Concatenation of Pre-Trained Convolutional Neural Networks for Enhanced COVID-19 Screening Using Transfer Learning Technique
Abstract
:1. Introduction
- The development and implementation of an accurate medical diagnosis support system for detecting coronavirus disease in chest X-ray images using a concatenation technique and TL-based models;
- The collection of a balanced medical X-ray image dataset includes four main classes (normal, viral pneumonia, tuberculosis, and coronavirus cases) for the training and testing the proposed system;
- The application of a parallel deep feature extraction approach based on TL models in the feature extraction phase of our concatenation-based models;
- The relative high accuracy of the proposed concatenation-based models performance.
2. Related Works
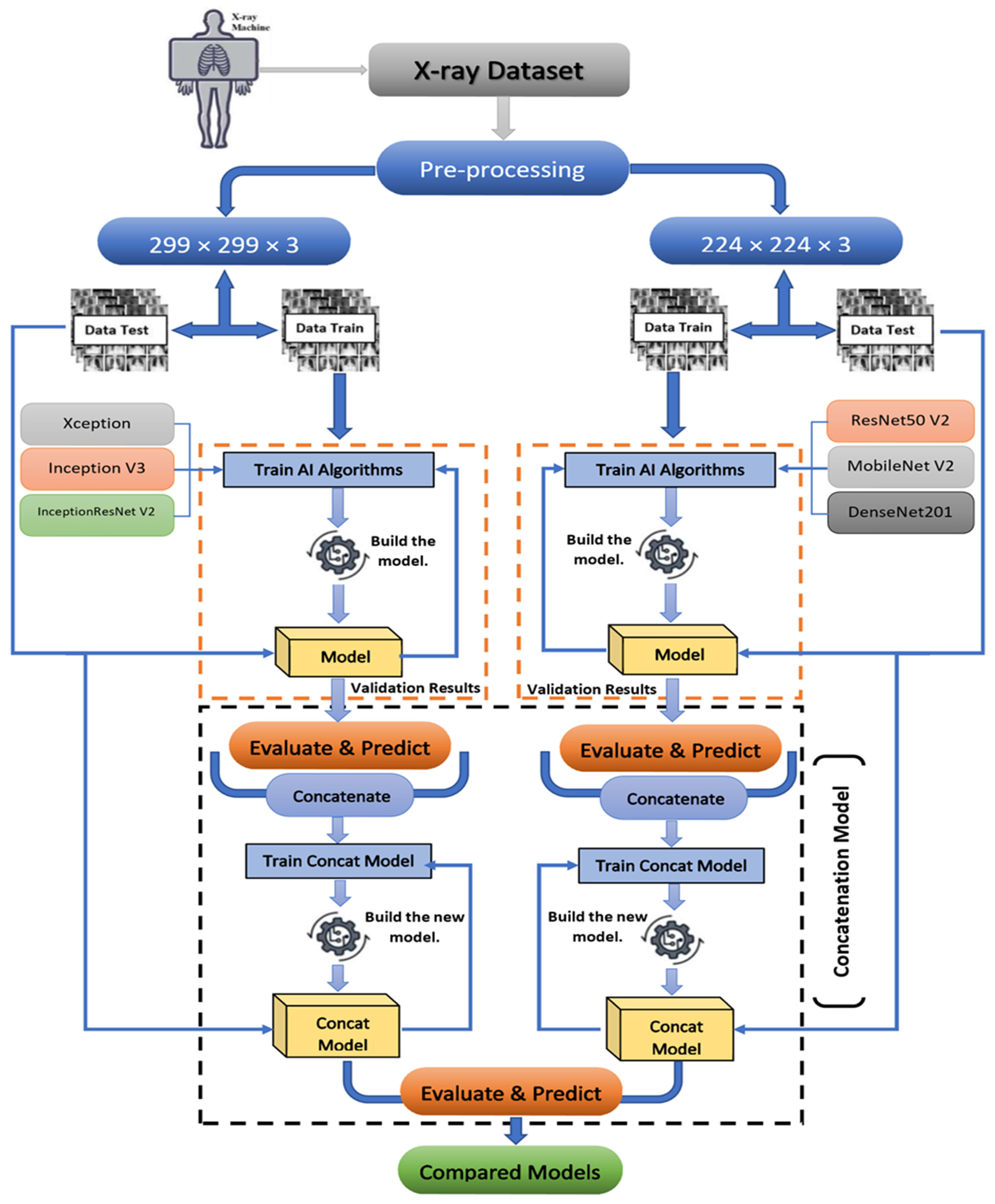
3. Resources and Procedures
3.1. Proposed Method Background
3.2. Dataset Description
3.3. Modified Pre-Trained Models Based on TL Technique
3.3.1. Modified Xception Based Model
3.3.2. Modified Inception V3 Based Model
3.3.3. Modified InceptionResNet V2 Based Model
3.3.4. Modified ResNet50 V2 Based Model
3.3.5. Modified MobileNet V2 Model
3.3.6. Modified DenseNet201 Based Model
3.3.7. Summary of the Modified Pre-Trained Models
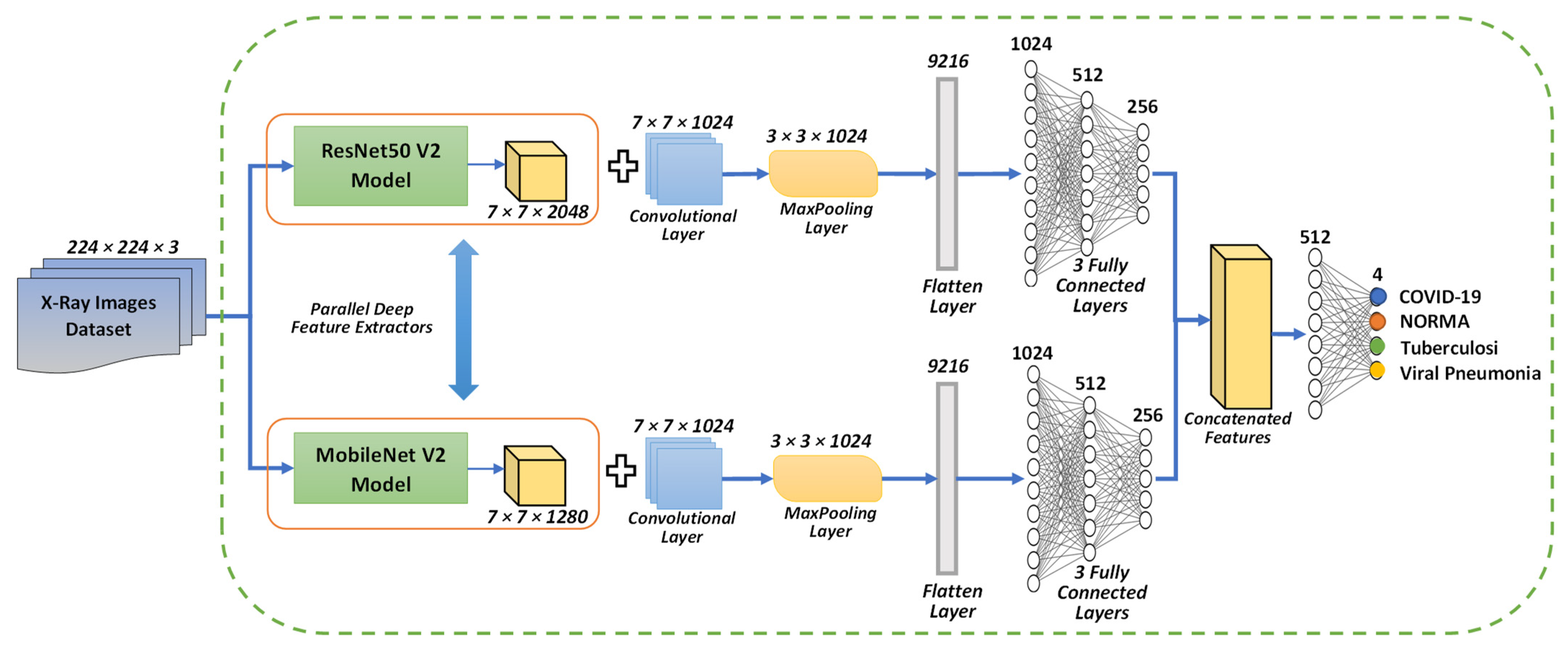
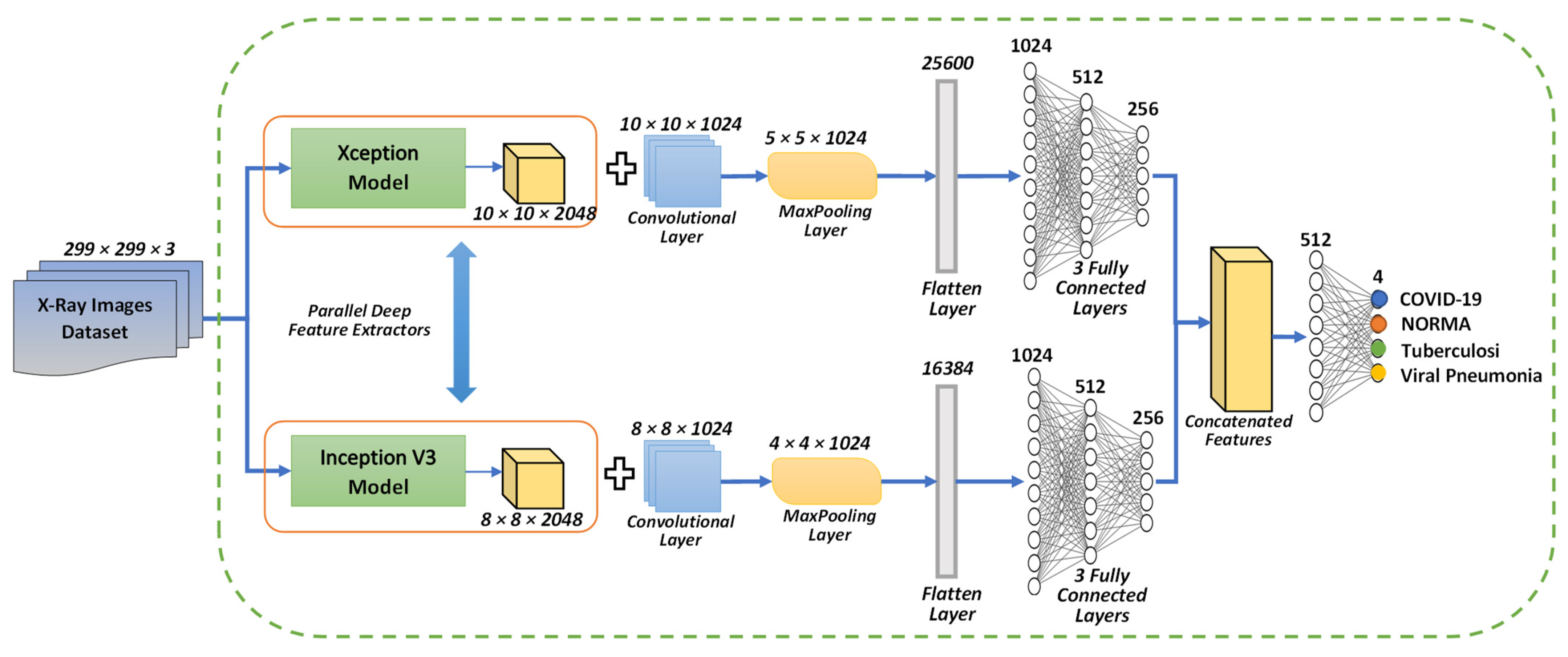
3.4. Proposed Concatenation Model Based on TL Technique
3.4.1. Concatenation Model Based on the Modified Resnet50 V2 and MobileNet V2 Models
3.4.2. Concatenation Model Based on the Modified Xception and Inception V3 Models
4. Results and Discussion
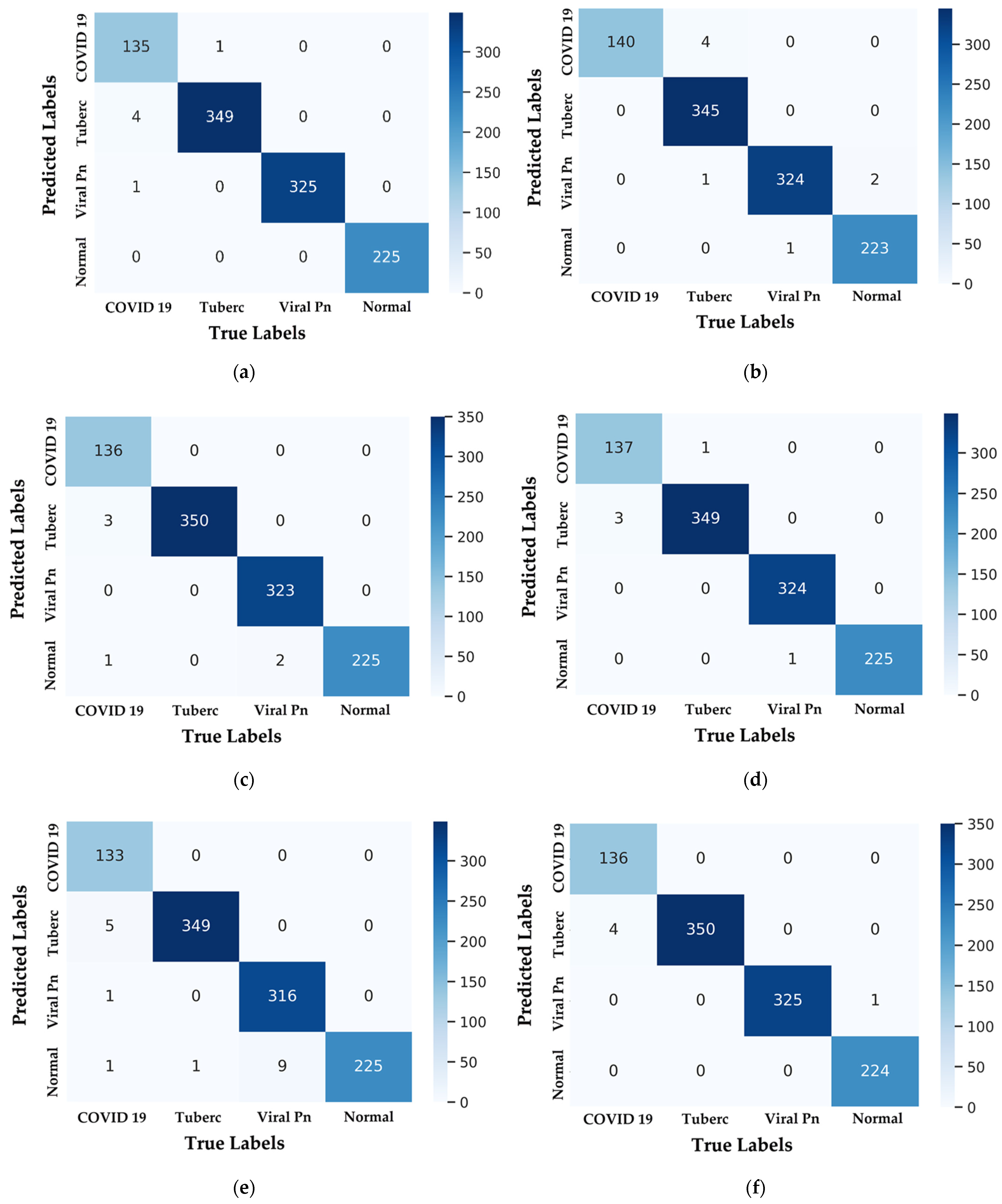
4.1. Confusion-Matrix and Metrics for Performance Evaluation
- The sample number successfully classified for a given class is denoted by the letter .
- The samples number of negatives that are mistakenly classified for another class is given by .
- The samples number of positives that are mistakenly classified for another class is denoted by the letter .
- The total of all samples is .
4.2. Experimental Results
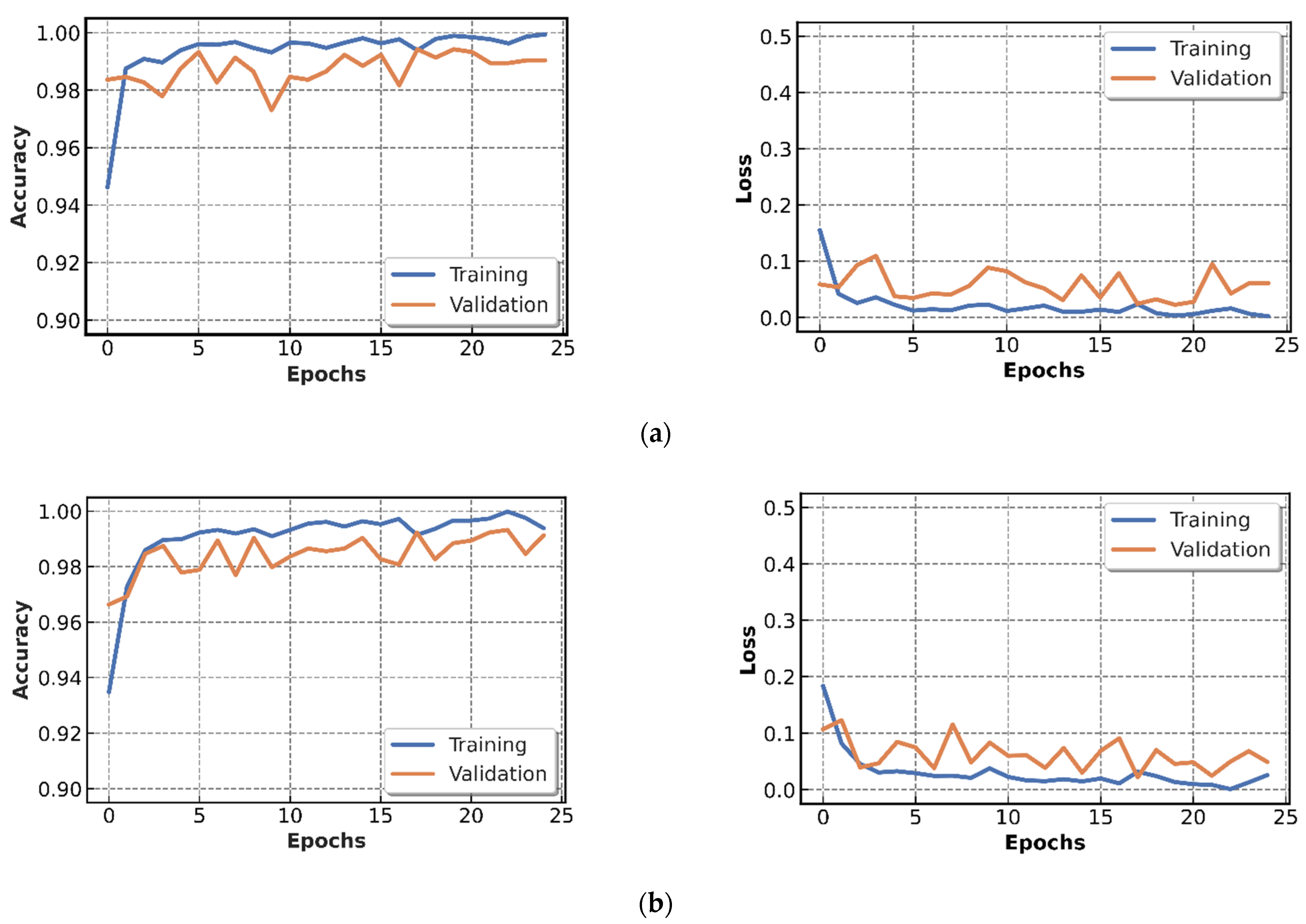
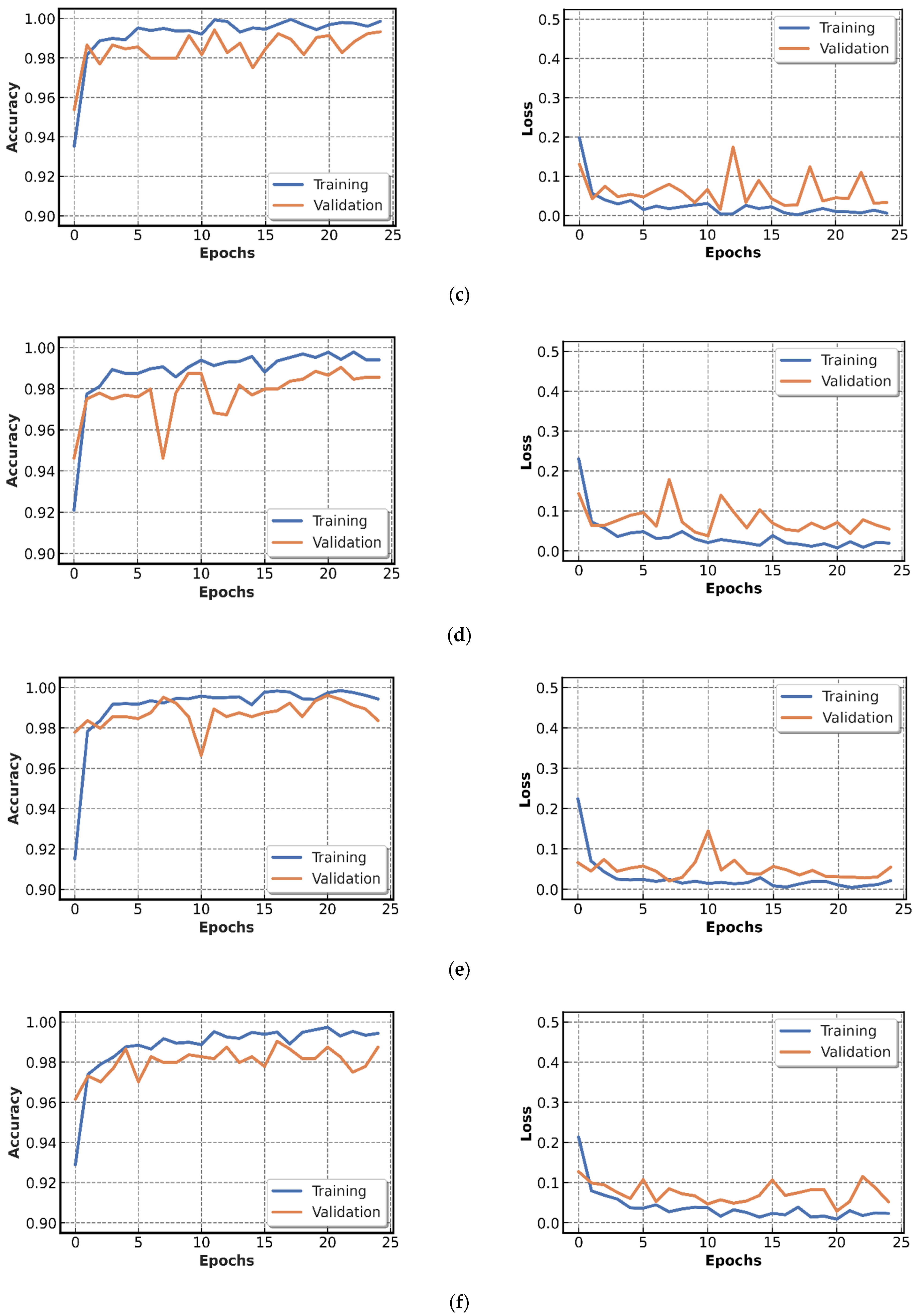
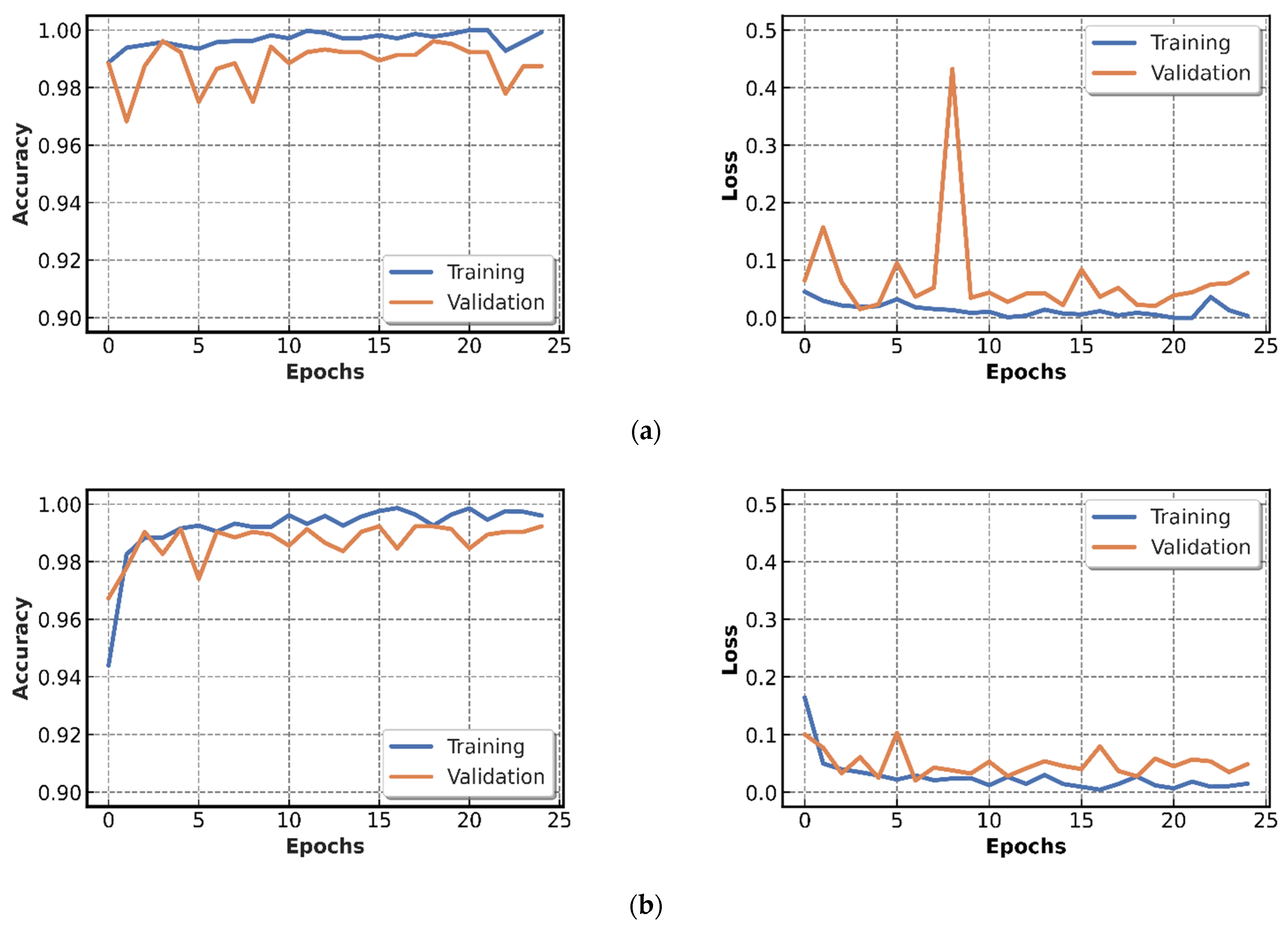
4.2.1. Results of CNN Models Training
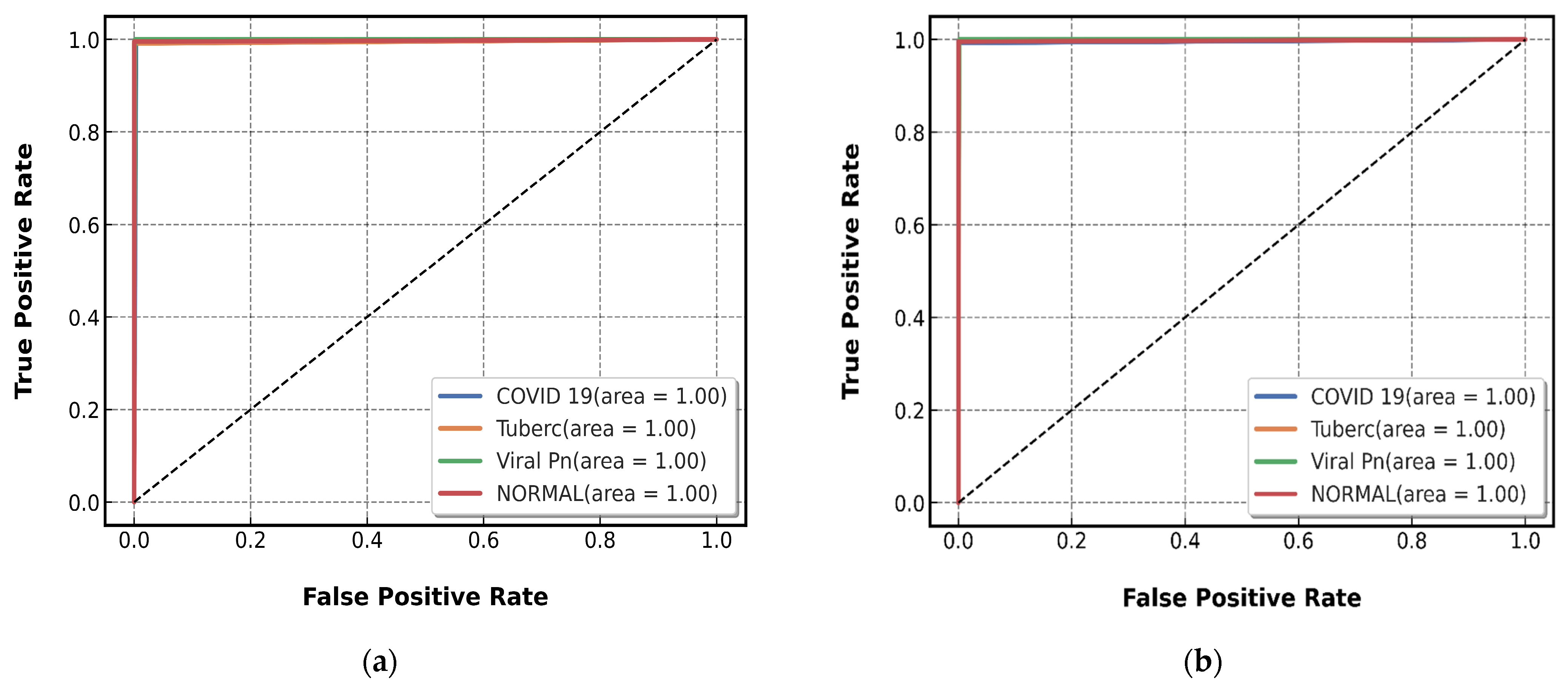
4.2.2. Testing of CNN Models Results
4.2.3. The Execution Time Results
4.3. Discussion
- Due to the lack of coronavirus images in comparison to the other diseases, six publicly available datasets were considered to construct a relatively equilibrated dataset. The enrichment of our COVID-19 images cases would ameliorate the proposed system accuracy.
- Despite the concatenation of pre-trained models contributing to the enhancement of the prediction accuracy, the computational complexity in terms of execution time increased considerably. This is an expected trade-off for enhanced accuracy, but the usage of parameter optimization in future work will ameliorate our results not only in terms of accuracy but also in terms of execution time.
5. Conclusions and Perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Astuti, I. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): An Overview of Viral Structure and Host Response. Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 407–412. [Google Scholar] [CrossRef]
- Sohrabi, C.; Alsafi, Z.; O’Neill, N.; Khan, M.; Kerwan, A.; Al-Jabir, A.; Iosifidis, C.; Agha, R. World Health Organization Declares Global Emergency: A Review of the 2019 Novel Coronavirus (COVID-19). Int. J. Surg. 2020, 76, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Larsen, J.R.; Martin, M.R.; Martin, J.D.; Kuhn, P.; Hicks, J.B. Modeling the Onset of Symptoms of COVID-19. Front. Public Health 2020, 8, 473. [Google Scholar] [CrossRef]
- Vetter, P.; Vu, D.L.; L’Huillier, A.G.; Schibler, M.; Kaiser, L.; Jacquerioz, F. Clinical Features of Covid-19. BMJ 2020, 369, m1470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- On behalf of the COVID-19 Commission of Accademia Nazionale dei Lincei, Rome; Forni, G.; Mantovani, A. COVID-19 Vaccines: Where We Stand and Challenges Ahead. Cell Death Differ. 2021, 28, 626–639. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, S.; Rashid, M.H.A.; Mridul, M.; Mohanty, C.; Swayamsiddha, S. Application of Artificial Intelligence in COVID-19 Drug Repurposing. Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 1027–1031. [Google Scholar] [CrossRef]
- Long, C.; Xu, H.; Shen, Q.; Zhang, X.; Fan, B.; Wang, C.; Zeng, B.; Li, Z.; Li, X.; Li, H. Diagnosis of the Coronavirus Disease (COVID-19): RRT-PCR or CT? Eur. J. Radiol. 2020, 126, 108961. [Google Scholar] [CrossRef] [PubMed]
- Hamida, S.; Gannour, O.E.; Cherradi, B.; Ouajji, H.; Raihani, A. Optimization of Machine Learning Algorithms Hyper-Parameters for Improving the Prediction of Patients Infected with COVID-19. In Proceedings of the 2020 IEEE 2nd International Conference on Electronics, Control, Optimization and Computer Science (ICECOCS), Kenitra, Morocco, 2–3 December 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1–6. [Google Scholar]
- Terrada, O.; Hamida, S.; Cherradi, B.; Raihani, A.; Bouattane, O. Supervised Machine Learning Based Medical Diagnosis Support System for Prediction of Patients with Heart Disease. Adv. Sci. Technol. Eng. Syst. J. 2020, 5, 269–277. [Google Scholar] [CrossRef]
- Terrada, O.; Cherradi, B.; Hamida, S.; Raihani, A.; Moujahid, H.; Bouattane, O. Prediction of Patients with Heart Disease Using Artificial Neural Network and Adaptive Boosting Techniques. In Proceedings of the 2020 3rd International Conference on Advanced Communication Technologies and Networking (CommNet), Marrakech, Morocco, 4–6 September 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1–6. [Google Scholar]
- Xie, J.; Jiang, J.; Wang, Y.; Guan, Y.; Guo, X. Learning an Expandable EMR-Based Medical Knowledge Network to Enhance Clinical Diagnosis. Artif. Intell. Med. 2020, 107, 101927. [Google Scholar] [CrossRef]
- Terrada, O.; Cherradi, B.; Raihani, A.; Bouattane, O. Atherosclerosis Disease Prediction Using Supervised Machine Learning Techniques. In Proceedings of the 2020 1st International Conference on Innovative Research in Applied Science, Engineering and Technology (IRASET), Meknes, Morocco, 16–19 April 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1–5. [Google Scholar]
- Sharma, P.; Choudhary, K.; Gupta, K.; Chawla, R.; Gupta, D.; Sharma, A. Artificial Plant Optimization Algorithm to Detect Heart Rate & Presence of Heart Disease Using Machine Learning. Artif. Intell. Med. 2020, 102, 101752. [Google Scholar] [CrossRef]
- Cherradi, B.; Terrada, O.; Ouhmida, A.; Hamida, S.; Raihani, A.; Bouattane, O. Computer-Aided Diagnosis System for Early Prediction of Atherosclerosis Using Machine Learning and K-Fold Cross-Validation. In Proceedings of the 2021 International Congress of Advanced Technology and Engineering (ICOTEN), Taiz, Yemen, 4–5 July 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 1–9. [Google Scholar]
- Terrada, O.; Cherradi, B.; Raihani, A.; Bouattane, O. A Novel Medical Diagnosis Support System for Predicting Patients with Atherosclerosis Diseases. Inform. Med. Unlocked 2020, 21, 100483. [Google Scholar] [CrossRef]
- Hamida, S.; Cherradi, B.; Terrada, O.; Raihani, A.; Ouajji, H.; Laghmati, S. A Novel Feature Extraction System for Cursive Word Vocabulary Recognition Using Local Features Descriptors and Gabor Filter. In Proceedings of the 2020 3rd International Conference on Advanced Communication Technologies and Networking (CommNet), Marrakech, Morocco, 4–6 September 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1–7. [Google Scholar]
- Alsaeedi, A.; Al-Sarem, M. Detecting Rumors on Social Media Based on a CNN Deep Learning Technique. Arab. J. Sci. Eng. 2020, 45, 10813–10844. [Google Scholar] [CrossRef]
- Vijh, M.; Chandola, D.; Tikkiwal, V.A.; Kumar, A. Stock Closing Price Prediction Using Machine Learning Techniques. Procedia Comput. Sci. 2020, 167, 599–606. [Google Scholar] [CrossRef]
- Madani, Y.; Erritali, M.; Bouikhalene, B. Using Artificial Intelligence Techniques for Detecting COVID-19 Epidemic Fake News in Moroccan Tweets. Results Phys. 2021, 25, 104266. [Google Scholar] [CrossRef]
- Meldo, A.; Utkin, L.; Kovalev, M.; Kasimov, E. The Natural Language Explanation Algorithms for the Lung Cancer Computer-Aided Diagnosis System. Artif. Intell. Med. 2020, 108, 101952. [Google Scholar] [CrossRef]
- Ait Ali, N.; Cherradi, B.; El Abbassi, A.; Bouattane, O.; Youssfi, M. GPU Fuzzy C-Means Algorithm Implementations: Performance Analysis on Medical Image Segmentation. Multimed. Tools Appl. 2018, 77, 21221–21243. [Google Scholar] [CrossRef]
- Ali, N.A.; Abbassi, A.E.; Cherradi, B. The Performances of Iterative Type-2 Fuzzy C-Mean on GPU for Image Segmentation. J. Supercomput. 2021, 77, 1–19. [Google Scholar] [CrossRef]
- Bouattane, O.; Cherradi, B.; Youssfi, M.; Bensalah, M.O. Parallel C-Means Algorithm for Image Segmentation on a Reconfigurable Mesh Computer. Parallel Comput. 2011, 37, 230–243. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Chen, P.; Li, Z.; Su, H.; Yang, L.; Zhong, D. Rule-Based Automatic Diagnosis of Thyroid Nodules from Intraoperative Frozen Sections Using Deep Learning. Artif. Intell. Med. 2020, 108, 101918. [Google Scholar] [CrossRef] [PubMed]
- Pereira, R.M.; Bertolini, D.; Teixeira, L.O.; Silla, C.N.; Costa, Y.M.G. COVID-19 Identification in Chest X-Ray Images on Flat and Hierarchical Classification Scenarios. Comput. Methods Programs Biomed. 2020, 194, 105532. [Google Scholar] [CrossRef]
- Vaishya, R.; Javaid, M.; Khan, I.H.; Haleem, A. Artificial Intelligence (AI) Applications for COVID-19 Pandemic. Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 337–339. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, S.; Chen, Y.; Luo, F.; Kang, Z.; Cai, S.; Zhao, W.; Liu, J.; Zhao, D.; Li, Y. A Deep-Learning-Based Framework for Severity Assessment of COVID-19 with CT Images. Expert Syst. Appl. 2021, 185, 115616. [Google Scholar] [CrossRef]
- Moujahid, H.; Cherradi, B.; Al-Sarem, M.; Bahatti, L.; Bakr Assedik Mohammed Yahya Eljialy, A.; Alsaeedi, A.; Saeed, F. Combining CNN and Grad-Cam for COVID-19 Disease Prediction and Visual Explanation. Intell. Autom. Soft Comput. 2022, 32, 723–745. [Google Scholar] [CrossRef]
- Hamida, S.; El Gannour, O.; Cherradi, B.; Raihani, A.; Moujahid, H.; Ouajji, H. A Novel COVID-19 Diagnosis Support System Using the Stacking Approach and Transfer Learning Technique on Chest X-Ray Images. J. Healthc. Eng. 2021, 2021, 9437538. [Google Scholar] [CrossRef] [PubMed]
- Schmidhuber, J. Deep Learning in Neural Networks: An Overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef] [Green Version]
- Yamashita, R.; Nishio, M.; Do, R.K.G.; Togashi, K. Convolutional Neural Networks: An Overview and Application in Radiology. Insights Imaging 2018, 9, 611–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Shen, C.; Hengel, A. van den Cross-Convolutional-Layer Pooling for Image Recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2305–2313. [Google Scholar] [CrossRef] [Green Version]
- Hutchison, D.; Kanade, T.; Kittler, J.; Kleinberg, J.M.; Mattern, F.; Mitchell, J.C.; Naor, M.; Nierstrasz, O.; Pandu Rangan, C.; Steffen, B.; et al. Evaluation of Pooling Operations in Convolutional Architectures for Object Recognition. In Artificial Neural Networks—ICANN 2010; (Lecture Notes in Computer, Science); Diamantaras, K., Duch, W., Iliadis, L.S., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 6354, pp. 92–101. ISBN 978-3-642-15824-7. [Google Scholar]
- Basha, S.H.S.; Dubey, S.R.; Pulabaigari, V.; Mukherjee, S. Impact of Fully Connected Layers on Performance of Convolutional Neural Networks for Image Classification. Neurocomputing 2020, 378, 112–119. [Google Scholar] [CrossRef] [Green Version]
- Moujahid, H.; Cherradi, B.; Al-Sarem, M.; Bahatti, L. Diagnosis of COVID-19 Disease Using Convolutional Neural Network Models Based Transfer Learning. In Innovative Systems for Intelligent Health Informatics (Lecture Notes on Data Engineering and Communications Technologies); Saeed, F., Mohammed, F., Al-Nahari, A., Eds.; Springer International Publishing: Cham, Switzerland, 2021; Volume 72, pp. 148–159. ISBN 978-3-030-70712-5. [Google Scholar]
- Morís, D.I.; de Moura Ramos, J.J.; Buján, J.N.; Hortas, M.O. Data Augmentation Approaches Using Cycle-Consistent Adversarial Networks for Improving COVID-19 Screening in Portable Chest X-Ray Images. Expert Syst. Appl. 2021, 185, 115681. [Google Scholar] [CrossRef]
- El Gannour, O.; Hamida, S.; Cherradi, B.; Raihani, A.; Moujahid, H. Performance Evaluation of Transfer Learning Technique for Automatic Detection of Patients with COVID-19 on X-Ray Images. In Proceedings of the 2020 IEEE 2nd International Conference on Electronics, Control, Optimization and Computer Science (ICECOCS), Kenitra, Morocco, 2–3 December 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1–6. [Google Scholar]
- Khan, M.; Mehran, M.T.; Haq, Z.U.; Ullah, Z.; Naqvi, S.R.; Ihsan, M.; Abbass, H. Applications of Artificial Intelligence in COVID-19 Pandemic: A Comprehensive Review. Expert Syst. Appl. 2021, 185, 115695. [Google Scholar] [CrossRef]
- Ashour, A.S.; Eissa, M.M.; Wahba, M.A.; Elsawy, R.A.; Elgnainy, H.F.; Tolba, M.S.; Mohamed, W.S. Ensemble-Based Bag of Features for Automated Classification of Normal and COVID-19 CXR Images. Biomed. Signal Process. Control 2021, 68, 102656. [Google Scholar] [CrossRef]
- Koyuncu, H.; Barstuğan, M. COVID-19 Discrimination Framework for X-Ray Images by Considering Radiomics, Selective Information, Feature Ranking, and a Novel Hybrid Classifier. Signal Process. Image Commun. 2021, 97, 116359. [Google Scholar] [CrossRef]
- Sharifrazi, D.; Alizadehsani, R.; Roshanzamir, M.; Joloudari, J.H.; Shoeibi, A.; Jafari, M.; Hussain, S.; Sani, Z.A.; Hasanzadeh, F.; Khozeimeh, F.; et al. Fusion of Convolution Neural Network, Support Vector Machine and Sobel Filter for Accurate Detection of COVID-19 Patients Using X-Ray Images. Biomed. Signal Process. Control 2021, 68, 102622. [Google Scholar] [CrossRef]
- Fan, Y.; Liu, J.; Yao, R.; Yuan, X. COVID-19 Detection from X-Ray Images Using Multi-Kernel-Size Spatial-Channel Attention Network. Pattern Recognit. 2021, 119, 108055. [Google Scholar] [CrossRef]
- Das, S.; Roy, S.D.; Malakar, S.; Velásquez, J.D.; Sarkar, R. Bi-Level Prediction Model for Screening COVID-19 Patients Using Chest X-Ray Images. Big Data Res. 2021, 25, 100233. [Google Scholar] [CrossRef]
- Thakur, S.; Kumar, A. X-Ray and CT-Scan-Based Automated Detection and Classification of Covid-19 Using Convolutional Neural Networks (CNN). Biomed. Signal Process. Control 2021, 69, 102920. [Google Scholar] [CrossRef]
- Ozcan, T. A New Composite Approach for COVID-19 Detection in X-Ray Images Using Deep Features. Appl. Soft Comput. 2021, 111, 107669. [Google Scholar] [CrossRef]
- Shorfuzzaman, M.; Masud, M.; Alhumyani, H.; Anand, D.; Singh, A. Artificial Neural Network-Based Deep Learning Model for COVID-19 Patient Detection Using X-Ray Chest Images. J. Healthc. Eng. 2021, 2021, 5513679. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.D.K.; Ahmed, S.; Abdullah, Z.M.E.; Monirujjaman Khan, M.; Anand, D.; Singh, A.; AlZain, M.; Masud, M. Deep Learning Approaches for Detecting Pneumonia in COVID-19 Patients by Analyzing Chest X-Ray Images. Math. Probl. Eng. 2021, 2021, 9929274. [Google Scholar] [CrossRef]
- Al-Rakhami, M.S.; Islam, M.M.; Islam, M.Z.; Asraf, A.; Sodhro, A.H.; Ding, W. Diagnosis of COVID-19 from X-Rays Using Combined CNN-RNN Architecture with Transfer Learning. Health Inform. 2020, 1–14. [Google Scholar] [CrossRef]
- Gaur, L.; Bhatia, U.; Jhanjhi, N.Z.; Muhammad, G.; Masud, M. Medical Image-Based Detection of COVID-19 Using Deep Convolution Neural Networks. Multimed. Syst. 2021, 27, 1–10. [Google Scholar] [CrossRef]
- Zulkifley, M.A.; Abdani, S.R.; Zulkifley, N.H.; Shahrimin, M.I. Residual-Shuffle Network with Spatial Pyramid Pooling Module for COVID-19 Screening. Diagnostics 2021, 11, 1497. [Google Scholar] [CrossRef]
- Barua, P.D.; Muhammad Gowdh, N.F.; Rahmat, K.; Ramli, N.; Ng, W.L.; Chan, W.Y.; Kuluozturk, M.; Dogan, S.; Baygin, M.; Yaman, O.; et al. Automatic COVID-19 Detection Using Exemplar Hybrid Deep Features with X-Ray Images. IJERPH 2021, 18, 8052. [Google Scholar] [CrossRef]
- Pan, S.J.; Yang, Q. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 2010, 22, 1345–1359. [Google Scholar] [CrossRef]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. arXiv 2017, arXiv:1610.02357,. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. arXiv 2015, arXiv:1512.00567. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. arXiv 2016, arXiv:1602.07261. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity Mappings in Deep Residual Networks. arXiv 2016, arXiv:1603.05027. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. arXiv 2019, arXiv:1801.04381. [Google Scholar]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. arXiv 2018, arXiv:1608.06993. [Google Scholar]
- Ren, X.; Du, S.; Zheng, Y. Parallel RCNN: A Deep Learning Method for People Detection Using RGB-D Images. In Proceedings of the 2017 10th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI), Shanghai, China, 14–16 October 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 1–6. [Google Scholar]
- Laghmati, S.; Cherradi, B.; Tmiri, A.; Daanouni, O.; Hamida, S. Classification of Patients with Breast Cancer Using Neighbourhood Component Analysis and Supervised Machine Learning Techniques. In Proceedings of the 2020 3rd International Conference on Advanced Communication Technologies and Networking (CommNet), Marrakech, Morocco, 4–6 September 2020; IEEE: Piscataway, NJ, USA, 2020. [Google Scholar]
- Ouhmida, A.; Raihani, A.; Cherradi, B.; Terrada, O. A Novel Approach for Parkinson’s Disease Detection Based on Voice Classification and Features Selection Techniques. Int. J. Online Eng. 2021, 17, 111. [Google Scholar] [CrossRef]
- Ouhmida, A.; Terrada, O.; Raihani, A.; Cherradi, B.; Hamida, S. Voice-Based Deep Learning Medical Diagnosis System for Parkinson’s Disease Prediction. In Proceedings of the 2021 International Congress of Advanced Technology and Engineering (ICOTEN), Taiz, Yemen, 4–5 July 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 1–5. [Google Scholar]
- Lopes, R.R.; Bleijendaal, H.; Ramos, L.A.; Verstraelen, T.E.; Amin, A.S.; Wilde, A.A.M.; Pinto, Y.M.; de Mol, B.A.J.M.; Marquering, H.A. Improving Electrocardiogram-Based Detection of Rare Genetic Heart Disease Using Transfer Learning: An Application to Phospholamban p.Arg14del Mutation Carriers. Comput. Biol. Med. 2021, 131, 104262. [Google Scholar] [CrossRef] [PubMed]
- Zucker, E.J.; Barnes, Z.A.; Lungren, M.P.; Shpanskaya, Y.; Seekins, J.M.; Halabi, S.S.; Larson, D.B. Deep Learning to Automate Brasfield Chest Radiographic Scoring for Cystic Fibrosis. J. Cyst. Fibros. 2020, 19, 131–138. [Google Scholar] [CrossRef] [Green Version]
- Altan, G.; Kutlu, Y.; Allahverdi, N. Deep Learning on Computerized Analysis of Chronic Obstructive Pulmonary Disease. IEEE J. Biomed. Health Inform. 2020, 24, 1344–1350. [Google Scholar] [CrossRef]
- Oniani, S.; Marques, G.; Barnovi, S.; Pires, I.M.; Bhoi, A.K. Artificial Intelligence for Internet of Things and Enhanced Medical Systems. In Bio-inspired Neurocomputing; (Studies in Computational Intelligence); Bhoi, A.K., Mallick, P.K., Liu, C.-M., Balas, V.E., Eds.; Springer: Singapore, 2021; Volume 903, pp. 43–59. ISBN 9789811554940. [Google Scholar]
- Sun, L.; Jiang, X.; Ren, H.; Guo, Y. Edge-Cloud Computing and Artificial Intelligence in Internet of Medical Things: Architecture, Technology and Application. IEEE Access 2020, 8, 101079–101092. [Google Scholar] [CrossRef]
- Talukder, A.; Haas, R. AIoT: AI Meets IoT and Web in Smart Healthcare. In Proceedings of the 13th ACM Web Science Conference 2021, Virtual Event, 21–25 June 2021; Association for Computing Machinery: New York, NY, USA, 2021; pp. 92–98. [Google Scholar]




| Ref. | No. of Classes | Models | Accuracy | Precision | Sensitivity | Specificity |
|---|---|---|---|---|---|---|
| [39] | Coronavirus | BoF | 98.6% | 97.7% | 99.4% | N.A |
| Normal | ||||||
| [40] | Coronavirus | Hybrid Classifier | 99.53% | N.A | N.A | N.A |
| Normal | ||||||
| Bac-pneumonia | ||||||
| [41] | Coronavirus | CNN-SVM + Sobel | 99.61% | N.A | 99.80% | 99.56% |
| Normal | ||||||
| [42] | Coronavirus | MKSC | 98.2% | 97.52% | 98.05% | 97.82% |
| Normal | ||||||
| [43] | Coronavirus | Bi-level classification | 99.26% | 99.23% | 99.26% | N.A |
| Normal | ||||||
| Pneumonia | ||||||
| [44] | Coronavirus | CNN | 99.64% | 99.56% | 99.58% | N.A |
| Normal | ||||||
| Coronavirus | 98.28% | 98.22% | 98.25% | N.A | ||
| Normal | ||||||
| Pneumonia | ||||||
| [45] | Coronavirus | SLB1 | 84.71% | 87.75% | 86.13% | 90.91% |
| SLB2 | 80.89% | 83.79% | 81.67% | 88.71% | ||
| SLB3 | 80.00% | 84.28% | 80.60% | 88.08% | ||
| Normal | SLB4 | 84.44% | 87.80% | 84.73% | 90.73% | |
| FFB1 | 83.82% | 87.32% | 85.07% | 90.35% | ||
| FFB2 | 84.27% | 87.65% | 85.60% | 90.62% | ||
| FFB3 | 87.64% | 90.87% | 88.13% | 92.59% | ||
| Pneumonia | FFB4 | 79.91% | 83.28% | 80.93% | 88.11% | |
| FFB5 | 82.13% | 85.90% | 83.40% | 89.36% | ||
| FFB6 | 81.87% | 85.90% | 82.40% | 89.18% | ||
| [46] | Coronavirus | ResNet50 V2 | 95.49% | 96.85% | 99.19% | 98.27% |
| Normal | VGG16 | 92.70% | 97.50% | 94.35% | 98.69% | |
| Pneumonia | Inception V3 | 92.97% | 97.60% | 98.39% | 98.67% | |
| [47] | Coronavirus Normal Pneumonia | VGG16 | 91.69% | 92.33% | 95.92% | 100% |
| [48] | Coronavirus Normal Pneumonia | VGG19-RNN | 99.85% | N.A | N.A | N.A |
| [49] | Coronavirus | VGG16 | 87.84% | 82.00% | 82.33% | 91.20% |
| Normal | Inception-V3 | 91.32% | 87.54% | 89.00% | 94.00% | |
| Viral-pneumonia | EfficientNetB0 | 92.93% | 88.30% | 90.00% | 95.00% | |
| [50] | Coronavirus | Residual-Shuffle-Net | 97.39% | 97.40% | 97.39% | 98.69% |
| Usual | ||||||
| Pneumonia | ||||||
| [51] | Control Bacterial Pneumonia Viral Pneumonia Coronavirus | Exemplar COVID-19FclNet9 | 97.60% | N.A | N.A | N.A |
| Sources of Datasets | Coronavirus | Normal | Viral-Pneumonia | Tuberculosis |
|---|---|---|---|---|
| Coronavirus Radiography Database 1 | 1200 | 1341 | 1345 | - |
| Coronavirus Detection X-ray Dataset 2 | 60 | 880 | 412 | - |
| Coronavirus Patients Lungs X-ray Images 3 | 70 | 28 | - | - |
| Coronavirus X-rays 4 | 70 | - | - | - |
| Viral-Pneumonia vs. Bacteria-Pneumonia 5 | - | - | 1493 | - |
| Chest X-ray Database of Tuberculosis (TB) 6 | - | - | - | 3500 |
| Resulting Dataset | 1400 | 2249 | 3250 | 3500 |
| Category | Training (80%) | Validation and Testing (20%) | Total (100%) |
|---|---|---|---|
| Normal | 1799 | 450 | 2249 |
| Tuberculosis | 2800 | 700 | 3500 |
| Coronavirus | 1120 | 280 | 1400 |
| Viral-Pneumonia | 2600 | 650 | 3250 |
| Total | 8319 | 2080 | 10,399 |
| Designation | Output-Shape | Parameters Number | |
|---|---|---|---|
| Original Model | xception_v2 | 10 × 10 × 2048 | 20,861,480 |
| Additional Layers | conv2d_1 (Conv2D) | 10 × 10 × 1024 | 2,098,176 |
| mp_1 (MaxPooling2D) | 5 × 5 × 1024 | 0 | |
| flatten (Flatten) | 25,600 | 0 | |
| Dense-1 | 1024 | 26,215,424 | |
| Dense-2 | 512 | 524,800 | |
| Dense-3 | 256 | 131,328 | |
| Dense-4 | 4 | 1028 |
| Designation | Output Shape | Parameters Number | |
|---|---|---|---|
| Original Model | Inception-V3 | 8 × 8 × 2048 | 21,802,784 |
| Additional Layers | conv2d_1 (Conv2D) | 8 × 8 × 1024 | 2,098,176 |
| mp_1 (MaxPooling2D) | 4 × 4 × 1024 | 0 | |
| flatten (Flatten) | 16,384 | 0 | |
| Dense-1 | 1024 | 16,778,240 | |
| Dense-2 | 512 | 524,800 | |
| Dense-3 | 256 | 13,128 | |
| Dense-4 | 4 | 1028 |
| Designation | Output Shape | Parameters Number | |
|---|---|---|---|
| Original Model | Inceptionresnet-V2 | 8 × 8 × 1536 | 54,336,736 |
| Additional Layers | conv2d_1 (Conv2D) | 8 × 8 × 1024 | 1,573,888 |
| mp_1 (MaxPooling2D) | 4 × 4 × 1024 | 0 | |
| flatten (Flatten) | 16,384 | 0 | |
| Dense-1 | 1024 | 16,778,240 | |
| Dense-2 | 512 | 524,800 | |
| Dense-3 | 256 | 131,328 | |
| Dense-4 | 4 | 1028 |
| Designation | Output Shape | Parameter Number | |
|---|---|---|---|
| Original Model | resnet50_v2 | 7 × 7 × 2048 | 23,564,800 |
| Additional Layers | conv2d_1 (Conv2D-1) | 7 × 7 × 1024 | 2,098,176 |
| mp_1 (MaxPooling2D) | 3 × 3 × 1024 | 0 | |
| flatten (Flatten) | 9216 | 0 | |
| Dense-1 | 1024 | 9,438,208 | |
| Dense-2 | 512 | 524,800 | |
| Dense-3 | 256 | 131,328 | |
| Dense-4 | 4 | 1028 |
| Designation | Output Shape | Parameter Number | |
|---|---|---|---|
| Original Model | mobilenet_v2 | 7 × 7 × 1280 | 2,257,984 |
| Additional Layers | conv2d_1 (Conv2D) | 7 × 7 × 1024 | 1,311,744 |
| mp_1 (MaxPooling2D) | 3 × 3 × 1024 | 0 | |
| flatten (Flatten) | 9216 | 0 | |
| dense_1 (Dense) | 1024 | 9,438,208 | |
| dense_2 (Dense) | 512 | 524,800 | |
| dense_3 (Dense) | 256 | 131,328 | |
| dense_4 (Dense) | 4 | 1028 |
| Designation | Output Shape | Parameter Number | |
|---|---|---|---|
| Original Model | DenseNet201 | 7 × 7 × 1920 | 18,321,984 |
| Additional Layers | conv2d_1 (Conv2D) | 7 × 7 × 1024 | 1,967,104 |
| mp_1 (MaxPooling2D) | 3 × 3 × 1024 | 0 | |
| flatten (Flatten) | 9216 | 0 | |
| dense_1 (Dense) | 1024 | 9,438,208 | |
| dense_2 (Dense) | 512 | 524,800 | |
| dense_3 (Dense) | 256 | 131,328 | |
| dense_4 (Dense) | 4 | 1028 |
| Modified Models | Trainable Parameters | No Trainable Parameters | Total Parameters |
|---|---|---|---|
| Xception based model | 49,777,708 | 54,528 | 49,832,236 |
| Inception V3 based model | 41,301,924 | 34,432 | 41,336,356 |
| InceptionResNet V2 based model | 73,285,476 | 60,544 | 73,346,020 |
| ResNet50 V2 based model | 35,712,900 | 45,440 | 35,758,340 |
| MobileNet V2 based model | 13,896,568 | 34,112 | 13,930,680 |
| DenseNet201 based model | 30,155,396 | 229,052 | 30,384,452 |
| Proposed Concatenation Model Based on Resnet50 V2 and MobileNet V2 | |||
|---|---|---|---|
| TL1 Layer-type | Parameters | TL1 Layer-type | Parameters |
| TL1_resnet50v2 | 23,564,800 | TL2_mobilenetv2 | 2,257,984 |
| TL1_Conv2D-1 | 2,098,176 | TL1_Conv2D-1 | 1,311,744 |
| TL1_MaxPooling2D-1 | 0 | TL1_MaxPooling2D-1 | 0 |
| TL1_Flatten | 0 | TL1_Flatten | 0 |
| TL1_Dense-1 | 9,438,208 | TL1_Dense-1 | 9,438,208 |
| TL1_Dense-2 | 524,800 | TL1_Dense-2 | 524,800 |
| TL1_Dense-3 | 131,328 | TL1_Dense-3 | 131,328 |
| Concatenate Layer | Output Shape | Parameters | |
| concatenate_model (concatenate) | 512 | 0 | |
| concatenate_dense_1 (Dense) | 4 | 2052 | |
| Proposed Concatenate Model Based on Xception and Inception V3 | |||||
|---|---|---|---|---|---|
| TL1 Layer type | Output Shape | Parameters | TL1 Layer type | Output Shape | Parameters |
| TL1_xception | 10 × 10 × 2048 | 20,861,480 | TL2_inceptionv3 | 8 × 8 × 2048 | 21,802,784 |
| TL1_Conv2D-1 | 10 × 10 × 1024 | 2,098,176 | TL2_conv2d_1 (Conv2D) | 8 × 8 × 1024 | 2,098,176 |
| TL1_MaxPooling2D-1 | 5 × 5 × 1024 | 0 | TL2_mp_1 (MaxPooling2D) | 4 × 4 × 1024 | 0 |
| TL1_Flatten | 25,600 | 0 | TL2_flatten (Flatten) | 16,384 | 0 |
| TL1_Dense-1 | 1024 | 26,215,424 | TL2_dense_1 (Dense) | 1024 | 16,778,240 |
| TL1_Dense-2 | 512 | 524,800 | TL2_dense_2 (Dense) | 512 | 524,800 |
| TL1_Dense-3 | 256 | 131,328 | TL2_dense_3 (Dense) | 256 | 131,328 |
| Concatenate Layer | Output Shape | Parameters | |||
| concatenate_model (concatenate) | 512 | 0 | |||
| concatenate_dense_1 (Dense) | 4 | 2052 | |||
| Models | All Modified Models | Proposed Model 1 | Proposed Model 2 | |
|---|---|---|---|---|
| Activation Functions | Hidden 1 | Relu | ||
| Hidden 2 | ||||
| Hidden 3 | ||||
| Output | Softmax | |||
| Optimizer | Adam | |||
| Learning Rate | 1 × 10−4 | |||
| Batch Size | 16 | |||
| Loss Function | Categorical cross-entropy | |||
| Epochs | 25 | |||
| Modified Models | Accuracy | Loss | Precision | Sensitivity | Specificity | NPV |
|---|---|---|---|---|---|---|
| Xception based Model | 99.42% | 0.0342 | 99.46% | 99.04% | 99.79% | 99.83% |
| Inception V3 based Model | 99.23% | 0.0309 | 98.96%% | 99.34% | 99.75% | 99.72% |
| InceptionResNet V2 based Model | 99.42% | 0.0268 | 99.46% | 99.13% | 99.80% | 99.82% |
| ResNet50 V2 based Model | 99.51% | 0.0219 | 99.50% | 99.32% | 99.83% | 99.85% |
| MobileNet V2 based Model | 98.36% | 0.0518 | 98.40% | 97.99% | 99.45% | 99.46% |
| DenseNet201 based Model | 99.51% | 0.0329 | 99.64% | 99.17% | 99.82% | 99.86% |
| Proposed Model 1 | 99.71% | 0.015 | 99.40% | 99.67% | 99.88% | 99.86% |
| Proposed Model 2 | 99.80% | 0.005 | 99.85% | 99.71% | 99.93% | 99.94% |
| Modified Models | Accuracy | Runtime | Time/Epoch | Total Parameters (Millions) |
|---|---|---|---|---|
| Xception Based Model | 99.42% | 1 h 25 min 46 s | 206 s | 49 |
| Inception V3 Based Model | 99.23% | 33 min 7 s | 79 s | 41 |
| InceptionResNet V2 Based Model | 99.42% | 1 h 14 min 37 s | 178 s | 73 |
| ResNet50 V2 Based Model | 99.51% | 20 min 59 s | 50 s | 35 |
| MobileNet V2 Based Model | 98.36% | 19 min 50 s | 47 s | 13 |
| DenseNet201 Based Model | 99.51% | 42 min 31 s | 101 s | 30 |
| Proposed Model 1 | 99.71% | 2 h 36 min 45 s | 386 s | 49 |
| Proposed Model 2 | 99.80% | 3 h 1 min 36 s | 434 s | 91 |
| Works | Methods | Accuracy | Precision | Sensitivity | Specificity |
|---|---|---|---|---|---|
| [39] | BoF | 98.6% | 97.7% | 99.4% | N.A |
| [40] | Hybrid Classifier | 99.53% | N.A | N.A | N.A |
| [41] | CNN-SVM + Sobel | 99.61% | N.A | 99.80% | 99.56% |
| [42] | MKSC | 98.2% | 97.52% | 98.05% | 97.82% |
| [43] | Bi-level classification | 99.26% | 99.23% | 99.26% | N.A |
| [44] | CNN | 98.28% | 98.22% | 98.25% | N.A |
| [45] | FFB3 | 87.64% | 90.87% | 88.13% | 92.59% |
| [46] | ResNet50 V2 | 95.49% | 96.85% | 99.19% | 98.27% |
| [47] | VGG16 | 91.69% | 92.33% | 95.92% | 100% |
| [49] | EfficientNetB0 | 92.93% | 88.30% | 90.00% | 95.00% |
| [50] | Residual-Shuffle-Net | 97.39% | 97.40% | 97.39% | 98.69% |
| Proposed models | Con RN50V2_MNV2 1 | 99.71% | 99.40% | 99.67% | 99.88% |
| Con Xcep_IncepV3 2 | 99.80% | 99.85% | 99.71% | 99.93% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El Gannour, O.; Hamida, S.; Cherradi, B.; Al-Sarem, M.; Raihani, A.; Saeed, F.; Hadwan, M. Concatenation of Pre-Trained Convolutional Neural Networks for Enhanced COVID-19 Screening Using Transfer Learning Technique. Electronics 2022, 11, 103. https://doi.org/10.3390/electronics11010103
El Gannour O, Hamida S, Cherradi B, Al-Sarem M, Raihani A, Saeed F, Hadwan M. Concatenation of Pre-Trained Convolutional Neural Networks for Enhanced COVID-19 Screening Using Transfer Learning Technique. Electronics. 2022; 11(1):103. https://doi.org/10.3390/electronics11010103
Chicago/Turabian StyleEl Gannour, Oussama, Soufiane Hamida, Bouchaib Cherradi, Mohammed Al-Sarem, Abdelhadi Raihani, Faisal Saeed, and Mohammed Hadwan. 2022. "Concatenation of Pre-Trained Convolutional Neural Networks for Enhanced COVID-19 Screening Using Transfer Learning Technique" Electronics 11, no. 1: 103. https://doi.org/10.3390/electronics11010103
APA StyleEl Gannour, O., Hamida, S., Cherradi, B., Al-Sarem, M., Raihani, A., Saeed, F., & Hadwan, M. (2022). Concatenation of Pre-Trained Convolutional Neural Networks for Enhanced COVID-19 Screening Using Transfer Learning Technique. Electronics, 11(1), 103. https://doi.org/10.3390/electronics11010103










