Abstract
A WGAN-GP-based ECG signal expansion and an SE-ResNet1D-based ECG classification method are proposed to address the problem of poor modeling results due to the imbalanced sample distribution of ECG data sets. The network architectures of WGAN-GP and SE-ResNet1D are designed according to the characteristics of ECG signals so that they can be better applied to the generation and classification of ECG signals. First, ECG data were generated using WGAN-GP on the MIT-BIH arrhythmia database to balance the dataset. Then, the experiments were performed using the AAMI category and inter-patient data partitioning principles, and classification experiments were performed using SE-ResNet1D on the imbalanced and balanced datasets, respectively, and compared with three networks, VGGNet, DenseNet and CNN+Bi-LSTM. The experimental results show that using WGAN-GP to balance the dataset can improve the accuracy and robustness of the model classification, and the proposed SE-ResNet1D outperforms the comparison model, with a precision of 95.80%, recall of 96.75% and an F1 measure of 96.27% on the balanced dataset. Our methods have the potential to be a useful diagnostic tool to assist cardiologists in the diagnosis of arrhythmias.
1. Introduction
Cardiovascular disease is one of the leading causes of death in China, with approximately 330 million people now suffering from cardiovascular disease nationwide [1]. Electrocardiograms (ECGs) are routinely used in clinical practice to identify cardiac abnormalities. ECG is a bio-signal acquisition technique that uses an ECG machine to capture the electrical signals generated by the electrical activity cycle of the heart from the body’s surface. ECG is generated by different electrical activities of the heart through depolarisation and repolarisation. It is a vital tool for diagnosing early features of coronary heart disease, myocardial ischaemia and other non-cardiovascular diseases. However, ECG analysis and diagnosis are usually performed by specialist physicians, which consumes a lot of time and effort and squeezes out medical resources. Automated ECG diagnosis is now becoming commonplace, not only eliminating the possibility of human error, but also allowing algorithms to be embedded in on-the-go devices such as mobile phones to readily diagnose ECGs collected through wearable ECG monitoring devices.
Researchers have tried to develop automatic ECG signal classification methods, which require minimal human involvement, to detect cardiac abnormalities. Among these methods, Deep Neural Network (DNN) models have underpinned state-of-the-art performances [2,3], especially on small-scale ECG datasets such as the MIT-BIH arrhythmia database [4]. However, most existing methods of ECG classification suffer from the following problems:
- The training of the model relies on a large number of accurately annotated datasets, while ECG, as a sensitive medical dataset involving patient privacy, is difficult to obtain in large quantities, and the datasets need to be annotated and proofread by professional doctors, which will consume a lot of medical resources [5,6].
- ECGs have a predominantly normal heart rate and a low abnormal heart rate, so the sample size between each category of the ECG dataset is often highly imbalanced. The imbalance in the number of samples of ECG signals from different categories affects the performance of the classifier. Models trained on imbalanced datasets usually do not perform well, especially in the determination of abnormal categories [7,8].
- The experiments in most papers randomly disrupted the heartbeats of all patients and then divided the training and test sets, which resulted in high test accuracy but poor practical results. This is because ECG signals vary significantly from patient to patient, and therefore, the generalization performance of the model is reduced in this training approach [9].
In recent years, some researchers have taken note of these issues and have initiated some studies to generate ECGs [10,11,12,13,14] and use deep learning models to classify ECGs [15,16,17,18,19,20], but have not achieved good performance in multi-classification, especially in inter-patient. Currently, generation and classification tasks are the most prominent applications of supervised learning. WGAN-GP is widely used for generating time-series data [21,22]. SENet and ResNet have also achieved good performance in the area of image classification, but to our knowledge, it is rarely used in its one-dimensional form, especially for the classification of physiological signals. In this paper, we propose to use the improved WGAN-GP to generate ECG signals to balance the dataset and then classify the dataset before and after balancing using SE-ResNet1D, respectively, to compare the impact of the dataset before and after balancing on the classification performance of the model. Specifically, a more realistic inter-patient method is used for the classification of the dataset, resulting in a stronger generalization performance of the model. The contributions of our paper are as follows:
- We improve the structure of WGAN-GP. A Bi-GRU layer is embedded into the generator of the neurogenerative model to endorse the temporal constraint for better ECG synthesis.
- We redesigned the structure of SE-ResNet to a one-dimensional model to classify ECG signals.
- Our experimental dataset was divided using AAMI and inter-patient, which is more in line with the medical reality. Our experiments demonstrate that the above improvements can significantly improve the performance of arrhythmia classification.
The remainder of the paper is structured as follows. We present related work in Section 2 and background in Section 3. The proposed method is explained in detail in Section 4. In Section 5, the processing and division of the data set are described in detail. Then, the experimental results and analysis are presented in Section 6. Finally, Section 7 presents the conclusions of the paper and discusses the limitations of our method and future work.
2. Related Work
2.1. Deep Learning Network-Based ECG Classification
In recent years, DNN-based methods have achieved excellent performance in classifying small-scale ECG datasets. For instances, Maweu et al. [10] proposed a modular framework of CEFEs using Convolutional Neural Networks (CNN) to interpret ECG signals for interpretability. To classify the ECG signals without feature extraction, Chen et al. [11] developed a multi-channel multi-scale DNN model, which is an end-to-end structure. In order to enhance the performance of the DNN model, long short-term memory (LSTM) and attention were added along with convolutional layers, which were utilized to extract key features. An automatic MI detection system was proposed by Rai et al. [12]. CNN, hybrid CNN-LSTM, and ensemble techniques were used to analyze ECG signals in order to select the best performing model. To create the model, they employed 123,998 ECG beats from the PTB diagnostic database and MIT-BIH arrhythmia database. Based on the region of cardiac involvement, Jahmunah et al. [13] developed DenseNet and CNN models for the classification of healthy participants and patients with ten types of MI. The Physikalisch-Technische Bundesanstalt database was used to pre-process ECG signals, and an R peak detection technique was used to extract the ECG beats. Attallah et al. [14] suggested a new pipeline dubbed ECG-BiCoNet to explore the possibility of using ECG data for diagnosing COVID-19. Five deep learning models with various structural designs are used by ECG-BiCoNet. Each deep learning technique’s two separate layers are used to extract two levels of features for ECG-BiCoNet.
2.2. GAN Based ECG Signal Synthesize
The signal synthesis task is to learn the real data distribution from the dataset. One such deep learning method is Generative Adversarial Networks (GAN), which was initially proposed to generate realistic synthetic images [23]. Recently, some researchers have used it to synthesize ECG signals to expand the dataset for the purpose of improving model classification. For instance, Li et al. [15] conducted an automated myocardial infarction detection model SLC-GAN, which used generative adversarial networks (GAN) to create single-lead ECG data with high morphological similarity. CNNs containing real ECG data and artificial ECGs from the GAN were used to automatically diagnose MI. In order to produce multi-view ECG signals, Chen et al. [16] proposed a unique disease-aware generative adversarial network dubbed ME-GAN. ME-GAN acquires panoptic electrocardio representations tailored to heart disorders and projects the representations onto several standard perspectives. To address the data insufficiency issue, Rafi et al. [17] created a novel generative adversarial network-based deep learning technique dubbed HeartNet. A multi-head attention mechanism atop the CNN architecture compresses the deep learning technique that is being offered. By using GAN and creating more training samples, adversarial data synthesis was able to overcome the primary problem of insufficient data labels. In order to increase the amount of data in the arrhythmia dataset and address the issue of imbalanced arrhythmia data, Ma et al. [18] introduced ECG Deep Convolution Generative Adversarial Networks (ECG-DCGAN). Additionally, the ECG signals were automatically classified using the CNN model without the usage of artificial feature extraction. Ma et al. [19] converted the time series into Gramian angular summation field (GASF) images to aid the classifier in obtaining the rich information included in the ECG signals. They used the conditional Wasserstein generative adversarial network with gradient penalty CWGAN-GP model to amplify the minor categories in order to solve the imbalanced data problem. Wu et al. [20] used the Short Term Fourier Transform (STFT) based SpectroGAN and the Smooth Wavelet Transform (SWT) based WaveletGAN models to generate three ECG signals, left bundle branch block, right bundle branch block and normal heartbeat, and a new evaluation method to assess the reliability of the generated ECG signals, but the short length of the training samples involved in the experiments was not conducive to the generation of valid data for long segments.
3. Background
3.1. WGAN-GP
Generative Adversarial Networks (GAN) [24] was first proposed by Ian Goodfellow in 2014, and GAN was initially applied to generate realistic, non-existent images in the training set to learn the distribution of data in the training set by adversarial training.
The structure of the original GAN is not suitable for discrete data and one-dimensional time-series data. However, ECG signals are rich in diversity and are also one-dimensional time-series data, so the original GAN is not suitable for the task of generating ECGs. The WGAN proposed by Arjovsky et al. [25] improves the original GAN by replacing the JS scatter used in the original GAN with the Wasserstein distance. Gulrajani et al. [26] proposed WGAN-GP and pointed out the existence of weight clipping in WGAN leading to the problem of parameter concentration and proposed to solve it by applying gradient penalty to each sample individually. The discriminator in WGAN-GP can learn reasonable parameter values and thus solve the challenge of slow convergence of the network.
3.2. SE-ResNet
Deep residual network (ResNet) [27] was proposed by Kai-Ming He from Microsoft Asia Research Institute and won the ImageNet Large Scale Visual Recognition Challenge (ILSVRC) [28] in 2015. Squeeze Excitation Neural Networks (SENet) [29], the champion model of ILSVRC2017, proposes a structure of squeeze and excitation blocks that allows the network to perform channel-based recalibration of features.
SE-ResNet is a network model that introduces a squeeze block and incentive block structure on top of the ResNet model. It has been applied to image classification with good performance. Zhao et al. [30] applied its two-dimensional form to the classification of 12-lead ECGs, but this model is only suitable for large data sets.
4. Proposed Method
4.1. WGAN-GP Based ECG Synthesised Model
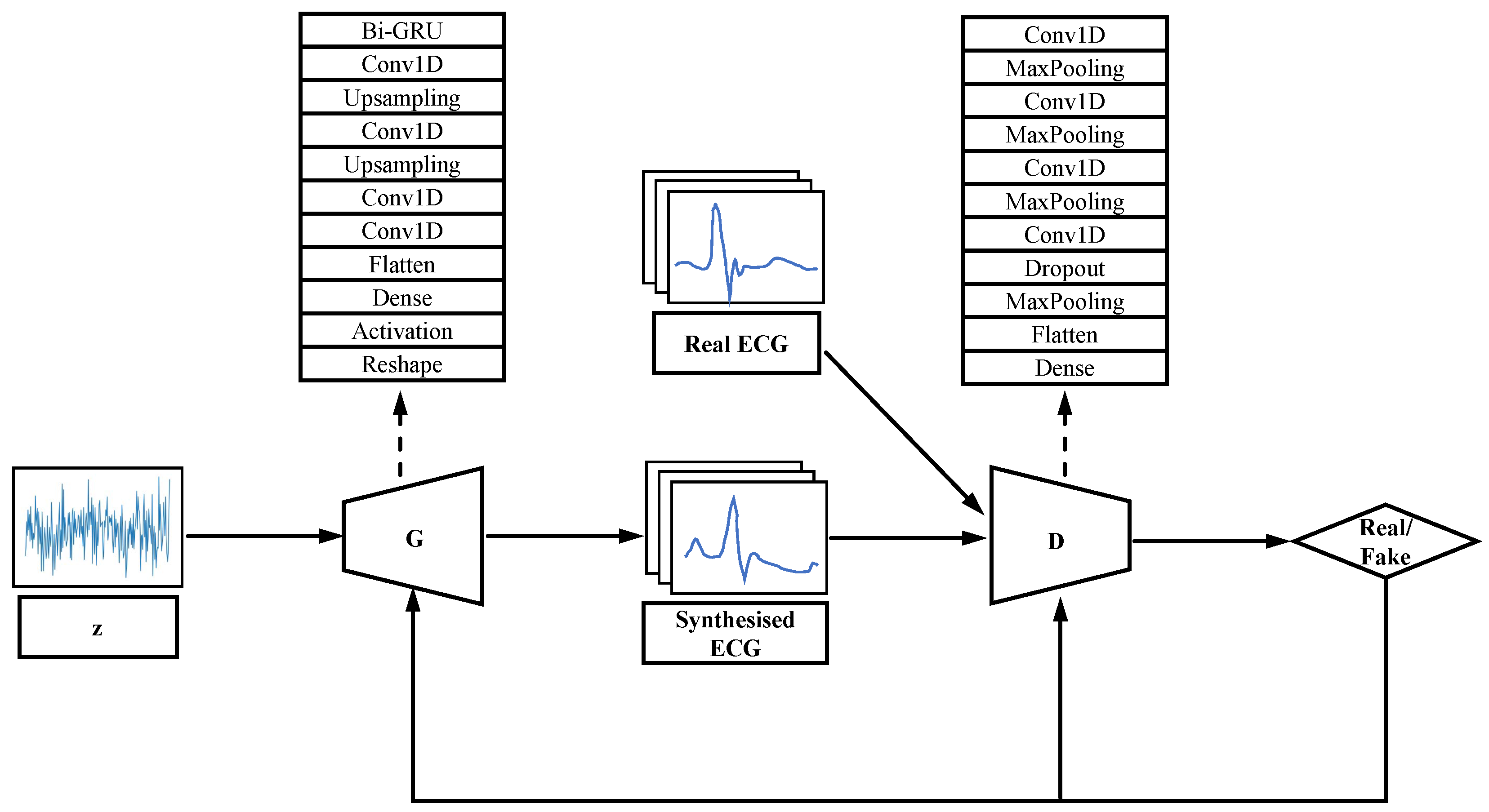
We redesigned the generator and discriminator of GAN according to the characteristics of one-dimensional continuity of ECG signals, and the architecture of WGAN-GP is shown in Figure 1. A one-dimensional convolutional layer with feature extraction function [23] and a Bi-directional Gate Recurrent Unit (Bi-GRU) was added to the generator to extract the features of the signal and learn the distribution of the real data. The discriminator consists of several one-dimensional convolutional layers, a one-dimensional pooling layer and a fully connected layer.
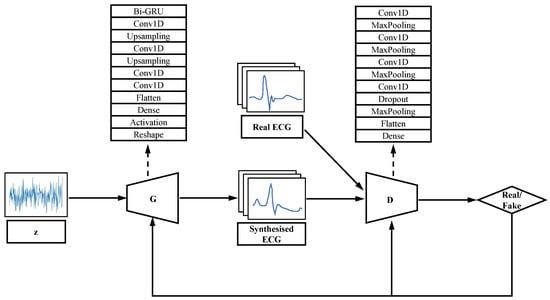
Figure 1.
Architecture of WGAN-GP.
Bi-GRU is an improved RNN that is more suitable for acquiring features of the ECG [31]. The Gate Recurrent Unit (GRU) consists of a memory unit, an input gate, an output gate and a forgetting gate. Each memory unit is cyclically connected to each other and the non-linear gating unit controls the input and output of data from each memory unit. However, the unidirectional GRU only processes the forward information and ignores the backward information, which is equally important in the ECG signal and affects the final output. The Bi-GRU model is a combination of a forward GRU and a backward GRU, with the output taking into account both the forward and backward input information, and in the task of this model, both the forward and backward band features can be taken into account. Where the reset gate is used to weaken features of low relevance in the previous unit and the update gate is used to determine the features that need to be passed from the previous unit to the next. The Bi-GRU model is implemented as follows.
where x denotes the input at the time t, denotes the activation function, and denotes the output of the reset gate and update gate, and denotes the candidate output and the actual output, respectively, and ⊙ denotes multiplication one by one.
The ultimate goal of training a GAN is to find the Nash equilibrium point of a zero-sum game. Both generators and discriminators want to minimize their cost functions, and it is often difficult to reach the Nash equilibrium point in the process of adversarial training. In addition, the adversarial training of GAN penalizes poorly generated samples and generators often choose to generate samples with known small penalties in order to avoid penalties, resulting in generating too homogeneous a variety of samples and eventually falling into pattern collapse [32]. In this paper, we add Batch Normalization between layers to prevent pattern collapse and improve training stability.
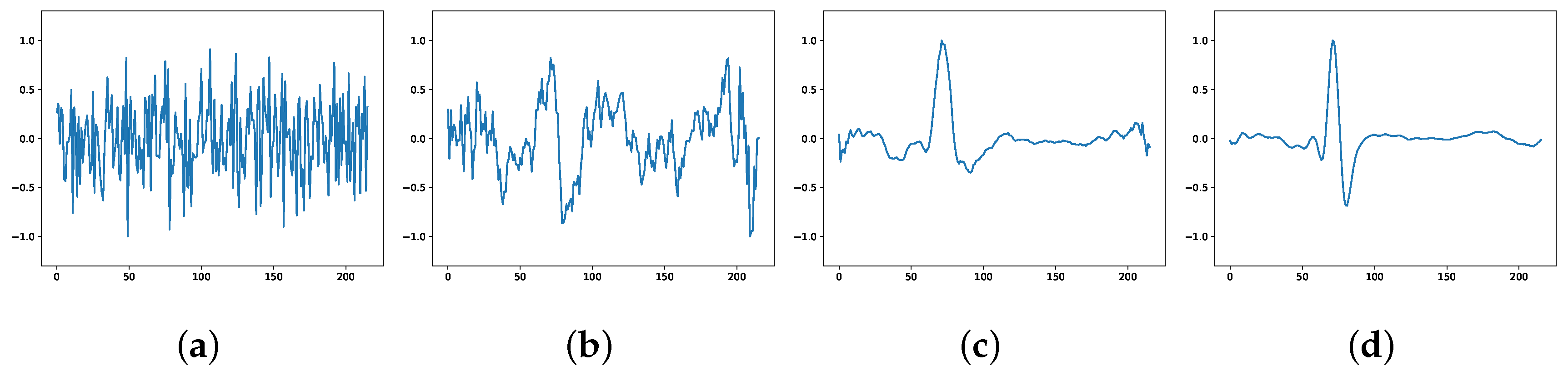
The ECG signals generated by the model at different stages are plotted as pictures, as shown in Figure 2. Figure 2a shows the high-dimensional random noise input at the beginning of training, and the output is also completely noisy at this time; Figure 2b shows the output at 100 rounds of training, where the output can be seen to have some of the morphological features of ECG, but the generated samples are still more noise biased; Figure 2c shows the output at 300 rounds of training, where the generated has the morphology of ECG, but it is not smooth enough; Figure 2d shows the output at 800 rounds of training when the generated ECG can successfully fool the discriminator and make it lose its judgment, and the adversarial training enters the Nash equilibrium state and the network converges.
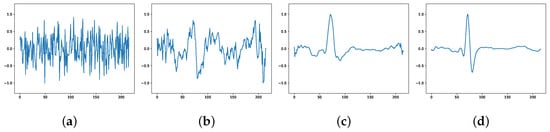
Figure 2.
ECG generated at different stages: (a) beginning of the training; (b) 100 rounds; (c) 300 rounds; (d) 800 rounds.
4.2. SE-ResNet1D Based ECG Classification Model
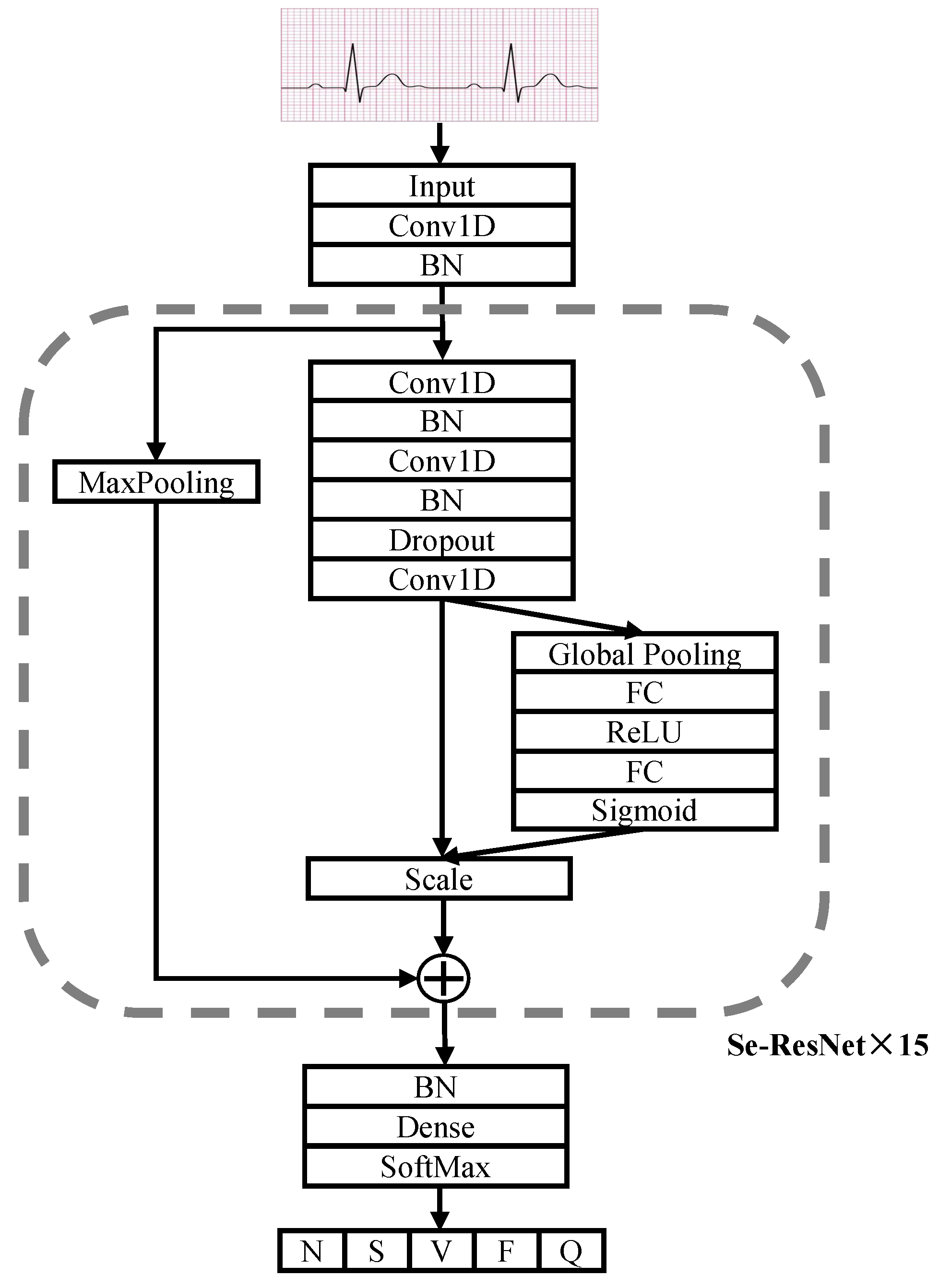
In this paper, we improve on the SE-ResNet model by replacing the 2D convolutional and 2D pooling layers originally used for image feature extraction with the 1D convolutional and 1D pooling layers used for temporal signal feature extraction to build the SE-ResNet1D ECG classification model, and redesign the overall network architecture and training parameters so that it can be better adapted to the task of arrhythmia classification.
The ECG SE-ResNet1D architecture is shown in Figure 3. The network has 17 modules, including a convolutional layer input module, a SoftMax output module and 15 blocks. Each connection block contains three convolutional layers for feature extraction and an SE module for modeling the spatio-temporal relationships between ECG channels. In the SE module, it can be seen that it first extracts features through the Global Pooling layer, reduces the feature dimensionality and then ramps back to the original dimensionality through two Fully Connected layers (FC). The two FC layers have more nonlinearity and can better fit the correlation within the ECG signal features, and the ECG features are enhanced by channel dynamic feature recalibration so that features that are clearly abnormal receive more weight. Finally, the weights are normalized by the Sigmoid activation function and then the features of each channel are weighted by the scale operation. The first convolutional layer and the initial cell of the SE-ResNet connection block have 64 convolutional filters, after which the number of filters is increased by a factor of two for every other connection block. A Dropout layer with a probability value of 0.5 is used between each convolutional layer, and L2 regularization is added to the weights of each layer to prevent overfitting of the model. Finally, the probability values of the five categories are output via the fully connected layers and the SoftMax activation function.
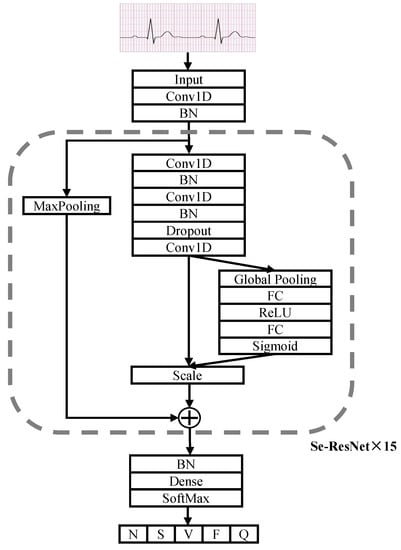
Figure 3.
Architecture of SE-ResNet1D.
The model was trained using random initialization weights for a total of 50 rounds. The optimization was performed using the Adam optimizer, with the initial learning rate set to 0.003 and the batch size set to 64.
5. Datasets and Data Pre-Processing
5.1. Introduction to the Data Set
The experimental dataset we used was the MIT-BIH arrhythmia database [4], open-sourced by MIT, which has been widely used by the academic community since it was made public in 1978 and has become the most widely used small ECG dataset in academic research today. The MIT-BIH Arrhythmia Database contains 48 half-hour excerpts of two-channel ambulatory ECG recordings, obtained from 47 subjects studied by the BIH Arrhythmia Laboratory. Twenty-three recordings were chosen at random from a set of 4000 24-h ambulatory ECG recordings collected from a mixed population of inpatients (about 60%) and outpatients (about 40%) at Boston’s Beth Israel Hospital; the remaining 25 recordings were selected from the same set to include less common but clinically significant arrhythmias that would not be well-represented in a small random sample. All ECGs are automatically annotated by the machine with the R-peak positions and calibrated by two cardiologists.
5.2. Data Pre-Processing
5.2.1. Heartbeat Segmentation
The annotation file of the MIT-BIH arrhythmia database indicates the R-peak location of each beat, but not all data sets are annotated with the R-peak location. In order to demonstrate the universality of the method, our experiment uses the QRS wave detection method proposed by Lourenco et al. [33] to find the R-peak, and after obtaining the R-peak location, the beats are intercepted by taking a section of sampling points forward and backward. The length of each intercepted heartbeat is 216 sampling points.
5.2.2. Wavelet Threshold Noise Reduction
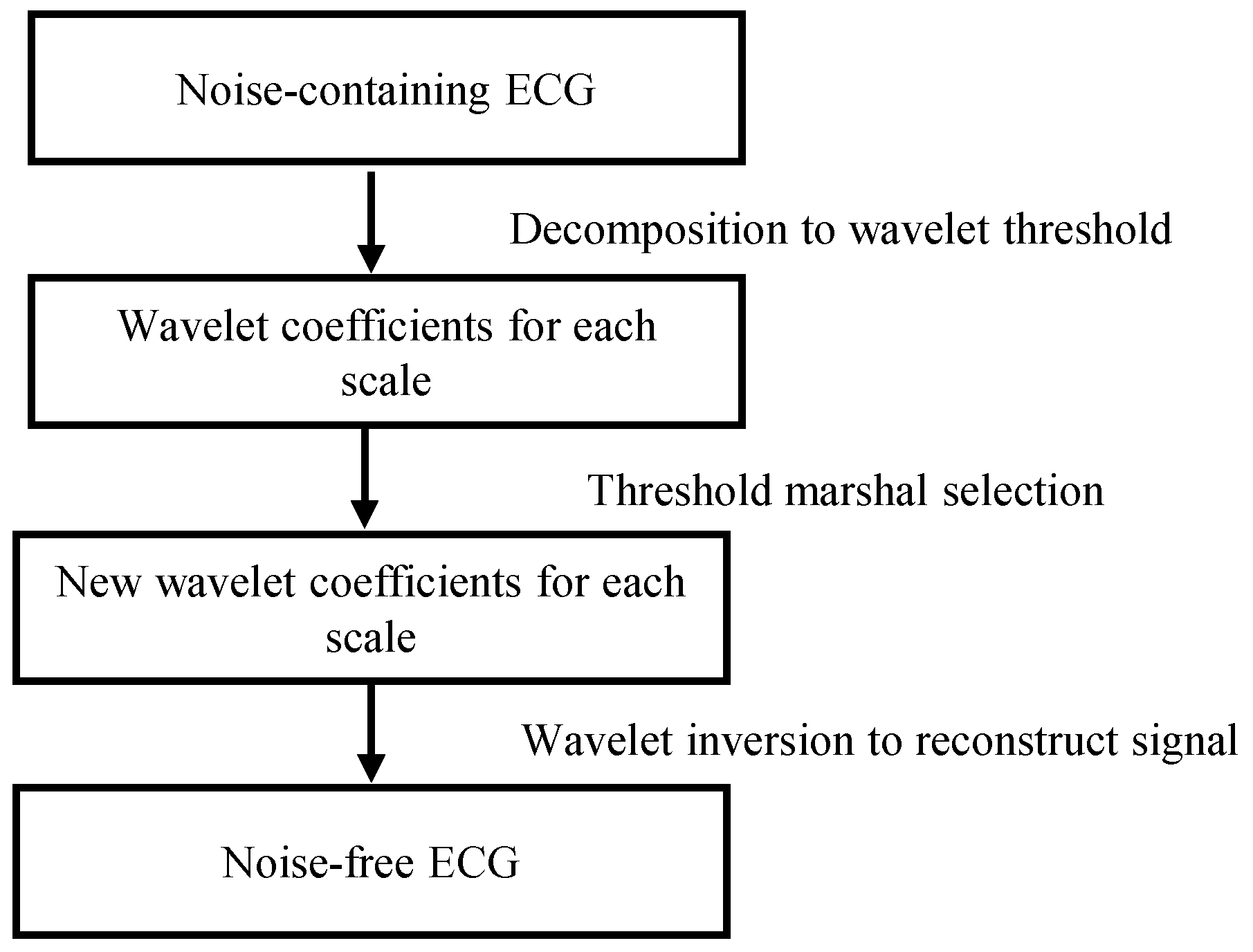
ECG signals are often disturbed by interference from EMG noise during acquisition, and the interference of these noises can make it difficult for the network to converge, so we use wavelet threshold denoising [34] to remove the noise, and the flow of the wavelet threshold denoising method is shown in Figure 4.
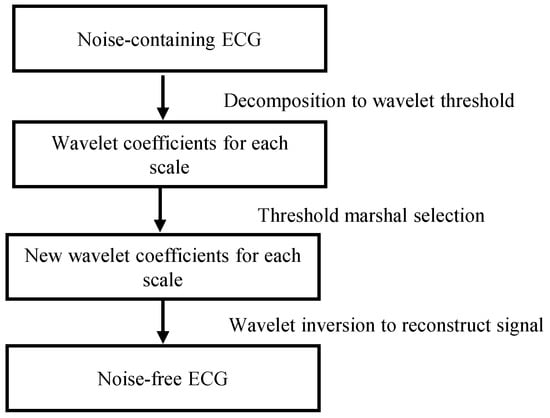
Figure 4.
Wavelet threshold denoising method.
The wavelet threshold function is shown in Equation (5) and Equation (6), where N denotes the length of the high-frequency coefficients for each scale of the wavelet and the scale is 9, respectively, denotes the threshold value, is a constant with a value of 0.674, w denotes the high-frequency wavelet coefficients.
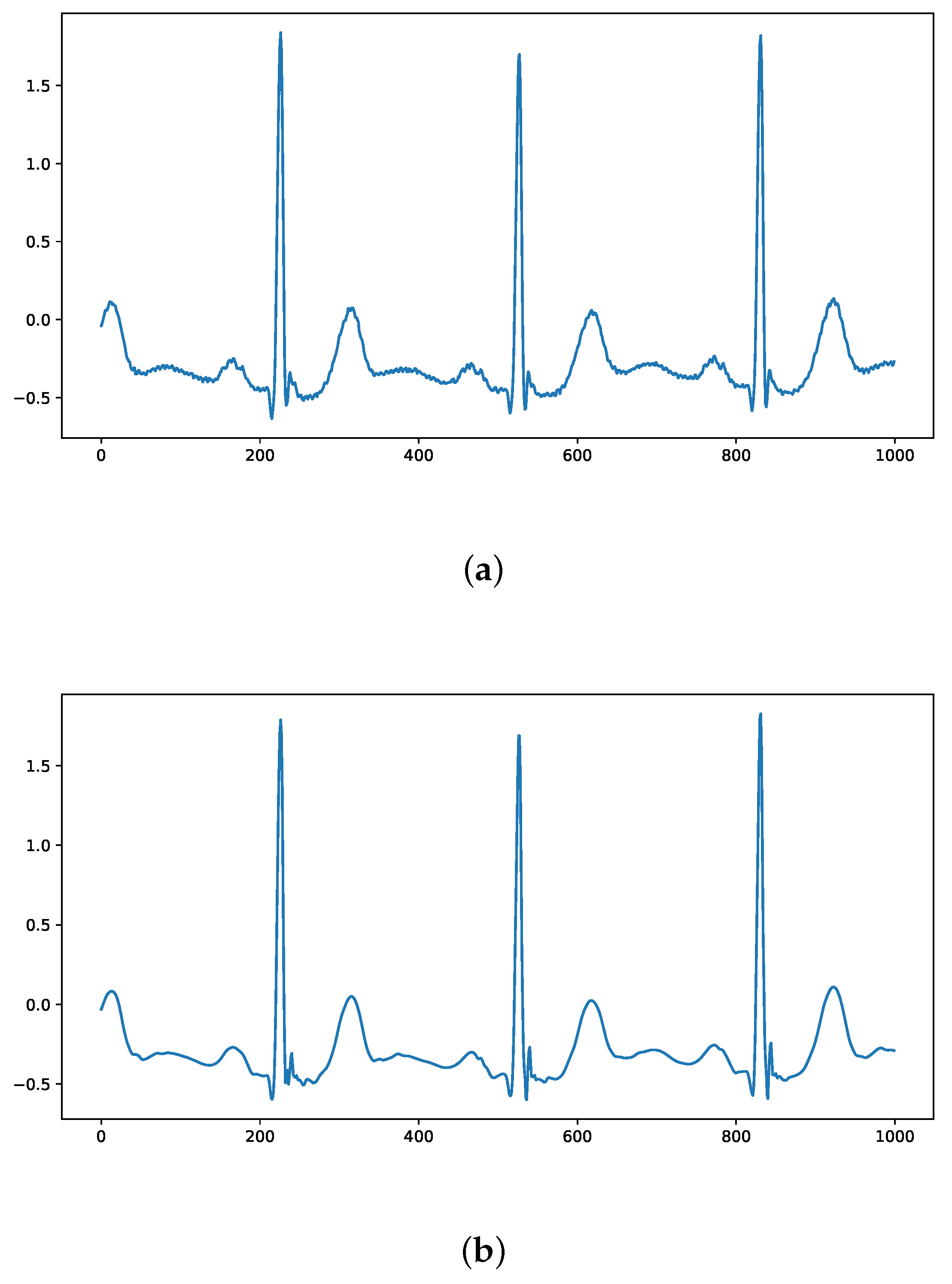
A comparison of the ECG signals in the MIT-BIH arrhythmia database before and after denoising using wavelet thresholding is shown in Figure 5, where it can be seen that the denoised ECG signal removes the interference of inotropic noise while maximizing the preservation of ECG characteristics.
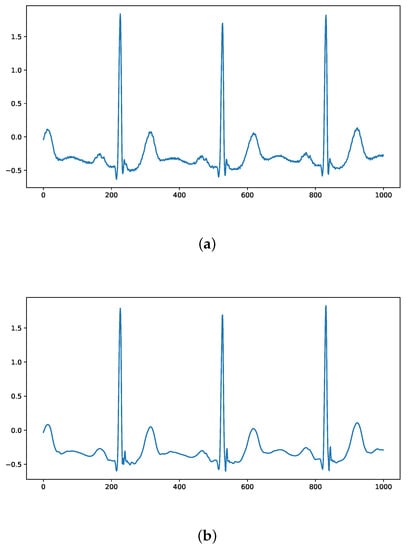
Figure 5.
Comparison of ECG signals before and after wavelet threshold denoising: (a) Original ECG signal; (b) Denoised signal.
5.2.3. Data Set Partitioning
There are 15 different heartbeat annotations in the MIT-BIH arrhythmia database, which are widely used in academic research to classify them into five categories using the American Association for the Advancement of Medical Instrumentation (AAMI) classification criteria [35], as shown in Table 1, which Both the original number of beats and the number after generating a beat-balanced dataset using WGAN-GP are shown in Table 1.

Table 1.
Category and number of the dataset.
Medical data has an inter-patient principle and an intra-patient principle when dividing the training and test sets [36]. The inter-patient principle separates the training and test sets by the patient. The intra-patient principle directly assigns patients randomly by breaking up their records. Most of the experiments in papers studying ECG use the intra-patient approach, which has high classification accuracy but it does so by training some of the heartbeats of the same patient and then testing the rest of the heartbeats of the same patient. This is not a rigorous approach, resulting in an inflated experimental result but poor generalization performance of the model, and this classification is not realistic. Our experiments follow the inter-patient principle and separate the test set and training set by the patient, i.e., the heartbeats of patients appearing in the training set will not appear in the test set, which is more realistic. The division of the training and test sets is shown in Table 2.

Table 2.
Train set and test set division under the inter-patient.
6. Experiments
6.1. Experimental Settings
The experiments were deployed on a tower workstation with a Core i9-10920X processor (3.5 GHz), Quadro RTX6000 graphics card (24 GB of video memory), running on 64 G RAM, and a deep learning environment using Window Sever 2019 + Python 3.8 + Keras 2.2. Classification experiments were performed using SE-ResNet1D was used to do classification experiments on imbalanced and balanced datasets, respectively, and compared with three networks, DenseNet [37], VGGNet [38] and CNN+Bi-LSTM [39]. For each experiment, 80% of the samples were randomly selected for training and 20% for testing. A 10-fold cross-validation was used during the training process, dividing all the samples into 10 copies, with 9 of them randomly selected as the training set and the remaining 1 as the validation set, thus improving the confidence of the experimental results.
For the model evaluation, Precision (P), Recall (R) and F1 Measure (F1) are used as metrics to compare our method with benchmark models.
6.2. Experimental Results and Analysis
In our work, we conducted two groups of comparison experiments, and the experimental results are shown on the left of Table 3. First, the classification performance was tested using SE-ResNet on the imbalanced MIT-BIH arrhythmia database and compared with three deep learning algorithms, DenseNet, VGGNet and CNN+Bi-LSTM. As can be seen from Table 3, with an imbalanced number of categories in the dataset, SE-Resnet1D achieved a precision rate of 93.16%, a recall rate of 93.31% and an F1 value of 93.23%, with the classification performance outperforming the other three methods in different evaluation metrics.

Table 3.
Comparison of experimental results.
Then, based on the above experimental results, we generate ECG signals to balance the MIT-BIH arrhythmia database by WGAN-GP and then conduct further experiments using the same four algorithms. As can be seen from Table 3, after data balancing with WAGN-GP, the classification performance of all four algorithms was significantly improved, especially the SE-ResNet1D effect was significantly improved, achieving a precision rate of 95.8%, a recall rate of 96.7% and an F1 value of 96.2% on the balanced dataset. Compared to the imbalanced results, the precision rate improved by 2.64%, the recall rate by 3.44% and the F1 value by 3.04%. The experimental results show that for imbalanced medical data, eliminating data imbalance by generating data with WGAN-GP can effectively improve the classification results.
We further compared the results of the proposed method with other inter-patient studies in recent years. As shown in Table 4, the proposed method obtained the best performance compared to previous studies of five Classifications [40,41,42]. Liu et al. proposed ECVT-Net [43] has better performance, but can only classify normal and abnormal classes.

Table 4.
Performance comparison with previous studies.
7. Discussion and Conclusions
Deep learning has achieved good performance in ECG automatic classification and diagnosis, yet there are still problems such as high annotation cost, imbalanced datasets and unreasonable dataset partitioning during training. In order to solve the above problems and improve the performance of the model for arrhythmia classification on imbalanced datasets, we propose an algorithmic model for balancing datasets by generating ECG signals via WGAN-GP, which adds a Bi-GRU layer to the generator based on the characteristics of one-dimensional continuity of ECG signals to compensate for the shortcomings of previous GAN-based algorithms for one-dimensional temporal signal generation. The experiments were validated on the MIT-BIH arrhythmia database, and we used SE-ResNet1D on a pre- and post-balanced dataset, respectively, and compared it with three other deep learning-based classification algorithms. The experimental results show that the WGAN-GP+SE-ResNet1D approach can achieve good results on datasets with a small sample size and imbalanced distribution.
Currently, there are two technical limitations to our model. First, our model cannot be successfully migrated from multiple training datasets to a completely different test dataset and the model trained using data from one lead does not work well for other leads either. To the best of our knowledge, very few works focus on cross-lead model issues. Therefore, the future work is a combination of migration learning and domain generation-based methods that can be used to improve the portability of the model, improve the accuracy of the model on ECG signals with different devices, different leads and different sampling rates, and make the model more robust. In addition, it is hard to explain the results of the presented model in the sense that we cannot determine the specific ECG morphology that results in the classification. Future work will involve expanding our model to incorporate improved explainability through prototype learning and attention.
Author Contributions
J.Q.: Project leader, the designer of the algorithm; F.G.: the implementation of code for experiments and the writing and revision of papers; Z.W.: the revision of theses; L.L.: ECG data evaluation; and C.J.: review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Youth Fund Project of the National Nature Fund of China under Grant 62002038.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Nie, Z.; Xu, H.; Chen, C.; Gan, Y.; Chen, G.; Wang, C.; Yue, W.; Yan, F.; Feng, Y.; Lu, Z. Population Attributable Risks of Potential Modifiable Factors for Atrial Fibrillation in China: A National Survey. Risk Manag. Healthc. Policy 2022, 15, 1215. [Google Scholar] [CrossRef] [PubMed]
- Mousavi, S.; Afghah, F. Inter-and intra-patient ecg heartbeat classification for arrhythmia detection: A sequence to sequence deep learning approach. In Proceedings of the ICASSP 2019—2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1308–1312. [Google Scholar]
- Salem, M.; Taheri, S.; Yuan, J.S. ECG arrhythmia classification using transfer learning from 2-dimensional deep CNN features. In Proceedings of the 2018 IEEE Biomedical Circuits and Systems Conference (BioCAS), Cleveland, OH, USA, 17–19 October 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 1–4. [Google Scholar]
- Moody, G.B.; Mark, R.G. The impact of the MIT-BIH arrhythmia database. IEEE Eng. Med. Biol. Mag. 2001, 20, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Strodthoff, N.; Wagner, P.; Schaeffter, T.; Samek, W. Deep learning for ECG analysis: Benchmarks and insights from PTB-XL. IEEE J. Biomed. Health Inform. 2020, 25, 1519–1528. [Google Scholar] [CrossRef]
- Petmezas, G.; Haris, K.; Stefanopoulos, L.; Kilintzis, V.; Tzavelis, A.; Rogers, J.A.; Katsaggelos, A.K.; Maglaveras, N. Automated atrial fibrillation detection using a hybrid CNN-LSTM network on imbalanced ECG datasets. Biomed. Signal Process. Control 2021, 63, 102194. [Google Scholar] [CrossRef]
- Murat, F.; Yildirim, O.; Talo, M.; Baloglu, U.B.; Demir, Y.; Acharya, U.R. Application of deep learning techniques for heartbeats detection using ECG signals-analysis and review. Comput. Biol. Med. 2020, 120, 103726. [Google Scholar] [CrossRef] [PubMed]
- Mar, T.; Zaunseder, S.; Martínez, J.P.; Llamedo, M.; Poll, R. Optimization of ECG classification by means of feature selection. IEEE Trans. Biomed. Eng. 2011, 58, 2168–2177. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Boulanger, P. A survey of heart anomaly detection using ambulatory Electrocardiogram (ECG). Sensors 2020, 20, 1461. [Google Scholar] [CrossRef]
- Maweu, B.M.; Dakshit, S.; Shamsuddin, R.; Prabhakaran, B. CEFEs: A CNN explainable framework for ECG signals. Artif. Intell. Med. 2021, 115, 102059. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Lin, Y.T.; Lee, S.J.; Tsai, W.C.; Huang, T.C.; Liu, Y.H.; Cheng, M.C.; Dai, C.Y. Automated ECG classification based on 1D deep learning network. Methods 2022, 202, 127–135. [Google Scholar] [CrossRef]
- Rai, H.M.; Chatterjee, K. Hybrid CNN-LSTM deep learning model and ensemble technique for automatic detection of myocardial infarction using big ECG data. Appl. Intell. 2022, 52, 5366–5384. [Google Scholar] [CrossRef]
- Jahmunah, V.; Ng, E.; Tan, R.S.; Oh, S.L.; Acharya, U.R. Explainable detection of myocardial infarction using deep learning models with Grad-CAM technique on ECG signals. Comput. Biol. Med. 2022, 146, 105550. [Google Scholar] [CrossRef]
- Attallah, O. ECG-BiCoNet: An ECG-based pipeline for COVID-19 diagnosis using Bi-Layers of deep features integration. Comput. Biol. Med. 2022, 142, 105210. [Google Scholar] [CrossRef]
- Li, W.; Tang, Y.M.; Yu, K.M.; To, S. SLC-GAN: An automated myocardial infarction detection model based on generative adversarial networks and convolutional neural networks with single-lead electrocardiogram synthesis. Inf. Sci. 2022, 589, 738–750. [Google Scholar] [CrossRef]
- Chen, J.; Liao, K.; Wei, K.; Ying, H.; Chen, D.Z.; Wu, J. ME-GAN: Learning panoptic electrocardio representations for multi-view ECG synthesis conditioned on heart diseases. In Proceedings of the International Conference on Machine Learning, PMLR, Baltimore, MD, USA, 17–23 July 2022; pp. 3360–3370. [Google Scholar]
- Rafi, T.H.; Woong-Ko, Y. HeartNet: Self Multi-Head Attention Mechanism via Convolutional Network with Adversarial Data Synthesis for ECG-based Arrhythmia Classification. IEEE Access 2022, 10, 2169–3536. [Google Scholar] [CrossRef]
- Ma, S.; Cui, J.; Chen, C.L.; Chen, X.; Ma, Y. An Effective Data Enhancement Method for Classification of ECG Arrhythmia. Measurement 2022, 203, 111978. [Google Scholar] [CrossRef]
- Ma, K.; Chang’an, A.Z.; Yang, F. Multi-classification of arrhythmias using ResNet with CBAM on CWGAN-GP augmented ECG Gramian Angular Summation Field. Biomed. Signal Process. Control. 2022, 77, 103684. [Google Scholar] [CrossRef]
- Wulan, N.; Wang, W.; Sun, P.; Wang, K.; Xia, Y.; Zhang, H. Generating electrocardiogram signals by deep learning. Neurocomputing 2020, 404, 122–136. [Google Scholar] [CrossRef]
- Jin, Q.; Lin, R.; Yang, F. E-WACGAN: Enhanced generative model of signaling data based on WGAN-GP and ACGAN. IEEE Syst. J. 2019, 14, 3289–3300. [Google Scholar] [CrossRef]
- Lee, J.; Lee, H. Improving SSH detection model using IPA time and WGAN-GP. Comput. Secur. 2022, 116, 102672. [Google Scholar] [CrossRef]
- Radford, A.; Metz, L.; Chintala, S. Unsupervised representation learning with deep convolutional generative adversarial networks. arXiv 2015, arXiv:1511.06434. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial networks. Commun. ACM 2020, 63, 139–144. [Google Scholar] [CrossRef]
- Arjovsky, M.; Chintala, S.; Bottou, L. Wasserstein generative adversarial networks. In Proceedings of the International Conference on Machine Learning, PMLR, Sydney, Australia, 6–11 August 2017; pp. 214–223. [Google Scholar]
- Gulrajani, I.; Ahmed, F.; Arjovsky, M.; Dumoulin, V.; Courville, A.C. Improved training of wasserstein gans. Adv. Neural Inf. Process. Syst. 2017, 30, 5769–5779. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. Imagenet large scale visual recognition challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar]
- Zhao, Z.; Fang, H.; Relton, S.D.; Yan, R.; Liu, Y.; Li, Z.; Qin, J.; Wong, D.C. Adaptive lead weighted resnet trained with different duration signals for classifying 12-lead ecgs. In Proceedings of the 2020 Computing in Cardiology, Rimini, Italy, 13–16 September 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1–4. [Google Scholar]
- Zhang, X.; Li, R.; Dai, H.; Liu, Y.; Zhou, B.; Wang, Z. Localization of myocardial infarction with multi-lead bidirectional gated recurrent unit neural network. IEEE Access 2019, 7, 161152–161166. [Google Scholar] [CrossRef]
- Salimans, T.; Goodfellow, I.; Zaremba, W.; Cheung, V.; Radford, A.; Chen, X. Improved techniques for training gans. Adv. Neural Inf. Process. Syst. 2016, 29, 2234–2242. [Google Scholar]
- Lourenço, A.; Silva, H.; Carreiras, C. Outlier detection in non-intrusive ECG biometric system. In Proceedings of the International Conference Image Analysis and Recognition; Springer: Berlin/Heidelberg, Germany, 2013; pp. 43–52. [Google Scholar]
- Shark, L.K.; Yu, C. Denoising by optimal fuzzy thresholding in wavelet domain. Electron. Lett. 2000, 36, 1. [Google Scholar] [CrossRef]
- Stergiou, G.S.; Alpert, B.; Mieke, S.; Asmar, R.; Atkins, N.; Eckert, S.; Frick, G.; Friedman, B.; Graßl, T.; Ichikawa, T.; et al. A universal standard for the validation of blood pressure measuring devices: Association for the Advancement of Medical Instrumentation/European Society of Hypertension/International Organization for Standardization (AAMI/ESH/ISO) Collaboration Statement. Hypertension 2018, 71, 368–374. [Google Scholar] [CrossRef]
- Garcia, G.; Moreira, G.; Menotti, D.; Luz, E. Inter-patient ECG heartbeat classification with temporal VCG optimized by PSO. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Lai, J.; Chen, Y.; Han, B.; Ji, L.; Shi, Y.; Huang, Z.; Yang, W.; Feng, Q. A DenseNet-based diagnosis algorithm for automated diagnosis using clinical ECG data. J. South. Med. Univ. 2019, 39, 69–75. [Google Scholar]
- Ali, O.M.A.; Kareem, S.W.; Mohammed, A.S. Comparative Evaluation for Two and Five Classes ECG Signal Classification: Applied Deep Learning. J. Algebr. Stat. 2022, 13, 580–596. [Google Scholar]
- Rahul, J.; Sharma, L.D. Automatic cardiac arrhythmia classification based on hybrid 1-D CNN and Bi-LSTM model. Biocybern. Biomed. Eng. 2022, 42, 312–324. [Google Scholar] [CrossRef]
- Li, Y.; Qian, R.; Li, K. Inter-patient arrhythmia classification with improved deep residual convolutional neural network. Comput. Methods Progr. Biomed. 2022, 214, 106582. [Google Scholar] [CrossRef]
- Kaur, A.; Kumar, S.; Agarwal, R.; Agarwal, A. Intra and inter-patient arrhythmia classification using feature fusion with novel feature set based on fractional-order and fibonacci series. Biomed. Signal Process. Control 2022, 72, 103365. [Google Scholar] [CrossRef]
- Niu, J.; Tang, Y.; Sun, Z.; Zhang, W. Inter-patient ECG classification with symbolic representations and multi-perspective convolutional neural networks. IEEE J. Biomed. Health Inform. 2019, 24, 1321–1332. [Google Scholar] [CrossRef]
- Liu, T.; Si, Y.; Yang, W.; Huang, J.; Yu, Y.; Zhang, G.; Zhou, R. Inter-Patient Congestive Heart Failure Detection Using ECG-Convolution-Vision Transformer Network. Sensors 2022, 22, 3283. [Google Scholar] [CrossRef]
- Liaqat, S.; Dashtipour, K.; Zahid, A.; Assaleh, K.; Arshad, K.; Ramzan, N. Detection of atrial fibrillation using a machine learning approach. Information 2020, 11, 549. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).