pathological patients with heart-valve defects (age and gender unavailable).
2.1. Preprocessing of HS Signal
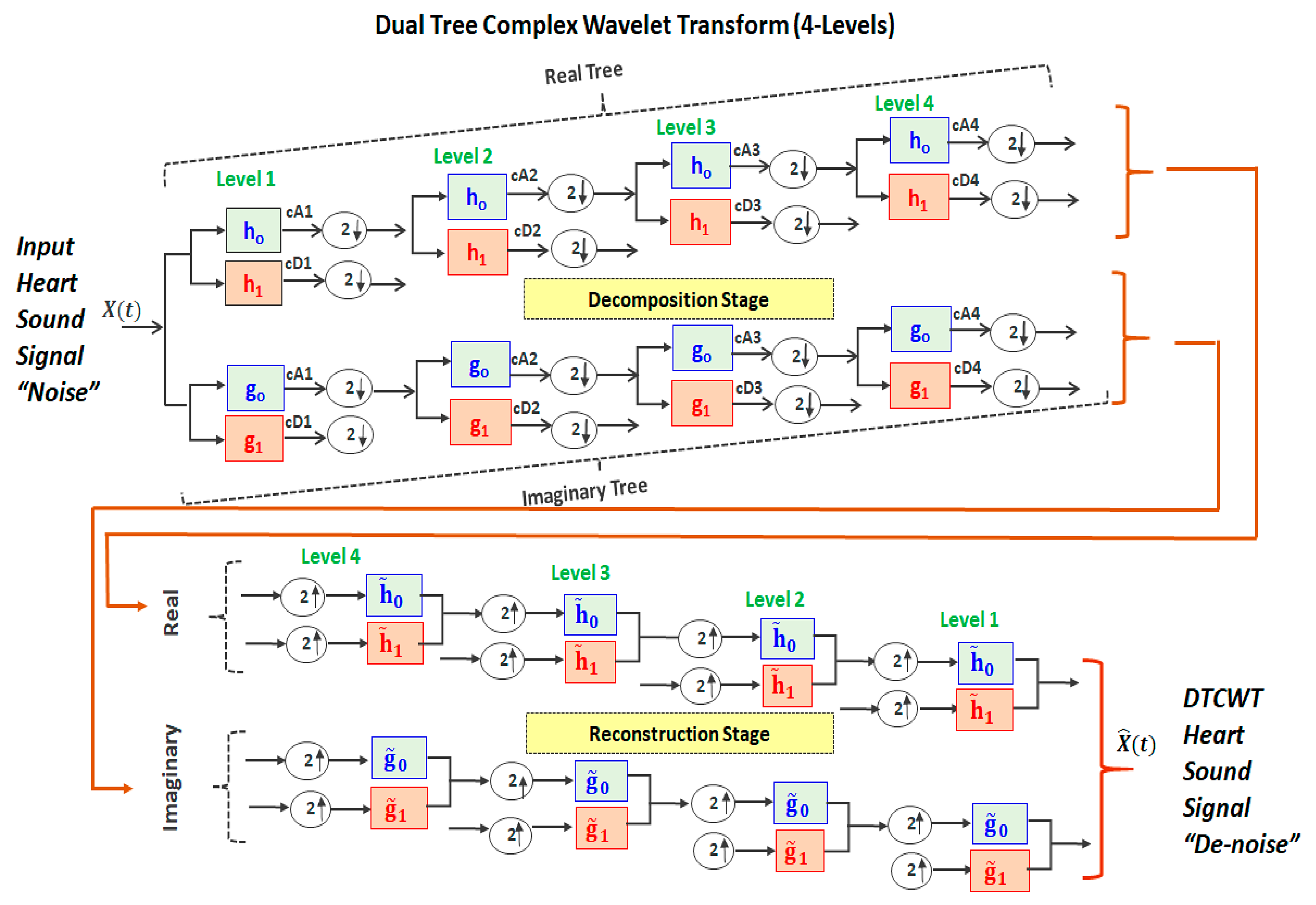
2.2. Dual-Tree Complex Wavelet Transform
- Implementing a perfect reconstruction (PR) to make the reconstructed signal identical to the original (input) signal X(t). This condition is achieved when the input signal’s noise is successively attenuated until the end of the number of decomposition levels. Then, the next process (mirror operation) successively synthesizes (i.e., reconstructs) the resulting signals after noise reduction.
- Implementing successive half-sample shifts (i.e., dividing the samples by factor of 2) of both the low-pass filters (h0 and g0) and high-pass filters (h1 and g1) on the real and imaginary parts (dual trees). This would avoid any disorder in satisfying the Hilbert pair condition.
- Implement an equal-sample shift to have the same range of frequencies through all the CTDWT levels during the decomposition and reconstruction stages.
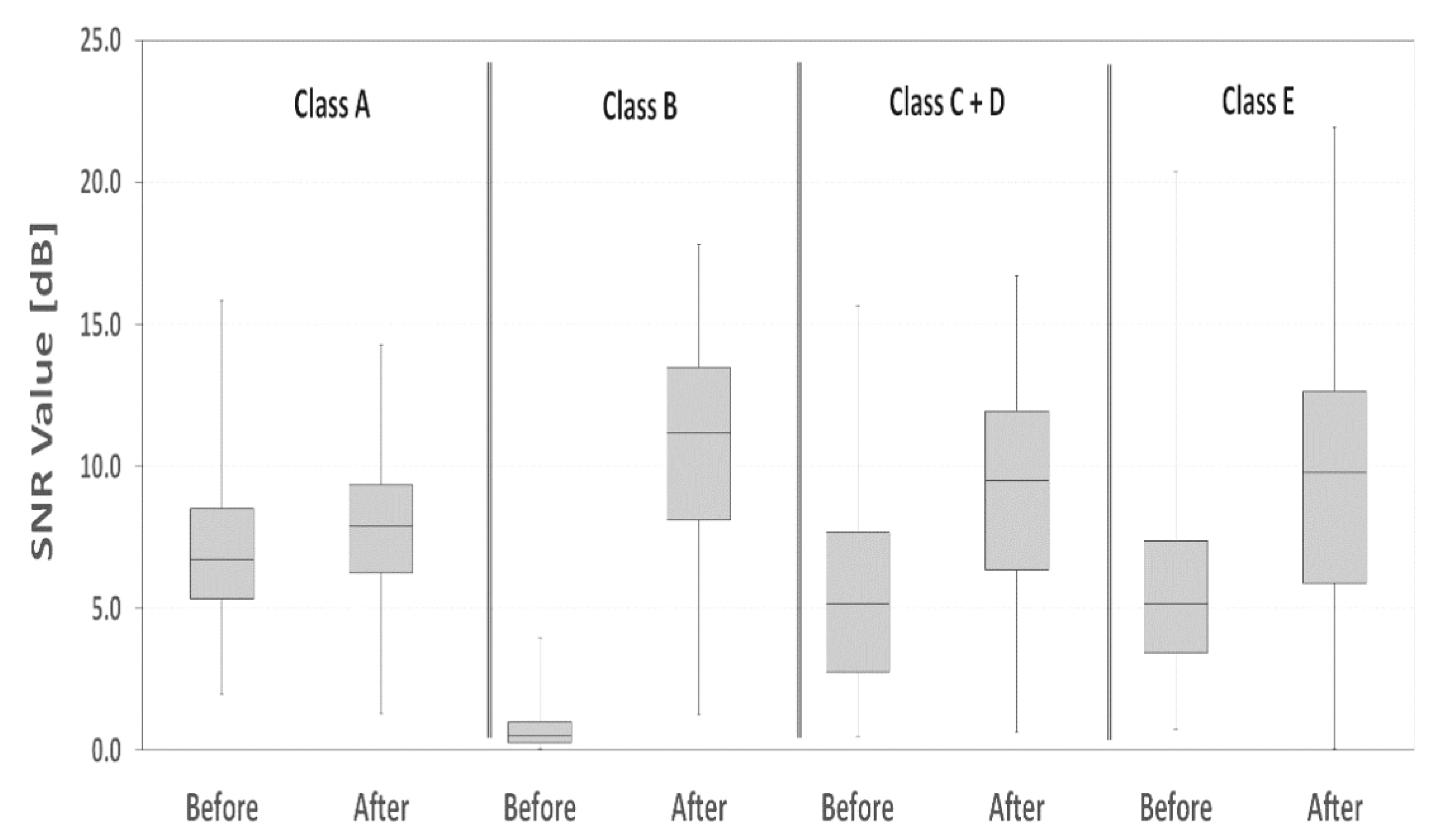
2.3. SNR Calculations
2.4. Feature Extractions
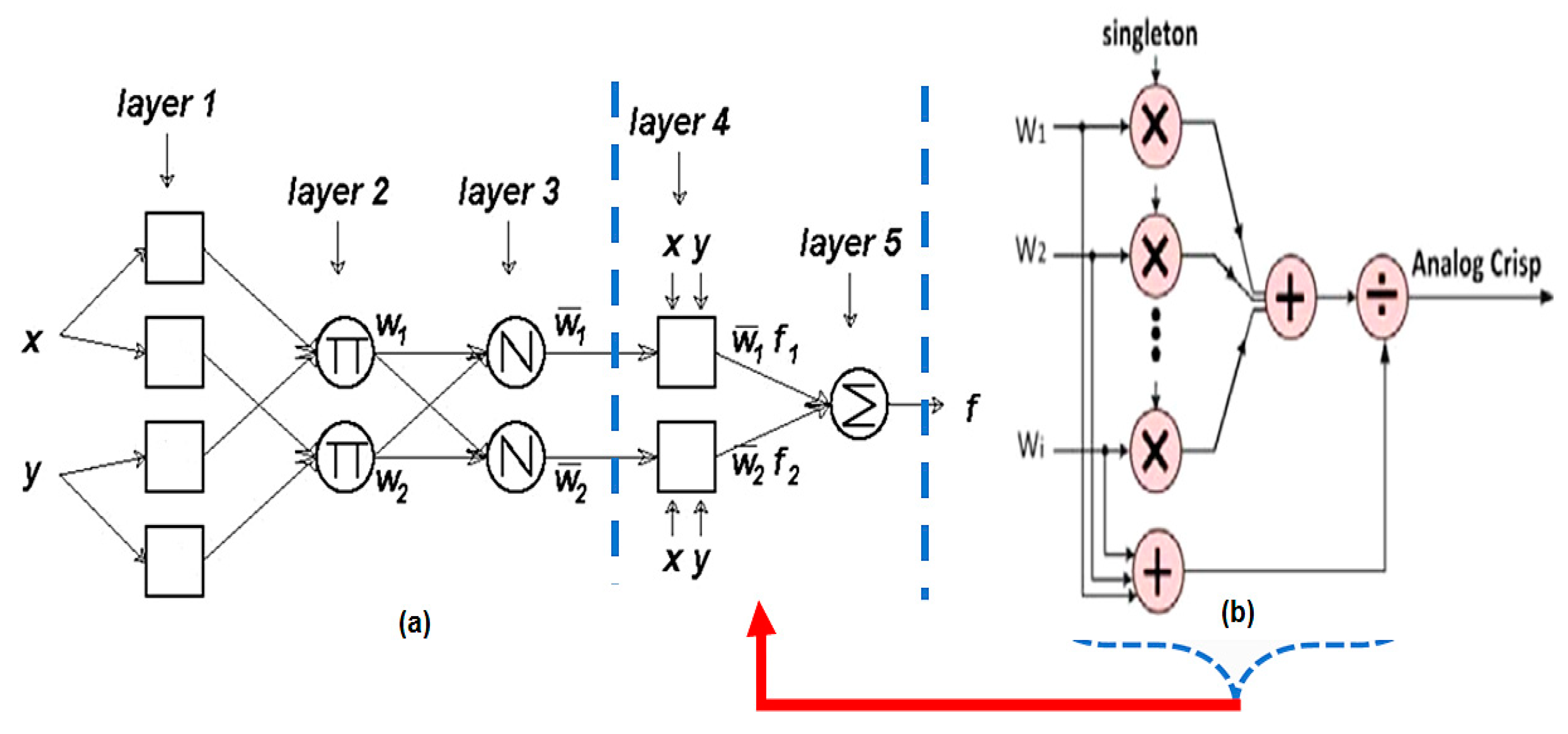
2.5. Adaptive Neuro-Fuzzy Inference System (ANFIS)
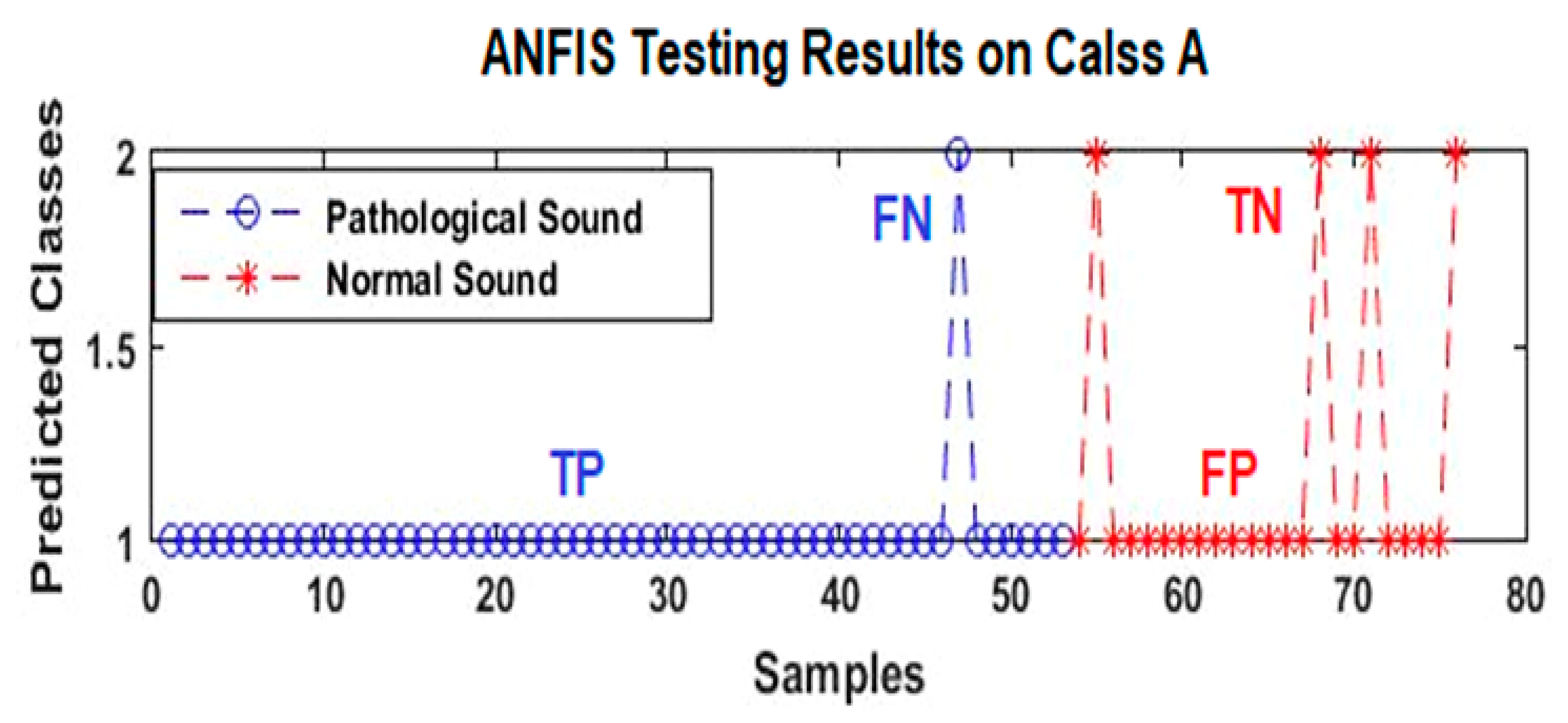
- -
- TP: true positive represents the HC-pathological samples detected correctly;
- -
- FP: false positive represents the NrHS-normal samples detected as abnormal;
- -
- TN: true negative represents the NrHS-normal samples detected correctly;
- -
- FN: false negative represents the HC samples detected as NrHS.
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gerbarg, D.S.; Taranta, A.; Spagnuolo, M.; Hofler, J.J. Computer analysis of phonocardiograms. Prog. Cardiovasc. Dis. 1963, 5, 393–405. [Google Scholar] [CrossRef]
- Dwivedi, A.K.; Imtiaz, S.A.; Rodriguez-Villegas, E. Algorithms for automatic analysis and classification of heart sounds—A systematic review. IEEE Access 2019, 7, 8316–8345. [Google Scholar] [CrossRef]
- Shub, C. Echocardiography or auscultation? How to evaluate systolic murmurs. Can. Fam. Physician 2003, 49, 163–167. [Google Scholar] [PubMed]
- Al-Naami, B.; Fraihat, H.; Gharaibeh, N.Y.; Al-Hinnawi, A.-R.M. A Framework classification of heart sound signals in physionet challenge 2016 using high order statistics and adaptive neuro-fuzzy inference system. IEEE Access 2020, 8, 224852–224859. [Google Scholar] [CrossRef]
- De Fazio, R.; De Vittorio, M.; Visconti, P. Innovative IoT solutions and wearable sensing systems for monitoring human biophysical parameters: A review. Electronics 2021, 10, 1660. [Google Scholar] [CrossRef]
- De Fazio, R.; Stabile, M.; De Vittorio, M.; Velázquez, R.; Visconti, P. An overview of wearable piezoresistive and inertial sensors for respiration rate monitoring. Electronics 2021, 10, 2178. [Google Scholar] [CrossRef]
- Gjoreski, M.; Gradisek, A.; Budna, B.; Gams, M.; Poglajen, G. Machine learning and end-to-end deep learning for the detection of chronic heart failure from heart sounds. IEEE Access 2020, 8, 20313–20324. [Google Scholar] [CrossRef]
- De Vos, J.P.; Blanckenberg, M.M. Automated pediatric cardiac auscultation. IEEE Trans. Biomed. Eng. 2007, 54, 244–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herzig, J.; Bickel, A.; Eitan, A.; Intrator, N. Monitoring cardiac stress using features extracted from s1 heart sounds. IEEE Trans. Biomed. Eng. 2015, 62, 1169–1178. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ren, Y.; Zhang, G.; Wang, R.; Cui, J.; Zhang, W. Detection and classification of abnormities of first heart sound using empirical wavelet transform. IEEE Access 2019, 7, 139643–139652. [Google Scholar] [CrossRef]
- Schmidt, S.; Graebe, M.; Toft, E.; Struijk, J. No evidence of nonlinear or chaotic behavior of cardiovascular murmurs. Biomed. Signal Process. Control 2011, 6, 157–163. [Google Scholar] [CrossRef]
- Grzegorczyk, I.; Solinski, M.; Lepek, M.; Perka, A.; Rosinski, J.; Rymko, J.; Stepien, K.; Gieraltowski, J. PCG Classification Using a Neural Network Approach. In Computing in Cardiology; IEEE Computer Society: Washington, DC, USA, 2016; Volume 43, pp. 1129–1132. [Google Scholar] [CrossRef]
- Chauhan, S.; Wang, P.; Lim, C.S.; Anantharaman, V. A computer-aided MFCC-based HMM system for automatic auscultation. Comput. Biol. Med. 2008, 38, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Saraçoğlu, R. Hidden Markov model-based classification of heart valve disease with PCA for dimension reduction. Eng. Appl. Artif. Intell. 2012, 25, 1523–1528. [Google Scholar] [CrossRef]
- Cheng, X.; Huang, J.; Li, Y.; Gui, G. Design and application of a laconic heart sound neural network. IEEE Access 2019, 7, 124417–124425. [Google Scholar] [CrossRef]
- Liu, C.; Springer, D.; Li, Q.; Moody, B.; Juan, R.A.; Chorro, F.J.; Castells, F.; Roig, J.M.; Silva, I.; Johnson, A.E.W.; et al. An open access database for the evaluation of heart sound algorithms. Physiol. Meas. 2016, 37, 2181–2213. [Google Scholar] [CrossRef] [PubMed]
- Beritelli, F.; Capizzi, G.; Sciuto, G.L.; Napoli, C.; Scaglione, F. Automatic heart activity diagnosis based on Gram polynomials and probabilistic neural networks. Biomed. Eng. Lett. 2017, 8, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Kay, E.; Agarwal, A. Drop Connected neural networks trained on time-frequency and inter-beat features for classifying heart sounds. Physiol. Meas. 2017, 38, 1645–1657. [Google Scholar] [CrossRef]
- Liu, C.; Springer, D.; Clifford, G.D. Performance of an open-source heart sound segmentation algorithm on eight independent databases. Physiol. Meas. 2017, 38, 1730–1745. [Google Scholar] [CrossRef] [PubMed]
- Homsi, M.N.; Warrick, P. Ensemble methods with outliers for phonocardiogram classification. Physiol. Meas. 2017, 38, 1631–1644. [Google Scholar] [CrossRef] [PubMed]
- Rubin, J.; Abreu, R.; Ganguli, A.; Nelaturi, S.; Matei, I.; Sricharan, K. Classifying heart sound recordings using deep convolutional neural networks and mel: Frequency cepstral coefficients. In Proceedings of the 2016 Computing in Cardiology Conference (CinC), Vancouver, BC, Canada, 11–14 September 2016; IEEE: Piscataway, NJ, USA, 2016. [Google Scholar] [CrossRef]
- Whitaker, B.M.; Suresha, P.B.; Liu, C.; Clifford, G.; Anderson, D.V. Combining sparse coding and time-domain features for heart sound classification. Physiol. Meas. 2017, 38, 1701–1713. [Google Scholar] [CrossRef]
- Plesinger, F.; Viscor, I.; Halamek, J.; Jurco, J.; Jurak, P. Heart sounds analysis using probability assessment. Physiol. Meas. 2017, 38, 1685–1700. [Google Scholar] [CrossRef] [PubMed]
- Vernekar, S.; Nair, S.; Vijayasenan, D.; Ranjan, R. A Novel approach for classification of normal/abnormal phonocardiogram recordings using temporal signal analysis and machine learning. In Proceedings of the 2016 Computing in Cardiology Conference (CinC), Vancouver, BC, Canada, 11–14 September 2016; IEEE: Piscataway, NJ, USA, 2016. [Google Scholar] [CrossRef]
- Maknickas, V.; Maknickas, A. Recognition of normal-abnormal phonocardiographic signals using deep convolutional neural networks and mel-frequency spectral coefficients. Physiol. Meas. 2017, 38, 1671–1684. [Google Scholar] [CrossRef] [PubMed]
- Tschannen, M.; Kramer, T.; Marti, G.; Heinzmann, M.; Wiatowski, T. Heart sound classification using deep structured features. In Proceedings of the 2016 Computing in Cardiology Conference (CinC), Vancouver, BC, Canada, 11–14 September 2016; IEEE: Piscataway, NJ, USA, 2016. [Google Scholar] [CrossRef]
- Nilanon, T.; Purushotham, S.; Liu, Y. Normal/abnormal heart sound recordings classification using convolutional neural network. In Proceedings of the 2016 Computing in Cardiology Conference (CinC), Vancouver, BC, Canada, 11–14 September 2016. [Google Scholar] [CrossRef]
- Potes, C.; Parvaneh, S.; Rahman, A.; Conroy, B. Ensemble of feature-based and deep learning-based classifiers for detection of abnormal heart sounds. In Proceedings of the 2016 Computing in Cardiology Conference (CinC), Vancouver, BC, Canada, 11–14 September 2016. [Google Scholar] [CrossRef]
- Bobillo, I.D. A Tensor approach to heart sound classification. In Proceedings of the 2016 Computing in Cardiology Conference (CinC), Vancouver, BC, Canada, 11–14 September 2016. [Google Scholar] [CrossRef]
- Zabihi, M.; Rad, A.B.; Kiranyaz, S.; Gabbouj, M.; Katsaggelos, A.K. Heart sound anomaly and quality detection using ensemble of neural networks without segmentation. In Proceedings of the 2016 Computing in Cardiology Conference (CinC), Vancouver, BC, Canada, 11–14 September 2016. [Google Scholar] [CrossRef]
- Xiao, B.; Xu, Y.; Bi, X.; Zhang, J.; Ma, X. Heart sounds classification using a novel 1-D convolutional neural network with extremely low parameter consumption. Neurocomputing 2020, 392, 153–159. [Google Scholar] [CrossRef]
- Li, F.; Tang, H.; Shang, S.; Mathiak, K.; Cong, F. Classification of heart sounds using convolutional neural network. Appl. Sci. 2020, 10, 3956. [Google Scholar] [CrossRef]
- Krishnan, P.T.; Balasubramanian, P.; Umapathy, S. Automated heart sound classification system from unsegmented phonocardiogram (PCG) using deep neural network. Phys. Eng. Sci. Med. 2020, 43, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Er, M.B. Heart sounds classification using convolutional neural network with 1D-local binary pattern and 1D-local ternary pattern features. Appl. Acoust. 2021, 180, 108152. [Google Scholar] [CrossRef]
- Deperlioglu, O. Heart sound classification with signal instant energy and stacked autoencoder network. Biomed. Signal Process. Control 2021, 64, 102211. [Google Scholar] [CrossRef]
- Deng, M.; Meng, T.; Cao, J.; Wang, S.; Zhang, J.; Fan, H. Heart sound classification based on improved MFCC features and convolutional recurrent neural networks. Neural Netw. 2020, 130, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Adiban, M.; BabaAli, B.; Shehnepoor, S. Statistical feature embedding for heart sound classification. J. Electr. Eng. 2019, 70, 259–272. [Google Scholar] [CrossRef] [Green Version]
- Han, W.; Xie, S.; Yang, Z.; Zhou, S.; Huang, H. Heart sound classification using the SNMFNet classifier. Physiol. Meas. 2019, 40, 105003. [Google Scholar] [CrossRef]
- Selesnick, I.; Baraniuk, R.G.; Kingsbury, N.G. The dual-tree complex wavelet transform. IEEE Signal Process. Mag. 2005, 22, 123–151. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Li, F.; Tang, S.; Xiong, W. A Review of Computer-aided heart sound detection techniques. BioMed Res. Int. 2020, 2020, 5846191. [Google Scholar] [CrossRef]
- Bianchi, G. Electronic Filter Simulation & Design, 1st ed.; McGraw-Hill Professional: New York, NY, USA, 2007. [Google Scholar]
- Al-Naami, B.; Owida, H.; Fraihat, H. Quantitative analysis signal-based approach using the dual tree complex wavelet transform for studying heart sound conditions. In Proceedings of the 2020 IEEE 5th Middle East and Africa Conference on Biomedical Engineering (MECBME), Amman, Jordan, 27–29 October 2020; pp. 1–4. [Google Scholar] [CrossRef]
- Vermaak, H.; Nsengiyumva, P.; Luwes, N. Using the dual-tree complex wavelet transform for improved fabric defect detection. J. Sens. 2016, 2016, 9794723. [Google Scholar] [CrossRef] [Green Version]
- Daubechies, I. Orthonormal bases of compactly supported wavelets. Commun. Pure Appl. Math. 1988, 41, 909–996. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Ji, Z. Application of the dual-tree complex wavelet transform in biomedical signal denoising. Bio-Med. Mater. Eng. 2014, 24, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, J.; Escalona, O.J.; Kodoth, V.; Manoharan, G. Efficacy of DWT denoising in the removal of power line interference and the effect on morphological distortion of underlying atrial fibrillatory waves in AF-ECG. In Proceedings of the World Congress on Medical Physics and Biomedical Engineering (IFMBE), Toronto, ON, Canada, 7–12 June 2015; Volume 51. [Google Scholar] [CrossRef]
- Van Drongelen, W. Signal averaging. In Signal Processing for Neuroscientists; Elsevier BV: Amsterdam, The Netherlands, 2007; pp. 55–70. [Google Scholar]
- Jang, J.-S.R. ANFIS: Adaptive-network-based fuzzy inference system. IEEE Trans. Syst. Man Cybern. 1993, 23, 665–685. [Google Scholar] [CrossRef]
- Goda, M.A.; Hajas, P. Morphological determination of pathological pcg signals by time and frequency domain analysis. In Computing in Cardiology; IEEE Computer Society: Washington, DC, USA, 2016; pp. 1133–1136. [Google Scholar] [CrossRef]
- Langley, P.; Murray, A. Abnormal Heart Sounds Detected from Short Duration Unsegmented Phonocardiograms by Wavelet Entropy. In Proceedings of the 2016 Computing in Cardiology Conference (CinC), Vancouver, BC, Canada, 11–14 September 2016; IEEE: Piscataway, NJ, USA, 2016. [Google Scholar] [CrossRef]
- Homsi, M.N.; Medina, N.; Hernandez, M.; Quintero, N.; Perpinan, G.; Quintana, A.; Warrick, P. Automatic Heart Sound Recording Classification using a Nested Set of Ensemble Algorithms. In Proceedings of the 2016 Computing in Cardiology Conference (CinC), Vancouver, BC, Canada, 11–14 September 2016; IEEE: Piscataway, NJ, USA, 2016. [Google Scholar] [CrossRef]
- Ghaffari, A.; Homaeinezhad, M.R.; Khazraee, M.; Daevaeiha, M.M. Segmentation of holter ECG waves via analysis of a discrete wavelet-derived multiple skewness-kurtosis based metric. Ann. Biomed. Eng. 2010, 38, 1497–1510. [Google Scholar] [CrossRef] [PubMed]
- Singh-Miller, N.; Singh-Miller, N. Using Spectral Acoustic Features to Identify Abnormal Heart Sounds. In Computing in Cardiology; IEEE Computer Society: Washington, DC, USA, 2016; pp. 557–560. [Google Scholar] [CrossRef]
- Mann, S.; Haykin, S. The chirplet transform: Physical considerations. IEEE Trans. Signal Process. 1995, 43, 2745–2761. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, S.K.; Ponnalagu, R.N.; Tripathy, R.K.; Acharya, U.R. Deep layer kernel sparse representation network for the detection of heart valve ailments from the time-frequency representation of PCG recordings. BioMed Res. Int. 2020, 2020, 8843963. [Google Scholar] [CrossRef]
- Shuvo, S.B.; Ali, S.N.; Swapnil, S.I.; Al-Rakhami, M.S.; Gumaei, A. CardioXNet: A novel lightweight deep learning framework for cardiovascular disease classification using heart sound recordings. IEEE Access 2021, 9, 36955–36967. [Google Scholar] [CrossRef]
- Popov, B.; Sierra, G.; Durand, L.-G.; Xu, J.; Pibarot, P.; Agarwal, R.; Lanzo, V. Automated extraction of aortic and pulmonary components of the second heart sound for the estimation of pulmonary artery pressure. In Proceedings of the 26th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, San Francisco, CA, USA, 1–5 September 2004; IEEE: Piscataway, NJ, USA, 2007; Volume 1, pp. 921–924. [Google Scholar]
- Djebbari, A.; Bereksi-Reguig, F.A. New Chirp-based wavelet for heart sounds time-frequency analysis. Int. J. Commun. Antenna Propag. 2011, 1, 92–102. [Google Scholar]

| Class | Sets | # Subjects | Abnormal HC | Normal HS |
|---|---|---|---|---|
| A | Training | 313 | 218 | 95 |
| Test | 78 | 54 | 24 | |
| B | Training | 356 | 72 | 284 |
| Test | 88 | 18 | 70 | |
| C + D | Training | 50 | 28 | 22 |
| Test | 12 | 7 | 5 | |
| E | Training | 1472 | 121 | 1351 |
| Test | 366 | 28 | 338 | |
| A, B, C, D, E | Training | 2191 | 439 | 1752 |
| Test | 544 | 107 | 437 | |
| Total HS Signals | 2735 | |||
| TYPE | SUGENO |
|---|---|
| FIS and Method | Prod |
| FIS or Method | Probor |
| FIS defuzzification Method | Wtaver is the weighted average performance of all rule outputs (i.e., WAM). |
| FIS implication Method | Prod |
| FIS aggregation Method | Sum |
| FIS inputs | 1 × 6 fisvar |
| FIS Outputs | 1 × 1 fisvar |
| FIS rules | 6 fis rule |
| Epoch number | 200 |
| Range of influence | 0.5 |
| FIS Creates a Sugeno FIS | Fis.Name = “sug41” |
| Dataset (before and after DTCWT) | Average SNR [dB] | SNR Standard Deviation [dB] | SNR Percentage Difference [dB] | Statistical Significance p-Value | |
|---|---|---|---|---|---|
| Class A | Before | 7.1 | 2.4 | +0.11 | <0.001 |
| After | 7.9 | 2.4 | |||
| Class B | Before | 0.7 | 0.6 | +13.96 | <0.001 |
| After | 10.8 | 3.5 | |||
| Class C + D | Before | 5.6 | 3.7 | +0.58 | <0.001 |
| After | 8.9 | 4.0 | |||
| Class E | Before | 5.6 | 3.0 | +0.65 | <0.001 |
| After | 9.3 | 4.8 | |||
| Data Set | Precision | Recall | F-Score | MAcc [%] |
|---|---|---|---|---|
| Class A | 0.73 | 0.98 | 0.84 | 85.5 |
| Class C + D | 0.86 | 0.87 | 0.86 | 86.0 |
| Class B | 0.55 | 0.89 | 0.68 | 72.0 |
| Class E | 0.56 | 0.51 | 0.53 | 53.5 |
| Average (A, B, C, D, E) | 0.68 | 0.81 | 0.74 | 74.5 |
| Author | Methodology | # Features | Neural Network Classifier | # HS Samples | Accuracy MAcc% |
|---|---|---|---|---|---|
| C. Potes et al. [28] | Time and frequency-domain features | 124 | CNN | 3240 | 86% |
| Whitaker, B. M. et al. [22] | Sparse coding | 20 | SVM | 2153 * | 86.5% |
| M. Tschannen et al. [26] | Wavelet deep convolutional neural network (CNN) | 25 | SVM | 1277 * | 81.2% |
| Goda et al. [49] | Wavelet envelope features | 128 | SVM | 3000 | 81.2% |
| Langley et al. [50] | Wavelet Entropy | Wavelet entropy | Classification algorithm | 400 * | 77% |
| Kay E. et al. [18] | Hidden semi-Markov model | 50 | Fully connected, two-hidden-layer neural network trained by error backpropagation | All datasets excluding Class E * | 74.8% |
| Grzegorczyk et al. [12] | Algorithm based on Hidden Markov Model. | 48 | Neural networks | 3000 | 79% |
| Nilanon et al. [27] | Spectrogram | Many time windows | SVM, CNN, and logistic regression (LR) | About 3000 | 68–80% |
| M. N. Homsi et al. [51] | Nested ensemble of algorithms | 131 | Random forest, LogitBoost, and a cost-sensitive classifier | 764 * | 84.48% |
| This work | DTCWT | 6 | ANFIS | 2735 | 74.5% |
| Dataset excluding Classes B, E | 86% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Naami, B.; Fraihat, H.; Al-Nabulsi, J.; Gharaibeh, N.Y.; Visconti, P.; Al-Hinnawi, A.-R. Assessment of Dual-Tree Complex Wavelet Transform to Improve SNR in Collaboration with Neuro-Fuzzy System for Heart-Sound Identification. Electronics 2022, 11, 938. https://doi.org/10.3390/electronics11060938
Al-Naami B, Fraihat H, Al-Nabulsi J, Gharaibeh NY, Visconti P, Al-Hinnawi A-R. Assessment of Dual-Tree Complex Wavelet Transform to Improve SNR in Collaboration with Neuro-Fuzzy System for Heart-Sound Identification. Electronics. 2022; 11(6):938. https://doi.org/10.3390/electronics11060938
Chicago/Turabian StyleAl-Naami, Bassam, Hossam Fraihat, Jamal Al-Nabulsi, Nasr Y. Gharaibeh, Paolo Visconti, and Abdel-Razzak Al-Hinnawi. 2022. "Assessment of Dual-Tree Complex Wavelet Transform to Improve SNR in Collaboration with Neuro-Fuzzy System for Heart-Sound Identification" Electronics 11, no. 6: 938. https://doi.org/10.3390/electronics11060938
APA StyleAl-Naami, B., Fraihat, H., Al-Nabulsi, J., Gharaibeh, N. Y., Visconti, P., & Al-Hinnawi, A.-R. (2022). Assessment of Dual-Tree Complex Wavelet Transform to Improve SNR in Collaboration with Neuro-Fuzzy System for Heart-Sound Identification. Electronics, 11(6), 938. https://doi.org/10.3390/electronics11060938








