Abstract
The study of Deoxyribonucleic Acid (DNA) methylation has allowed important advances in the understanding of genetic diseases related to abnormal cell behavior. DNA methylation analysis tools have become especially relevant in recent years. However, these tools have a high computational cost and some of them require the configuration of specific hardware and software, extending the time for research and diagnosis. In previous works, we proposed some tools for DNA methylation analysis and a new tool, called HPG-DHunter, for the detection and visualization of Differentially Methylated Regions (DMRs). Even though this tool offers a user-friendly interface, its installation and maintenance requires the information technology knowledge specified above. In this paper, we propose our tool as a web-based application, which allows biomedical researchers the use of a powerful tool for methylation analysis, even for those not specialized in the management of Graphics Processing Units (GPUs) and their related software. The performance evaluation results show that this web-based version of HPG-DHunter tool improves the response time offered to the user, also offering an improved interface and higher visualization quality, while showing the same efficiency in DMR identification than the standalone version.
1. Introduction
DNA methylation is a representative mechanism of epigenetics. Epigenetics refers to changes in DNA that do not change the actual DNA sequence, meaning that these changes are potentially reversible. In particular, DNA Methylation consists of the addition of a methyl group () to a cytosine, forming a link [1,2]. When this addition happens, that DNA position is methylated. DNA methylation inhibits the expression of certain genes by preventing the proteins responsible for DNA transcription to initiate this process [3]. In fact, it was originally proposed as a “silencing” epigenetic mark [4].
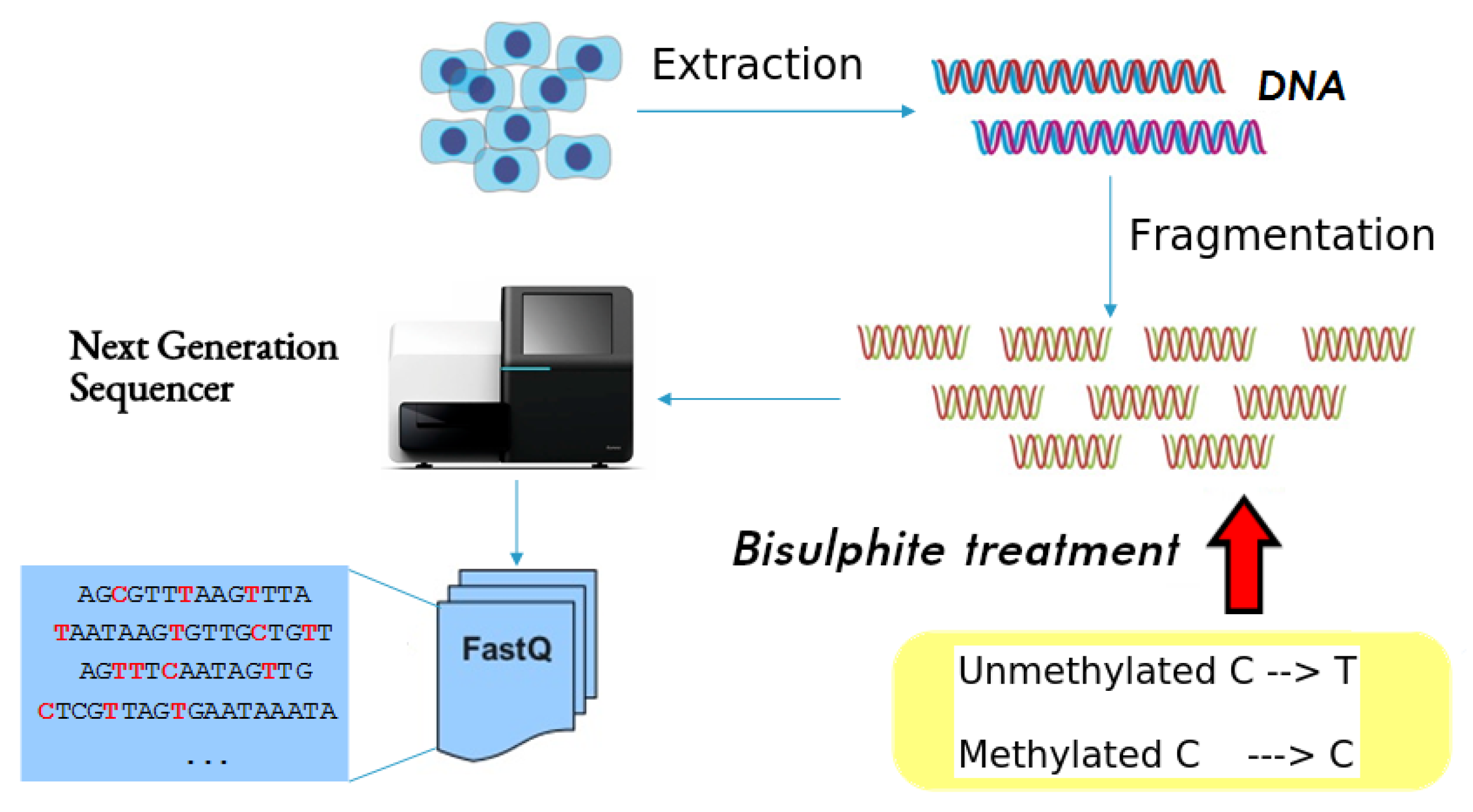
DNA methylation analysis requires a specific treatment of DNA that modifies its sequence, as well as software tools for their analysis. The methylation data can be obtained through bisulfite sequencing, which provides comprehensive DNA methylation maps at single-base pair resolution [5]. Figure 1 illustrates this process. The first step is the DNA extraction. Next, the Polymerase Chain Reaction (PCR) breaks up the DNA sequences into many fragments and makes multiple copies of these fragments, which are called reads and are the actual samples to be analyzed. At this point, bisulfite is added to the resulting DNA reads. Bisulfite treatment converts unmethylated cytosines (Cs) into thymines (Ts), which gives rise to C-to-T changes in DNA sequence after sequencing, while leaving methylated cytosines (5mCs) unchanged. Finally, the bisulfite-treated reads are processed by a sequencer, which reads each read and yields a text containing the sequence of nucleotides in the read, together with other meta-information such as the quality of the read, etc. (this is known as fastq format). Since the sequencer carries out this process with all the reads, the output of the sequencer is a text file in fastq format (usually huge) containing the sequence detected for each sample (read).
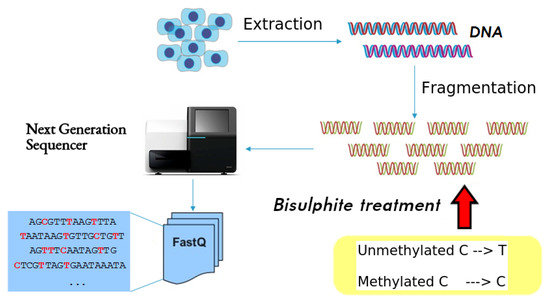
Figure 1.
Bisulfite sequencing process of a single biological sample: after DNA extraction, the Polymerase Chain Reaction (PCR) breaks up the DNA sequences into many fragments and makes multiple copies of these fragments, which are called reads. Then, bisulfite is added to the DNA reads. Finally, the bisulfite-treated reads are processed by a sequencer, which reads each read and yields a text containing the sequence of nucleotides in the read, together with other meta-information
By aligning and comparing bisulfite sequencing reads to the reference genomic DNA sequence, it is possible not only to align the read, but also to infer DNA methylation patterns at base-pair resolution. However, the data to be processed are huge, and a high computational power is required for an efficient operation. For example, the length of the DNA chain in the human genome is nucleotides, and each sample in a fastq file, whose size typically does not exceed hundreds of nucleotides, must be compared to the whole DNA chain of the genome to find the correct location (this is known as the alignment operation). Each fastq file coming from a next generation sequencer can easily contain tens or hundreds of millions reads. Also, typical study cases require the analysis and comparison of different biological samples coming from different tissues or different individuals, to detect different methylation levels in different genes, cells, tissues and/or individuals. These differences are called Differentially Methylated Regions (DMRs). There are a lot of software tools for the alignment of bisulfite-treated samples and their methylation study (Bismark [6], BS-Seeker [7], BRAT-BW [8], and HPG-Methyl [9]). These tools provide the user with single-base methylation results, indicating the absolute methylation level of each nucleotide found in the DNA of the analyzed sample. Most of them yield the results in Sequence Alignment Map (SAM) or Binary Alignment Map (BAM) files. Sequence Alignment Map is a text-based format originally designed for storing biological sequences aligned to a reference sequence, but it can also contain information about the methylation context of each methylated cytosine. The binary equivalent of a SAM file is a BAM file, which stores the same data in a compressed binary representation. Anyway, the results of a methylation analysis are a huge data file with the alignment and methylation results for every nucleotide in the samples.
Therefore, biomedical research of DNA methylation should compare the information of methylation level at different scales (DNA segments, encoding regions, DNA chromosomes, etc.), comparing the results across different biological samples, minimizing the inherent impact of biological variability. With that purpose, different tools have been proposed for the visualization of methylation level and the identification of differentially methylated regions (DMRs) [10,11,12,13,14,15,16]. However, many of these tools, such as BSmooth [10], DSS-single [13] or methylKit [17], are based on statistical techniques, adding a large computational workload to the typical large files with methylation data. As a result, the execution time required for these tools is very large, adding an excessive delay between the sample extraction from bisulfite DNA sequencers and the yielding of the results of the analysis. Moreover, the visualization of the methylation analysis results with the current tools is far from becoming user-friendly. Another disadvantage of these tools is that most of them are R scripts or some Python programs designed to be used through a command line terminal. This feature requires programming and/or computer systems administration knowledge from the users.
In previous works, we proposed the representation of methylated data as a methylation signal, and the implementation of the wavelet transform into the GPU [18], as well as a graphic tool called HPG-Dhunter [19] for an efficient detection and visualization of DMRs with a high level of usability. This tool can identify and display DMRs of different samples at different levels, and it is freely available at grev-uv github site. Nevertheless, HPG-Dhunter [19] was developed as a standalone tool to be installed on a high-performance platform server with new generation GPU devices. Therefore, this tool requires knowledge about GPU installation, setting up and maintenance. This requirement limits its use by biomedical researchers with little or no knowledge about this hardware and related software packages. To remove this limitation, in this paper we propose our tool as a web-based application, which allows biomedical researchers the use of a powerful tool for methylation analysis even for those not specialized in the management of GPUs and their related software. The web application is available at the URL https://fermidi1.informat.uv.es/hpgDhunter. The performance evaluation results show that this web-based version of HPG-DHunter tool improves the response time offered to the user, also offering an improved interface and higher visualization quality, while showing the same efficiency in DMR identification than the standalone version.
The rest of the paper is organized as follows: Section 2 shows the related work about web-based tools for DNA methylation analysis. Next, Section 3 describes the web tool architecture and use cases. Section 4 analyzes the performance of the proposed architecture under several stress tests. Finally, Section 5 present some concluding remarks.
2. Related Work
Most of the web-based existing tools for DMR detection and visualization [20,21,22] are R scripts [16,22], Python applications [21], or C/C++ applications, developed to be used into the RStudio environment or through a command line terminal. Curiously, this implies that the user of these applications needs programming skills. Nevertheless, R and its related packages are very common tools in the biological field, and most of the biological researchers have acquired these skills, so this requirement does not prevent these tools from being widely used.
On the other hand, there are many web-based applications for DNA analysis. Almost all of them are developed as a result of a new approach about a specific research. The rest of them are commercial software packages or software environments where the researcher can deploy new tools. For example, DBCAT [23] analyzes short fragments of DNA, but it does not analyze the whole genome nor the DMRs. Illumina Dragen Bio-IT on BaseSpace Sequence Hub [24] is a commercial package that provide genomic analysis of sequencing data, including methylation analysis and DMR identification. Qlucore Omics Explorer [25] is a visualization-based data analysis tool with built-in statistics that delivers immediate results and provides instant exploration and visualization of big data. In the same way, some other bioinformatics suites with different web-based tools are BABELOMICS [26], a suite of web tools for the functional profiling of genome scale experiments, or MetaCore [27], a web-based bioinformatics suite that allows researchers to upload data analysis results from experiments such as microarray, next generation sequencing, metabolic, SAGE, siRNA, microRNA, and screening. Galaxy [28] is a software system that provides support through a framework that gives experimentalists simple interfaces to powerful tools, while automatically managing the computational details.
Also, there are many specialized genomic tools. For example, for a Gene Ontology-term complex analysis, some visualization tools can be used: ProfCOM [29] is a unique tool with the ability to profile enrichment of not only available Gene Ontology (GO) terms but also of complex functions. GOEAST [30] is a toolkit providing easy to use, visualizable, comprehensive and unbiased Gene Ontology analysis for high-throughput experimental results, especially for results from microarray hybridization experiments. CARMAweb [31] is a tool that performs data preprocessing (background correction, quality control and normalization), detection of differentially expressed genes, cluster analysis, dimension reduction and visualization, classification, and Gene Ontology-term analysis.
There are other tools with only visualization purposes, such as DNAPlotter [32], which generates circular and linear representations of genomes. AutoGRAPH [33] is an interactive web server for automating and visualizing comparative genome maps. KEGGanim [34] is a tool for visualizing experimental data in the context of biological pathways. Another tool called CGHweb [35] generates a heatmap panel of segmented profiles. There are other tools with visualization capabilities which show the result of genome analysis, such as WebGeSTer DB [36], which is a transcription terminator database, or GenSAS [37], which is an online platform that provide a pipeline for whole genome structural and functional annotation for eukaryotes and prokaryotes.
A different kind of web-based tools are focused on alignment. BTW [38] suggests possible biological significance in certain positions in an optimal time warping alignment, computing the Boltzmann partition function and yielding Boltzmann pair probabilities. CRISPI [39] is a user-friendly web interface with many graphical tools and functions which allow the user to extract results, find Clustered Regularly Interspaced Palindromic Repeats (CRISPR) in personal sequences, or calculate sequence similarities with spacers. There are other web-based applications designed for annotation tasks, such as ARG-ANNOT [40], which was created to detect existing and putative new antibiotic resistance genes in bacterial genomes; FLAN [41], designed for genome annotation of influenza virus, or MAKER Web Annotation Service [42], a web-accessible graphical user interface for the MAKER annotation pipeline.
Finally, another example of web-based bioinformatics suite is PhenoGen [43], which is a comprehensive toolbox for storing, analyzing and integrating microarray data and related genotype and phenotype data.
3. Materials and Methods
In this section, we describe the main structure of the web tool that supports both the visualization of methylation signals and the identification of DMRs.
3.1. Infrastructure
We selected the client-server architecture [44] for the application infrastructure, in front of the Service Oriented Architecture (SOA) [45]. The main reason was the ease of use of that architecture for an adequate deployment of our tool. Additionally, we considered two commonly used formats to transfer data as XML [46] and JSON [46] (JavaScript Object Notation) interpreted natively by JavaScript [47].
The web application has been designed following the widespread criterion of separating the front end (user-side infrastructure, where the user interface and control logic are located) and the backend (the server-side infrastructure hosting the external services for processing the front-end requests). As any web-based application, the front end runs on the user’s browser, using HTML5 [48] and CSS [49] to format the content in the browser interface. It also uses JavaScript as the programming language to capture the events of the interface and establishing the communication with the backend when an external service is needed. To ensure the lowest possible latency when there is a request to transfer a large amount of data (biological data usually require huge datasets), we have selected the WebSocket protocol over a permanent TCP connection, because of its flexibility, bidirectional communication and high transfer rate. Also, we have chosen JSON for data exchange because of its simplicity, lightness and native processing speed within the JavaScript language.
We analyzed three frameworks to facilitate the design and logical control with JavaScript: Vue.JS [50], a light and framework for quick development; React [51], a view-oriented library, and Angular [52], based on a powerful compiler called TypeScript. We selected Angular as the most appropriate option, since it allows all the development in a single framework, and the parameterization of the internal architecture is hidden. In particular, we used Angular Material (https://material.angular.io/), the version for Angular from Material Design (https://material.io/design), which is the library created and used by Google for the interface of its applications. We used Material for our interface (organizing the different elements in the page, design of forms, windows, etc.) except the window for the graphical representation of data. For that window, we used ng2-charts (https://valor-software.com/ng2-charts/), a library for displaying data in different types of graphics.
Regarding the backend, the kernel of the whole system is HPG-Dhunter [19], which is programmed in C++ language. The wavelet transformation of the methylated signal in the GPU device and the identification of DMRs were developed with Qt [53] framework (LGPL3 Open-Source License). This framework can play the role of a server through the WebSockets protocol, which efficiently works with JSON format. Therefore, it was not necessary to suggest a migration of the kernel to another framework or programming language. However, the deployment of HPG-Dhunter as a web application required services for user access control and the loading of the input files with the methylated data. We decided to implement these functions in a separated module, to avoid a latency increase. Thus, the backend is shaped as a web-based application and two complementary services which solve the main needs of the proposed web application: the Qt server for the main logic of the service, a server for file management, and a server for user management. Since the front end is developed in JavaScript, we decided to use Node.JS [54] for file management.
Regarding user access control, we used Firebase Authentication [55], an authentication service developed and maintained by Google, since it was easily integrated within the interface through a secure https connection, and it allows federated identification, which reduces the user interaction.
Summarizing, the development of HPG-Dhunter as a web-based application relied on Angular, JavaScript, JSON, HTML5 and CSS for the front-end side and communications. The backend was based on Qt, Node.JS and Firebase Authentication, using WebSocket as a communication protocol over TCP within the client-server architecture. HPG-Dhunter was developed with the Qt framework, designing the user interface according to the MVC software development principle.
3.2. Architecture
To guarantee an adequate usability of this service, a new web interface based on the local version of HPG-Dhunter has been developed for the front end. Since the methylation samples are sensitive data, we have added a high degree of security to this service. To achieve this goal, individual access with a username and password is forced, establishing a secure connection between the user database, the interface and the rest of the system. The data to be analyzed for each user is stored in independent and isolated folders, hidden from the rest of the users. The user uploads the data in the output format of the HPG-Hmapper [56] tool if the upload option is enabled.
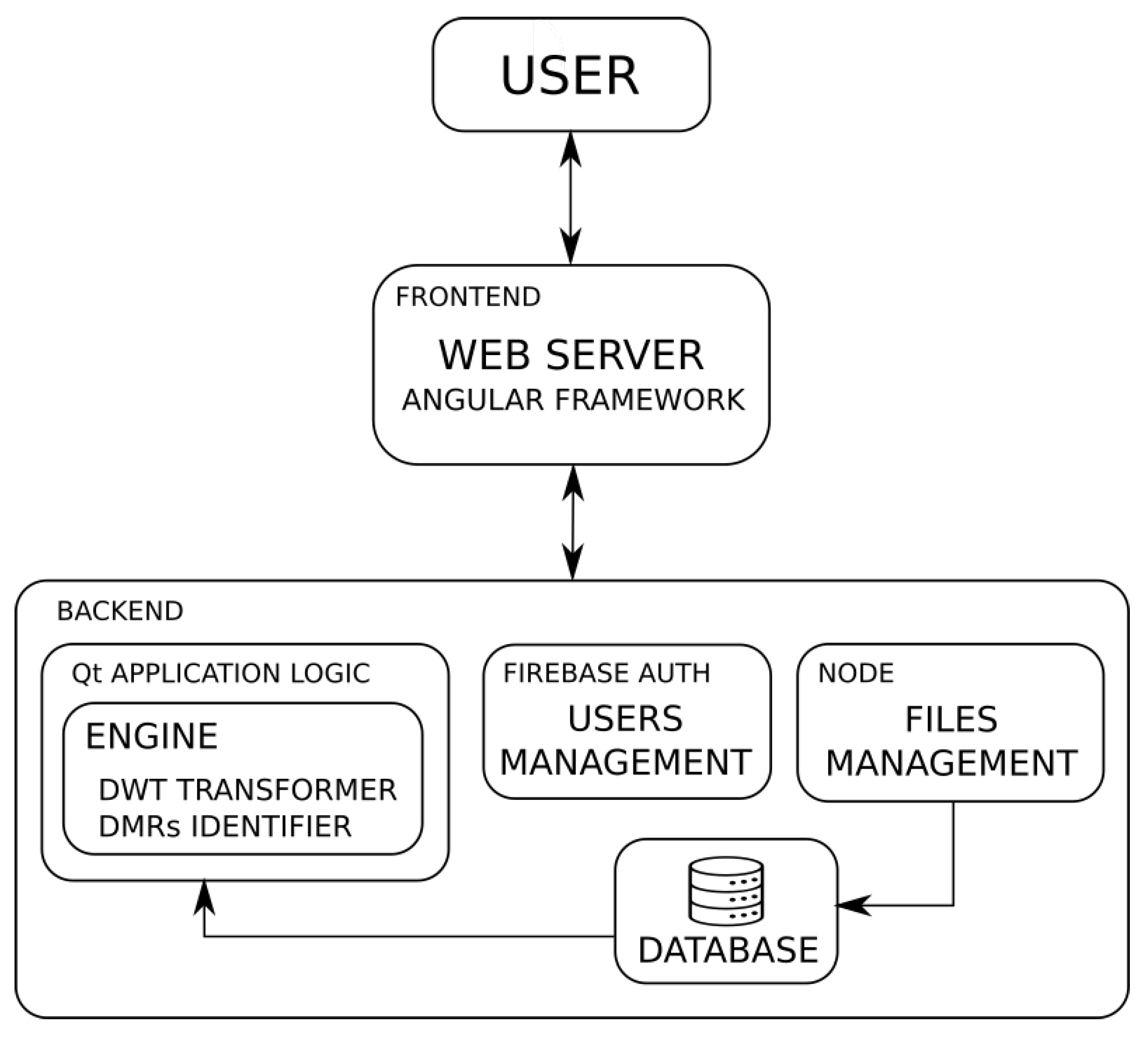
The basic architecture of the system can be seen in Figure 2. The user interacts with the system through the front end downloaded to the client browser. The front end is responsible for establishing communication with the backend servers, depending on the services requested by the user. The backend includes: the authentication service that will validate the user, the file manager that provides access to the data loaded by the user, and the main service of wavelet transformation and identification of DMRs. At the server side, the database module is responsible for the file management, and it provides the application logic module with the required data, according to the user requests.
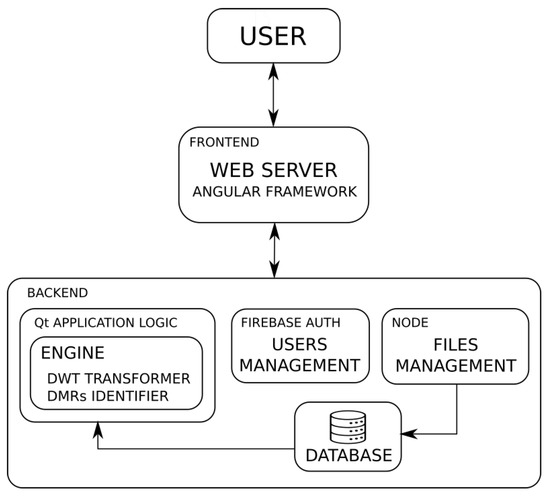
Figure 2.
Architecture of the web-based application.
The front-end interface is intended to provide a level of usability equal or greater than the standalone version of HPG-Dhunter. However, taking into account the important difference in data transmission rate between the standalone version (where data are transmitted along the PCI bus) and the web application (where the data are transmitted along a TCP connection through internet), the number of data to be displayed has been limited to 1024 points of the transformed signal. Depending on the transformation level, these 1024 points represent a different number of DNA locations, according to Equation (1), as explained below. Nevertheless, this limit can be adapted to the resolution of the user device. Additionally, after each request (assuming that the next user actions will require neighbor regions) the front end automatically makes a request for another 1024 adjacent data, so that if the user requires displaying these contiguous data, they will be already located (cached) in the client computer, improving the response time from the server. In this way, a buffer of 2048 points of the transformed signal is always available. The width of the display window is limited to 1024 points of the transformed signal. This width will represent a segment of the chromosome whose length is computed according to the expression (1), depending on the level of transformation set by the user.
The DMR identification is carried out after the desired analysis parameters have been set by the user. Like in the standalone version, these parameters are the transformation level, the minimum coverage, a validation threshold of the identified DMR, the percentage of samples per group with equal or higher coverage than the minimum coverage, and the minimum percentage of positions with minimum coverage per position window corresponding to the transformation level. Once the system has identified the DMRs, then the user can select any of them to be displayed in the front-end interface. The system can display any DMR with any methylation level of every sample uploaded by the user.
Figure 2 shows the three services hosted by the backend. The standalone tool HPG-DHunter performs the wavelet transformation of the data and identifies the DMRs among the data groups selected by the user. In the web application, there is no restriction on the number of samples per group, since that constraint was imposed in the standalone tool by the GPU memory size. However, the web application visualizes the data at the client web browser, and it uses the server GPU memory only for computation purposes. This feature allows the batch computation of Discrete Wavelet Transform (DWT) when it is needed. The system previously checks if the requested segment of all the samples to be processed in the GPU fits in the GPU memory. Then, a direct processing operation is performed on these data, and the results are dumped from the GPU’s shared memory to the server’s main memory, to be progressively transferred to the client side. Otherwise, a batch operation is planned, storing the intermediate results in a temporary cumulative matrix in the main memory of the server. This matrix is downloaded to the client browser at the end of the process. This solution is applied to both the visualization part and the identification of DMRs. In addition, a new strategy is proposed in the establishment of the lower and the higher end of the segment to be analyzed. Due to the particular way in which the multilevel wavelet transformation is computed, and to guarantee consistency in the results of the wavelet transformation, the starting and ending point of the segment requested by the user will be modified according to the expressions (2) and (3).
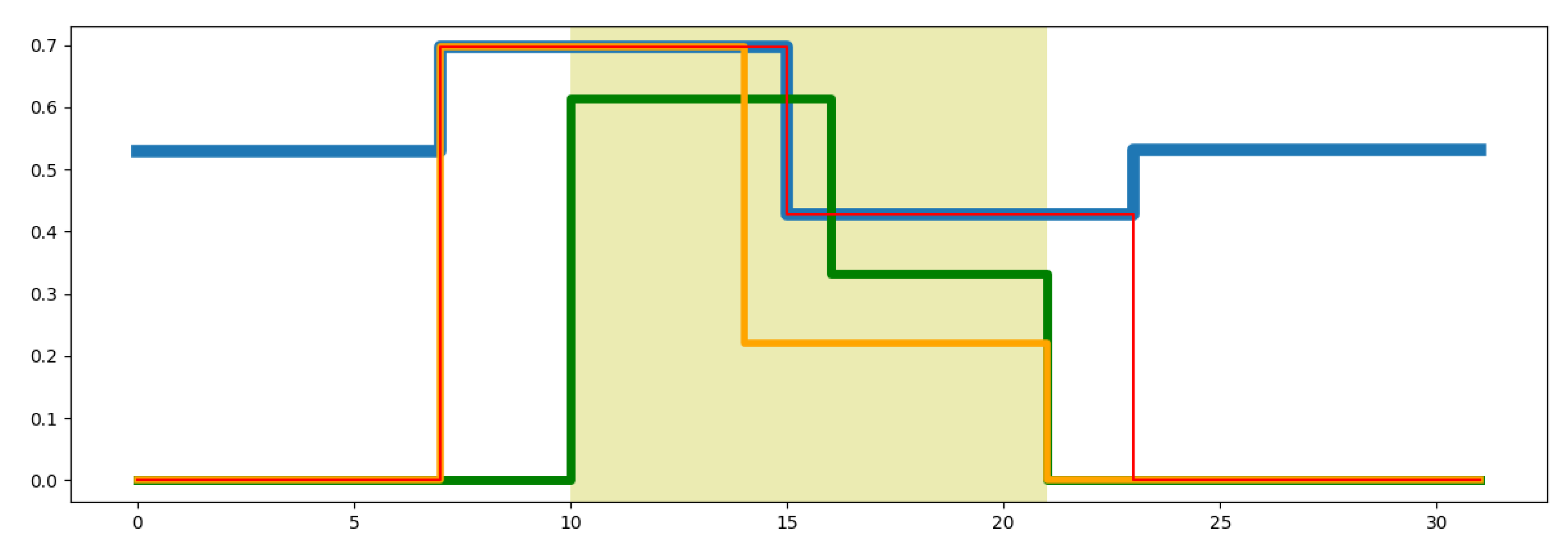
Thus, the starting and ending point of the region where the wavelet transformation is applied will always be the closest exponential position to the lower and upper positions requested by the user. Figure 3 illustrates an example of how this strategy makes a consistent visualization of the segments requested by users with those segments obtained from the DMR identification. Figure 3 shows an example of the limits for the wavelet transformation (WT) requested by the user (shaded region) at level three and the actual limits computed by the system. The blue line represents the methylation signal used to identify DMRs. The green line represents the WT of the requested region by the user. The orange line represents the WT region performed by the system after the adjustment of the lower limit. The red lines correspond to the portion of the methylation signal over which the wavelet transformation has been applied. This Figure shows that there is a guard band between the red lines and the limits requested by the user (shaded region).
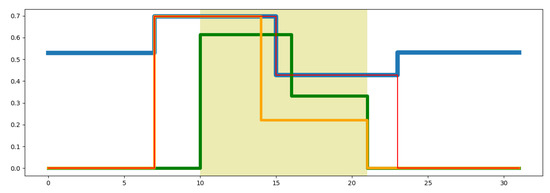
Figure 3.
Example of wavelet transform.
The backend is complemented with two more services. On the one hand, the system authenticates the user either as a guest user or as a registered user by email or by federated authentication with Google, Facebook or Twitter. This is supported by the authentication service of the Firebase framework. The communication between the front end and the authentication service is done under the https protocol, guaranteeing a secure communication between them. Also, the system offers a file management service with a simple drag and drop interface. This service is developed using NodeJS, and it establishes a secure communication with the front end using the https protocol. It must be noted that these samples must be uploaded in the HPG-Hmapper [56] file format, and they must be organized in folders. All the samples are saved in the server side in a directory whose access is only allowed to the corresponding user, remaining inaccessible to any other user, i.e., the proposed web application can be used with any other application for DNA methylation analysis (like Bismark, etc.) which yields SAM or BAM files. The only requirement is that the SAM or BAM files are pre-processed with HPG-HMapper (https://grev-uv.github.io/) before being uploaded.
3.3. Concurrent Access
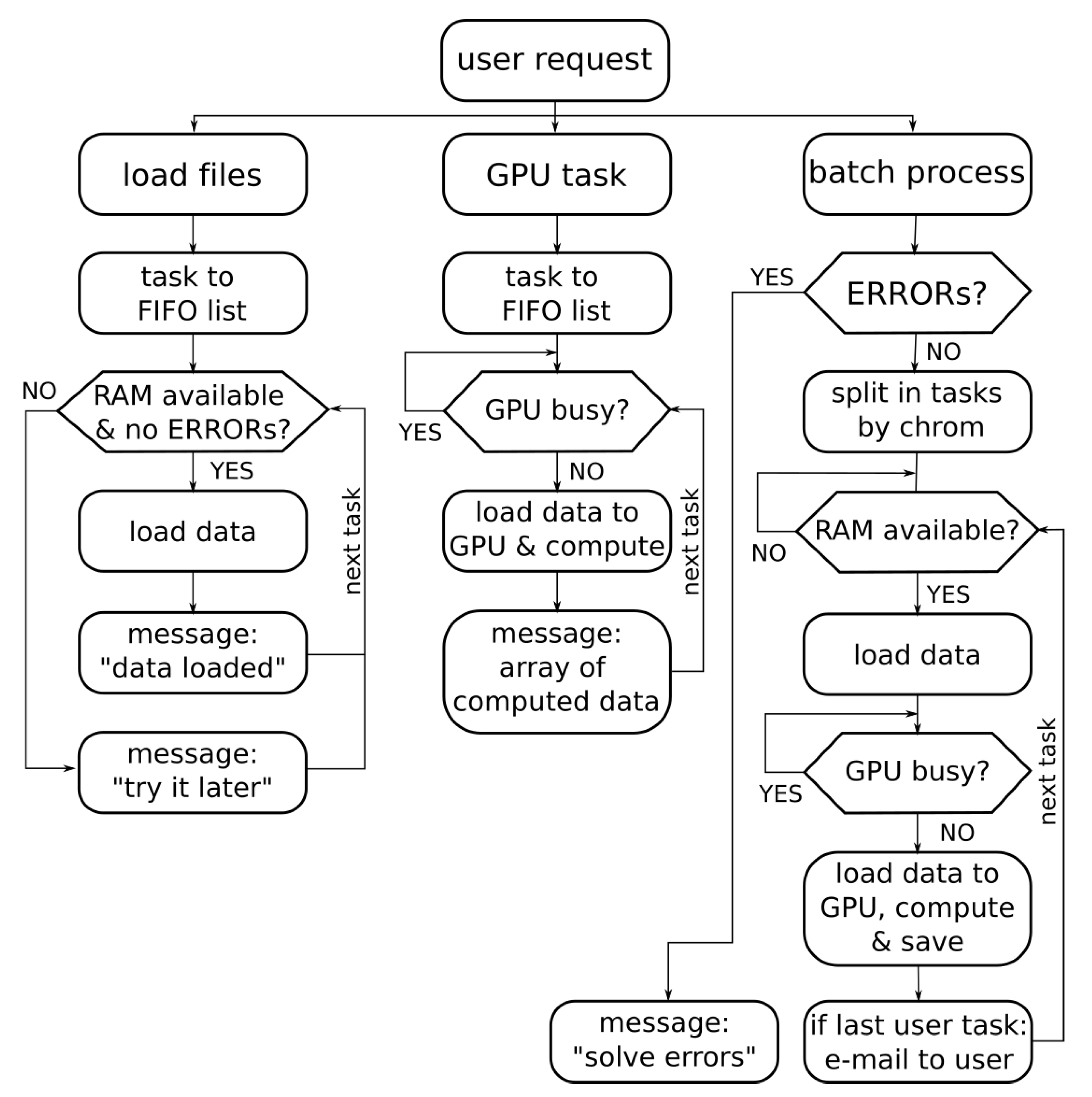
The proposed web-based application must be capable of managing the concurrency of several tasks requested by different users. Thus, the backend system must manage the RAM memory availability for loading the data from the files and the GPU state (busy or stopped) for the GPU computation tasks. Figure 4 shows the flow diagram for concurrent requests. The management of each request depends on the request type (load files, GPU computation or batch process). Each type of request has a different FIFO list of tasks. From these task lists, independent processes are launched. The procedure for loading files (in the visualization tab) do not need GPU availability but RAM memory, and therefore it is necessary to check the RAM memory before starting this process. The GPU tasks implicitly required in the visualization tab in turn require the use of the GPU, so the GPU state must be checked before loading the data array and computing the DWT of the methylation signal. Another independent process that can be requested by the user is the batching process (in the user interface tab labeled as “batch process”). This is a long and non-interactive procedure, so in this case the system sends an email to the user when the process ends, and the procedure is split into smaller tasks. All these strategies are focused on achieving a better performance in the GPU and RAM memory use when there are concurrent accesses to the server.
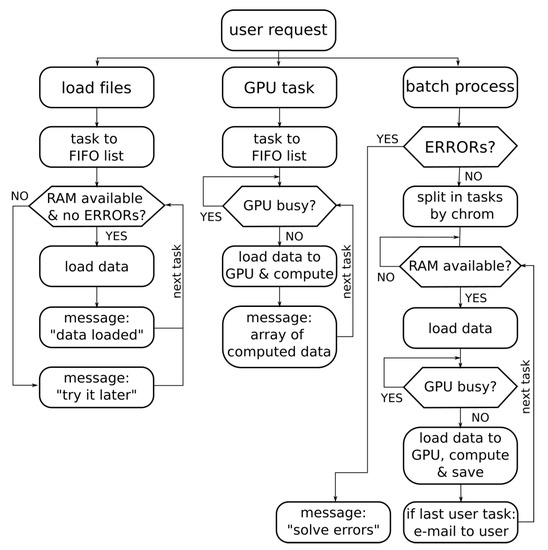
Figure 4.
Flow diagram of the concurrent requests management.
3.4. User Interface
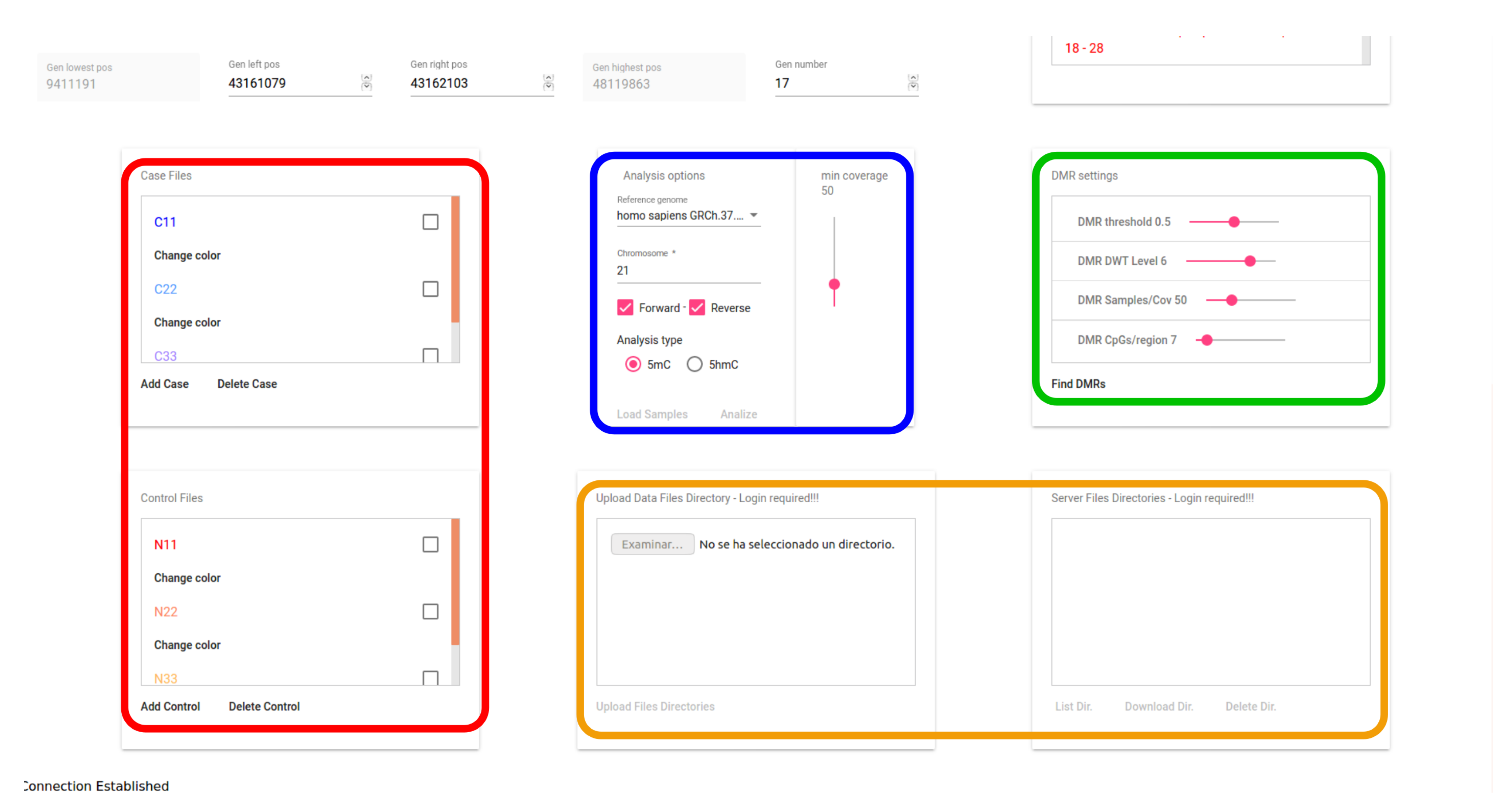
The user interface of the proposed web-based tool has been defined for two user roles: the guest role, which allows interaction with a demo database available in the system, and the registered user role, which allows a full interaction of the user with the system through the interface, using the WebSocket protocol. A registered user can upload any sample file to the server via secure https. Figure 5 shows a snapshot of the user interface, including the tab for selecting the files to be stored in a secure database. This figure shows, marked with a red line, the area including the tabs for selecting the files corresponding to the case and control samples to be visualized and analyzed. The initial parameters applied to the loaded samples are marked with a blue line. The interface area marked with a green line shows the controls for setting the parameters used for DMR detection. Finally, the area marked with an orange line highlights the area for the uploading service (only for registered users).
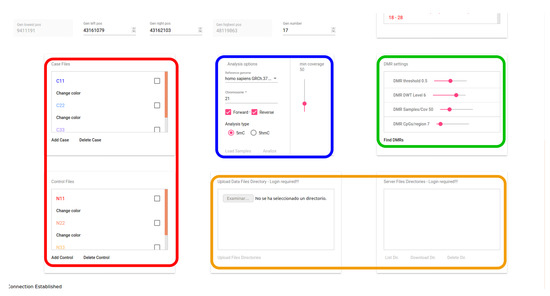
Figure 5.
Snapshot of the user interface with DMR parameters.
The main difference between a guest and a registered user is that a registered user can interact with his/her own data, while a guest user can only interact with the test dataset. Both users can select the files for the case and control groups, and both can also select the chromosome to be analyzed, if the data correspond to methylation or hydroxymethylation data, and which strand to visualize (forward, reverse or both). Once the selection has been made by the user, the system will display first the wavelet transformation level which corresponds to the expression (4), where the maximum transformation level is lowered by three units so there are two more levels available that can be immediately displayed.
At this point, the user can refine the visualized area, selecting the level of the wavelet transformation and the segment of the chromosome of the selected samples to be displayed. The user can also change the chromosome segment to be displayed at any time. Regarding the identification of DMRs, the user must set the desired parameters for the identification process (Figure 5, marked in green): the level of transformation to be used, the minimum threshold to characterize a DMR, the minimum coverage, the minimum percentage of samples per group with minimum coverage, and the minimum percentage of positions per window of positions with minimum coverage.
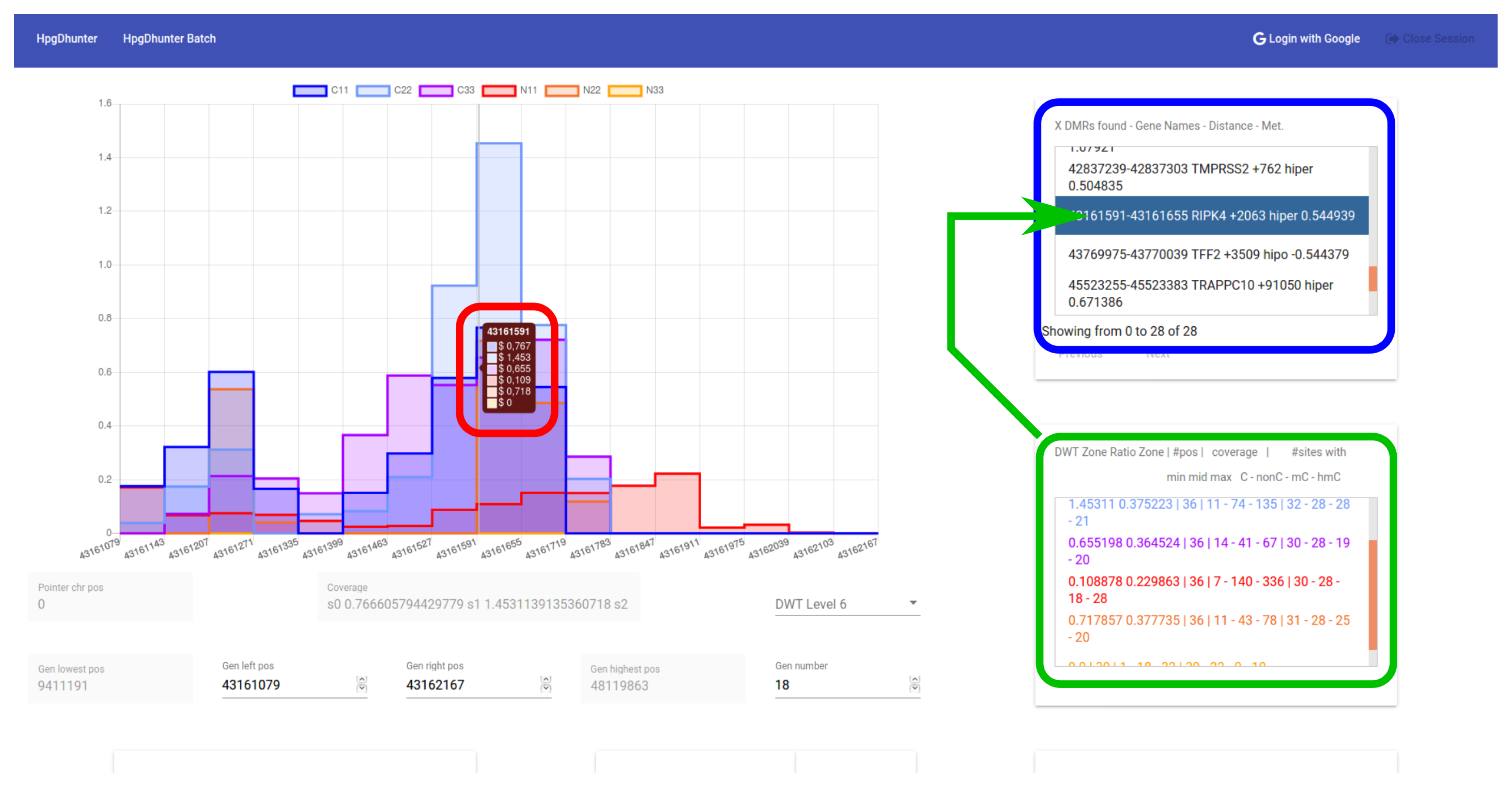
Figure 6 shows a snapshot of the user interface, including the list of segments returned by the server with the average methylation difference between the two groups (case and control), according to the parameters set by the user. The interface area marked in blue shows the list of all DMRs found by the server. The interface area marked in green shows information about the ratio, coverage and position of the selected DMR. Finally, the interface area marked in red shows the coverage ratio of each sample at that position and the wavelet transformation level. From the list of the DMRs detected, the user can select each one of them to visualize the methylation signals of the samples at that area. The user can interactively change the transformation level, zoom and scroll in the horizontal direction, focusing his/her attention on any area. Finally, the user can access to more specific information of any desired position by clicking on the left mouse button at that point of the graph. The user browser will open a new tab with the Ensemble Organization web page: grch37.ensembl.org, showing a 1000 bases width area centered at the clicked position.
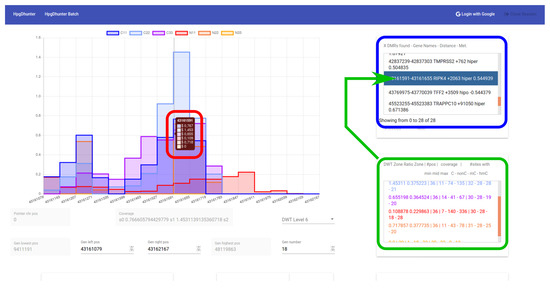
Figure 6.
DMR identification. Visualization of wavelet signals of samples from a selected DMR.
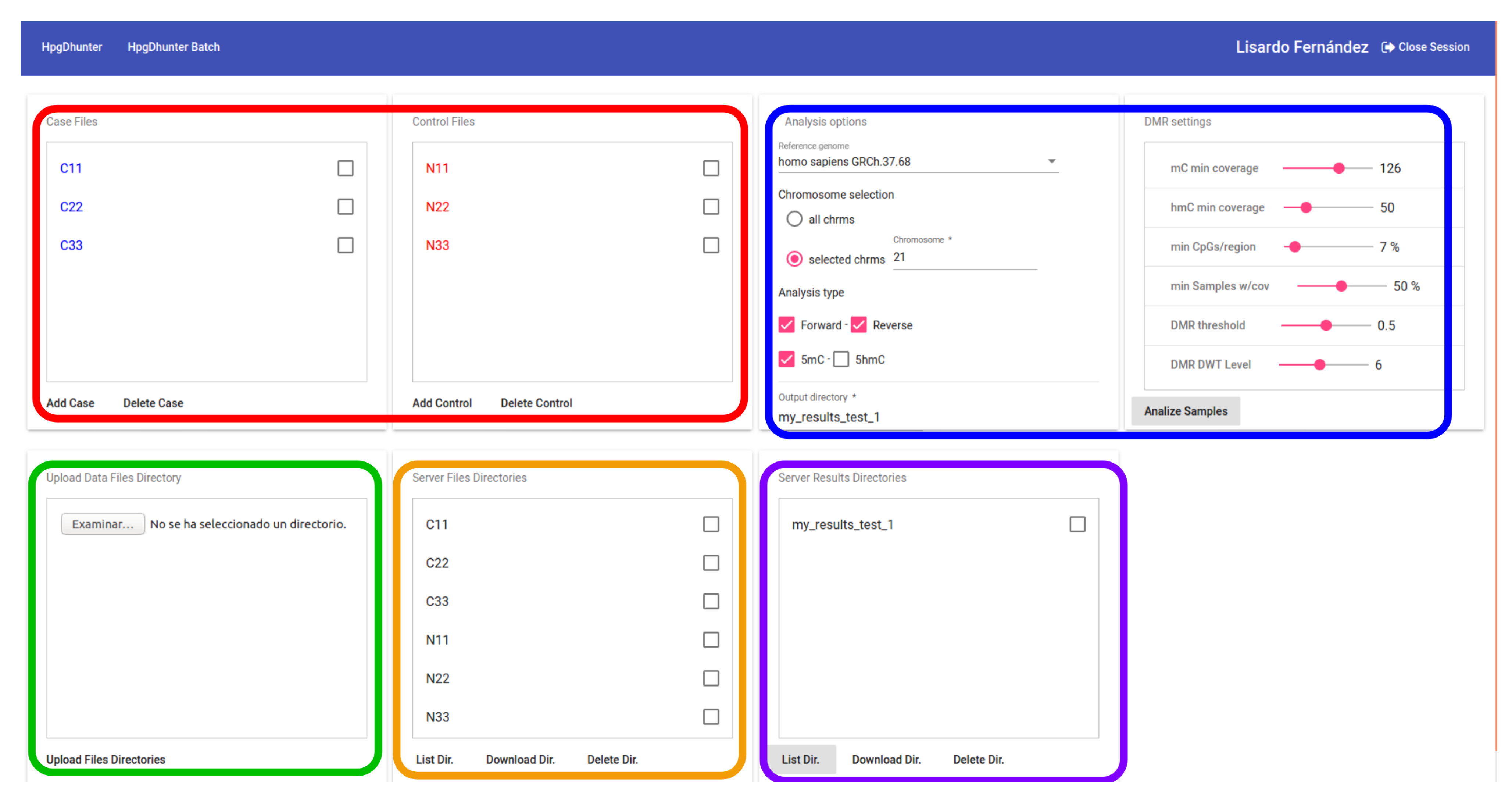
In addition, a registered user can request the identification of the DMRs of the whole genome by the batch option. Figure 7 shows a snapshot of the user interface for this option. In this figure, we have marked in green the area showing the local sample files to be uploaded. We have marked with an orange line the user directories uploaded to the system. The red line shows the interface area showing the control and case samples to be compared to identify DMRs. We have enclosed in a blue line the area showing the parameters for DMR identification. Finally, we have marked with a purple line the area showing the server directory where the results are saved (this tab window is not for visualization purposes, but for the identification of DMRs of all the chromosomes that the user needs, yielding the results in files ready to be downloaded). Since the batch DMR detection process may take a large time to be completed, the system shows a message informing the user that an email will be sent when the process is finished and the files containing the results are ready to be downloaded.
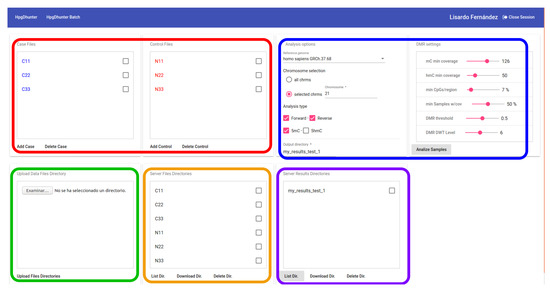
Figure 7.
DMR identification in a batch process. This option is only available by registered users.
4. Results and Discussion
In this section, we evaluate the performance of the web application in terms of latency, comparing the interactivity of this application against the standalone version, to prove that the web application can yield at least similar or better user experience. To avoid any interference with other applications and/or users, the performance evaluation has been carried out with a single user although the backend system for computation tasks is designed to manage concurrent users.
We have used two previously validated datasets for evaluation purposes: the first one is BsseqDataset [10,18], which contains six samples from different real patients. These samples are sorted in two groups, colon cancer tissues and normal tissues, with 380,000 methylated DNA positions of chromosome 21. The second dataset was provided by the “Instituto de Investigación Sanitaria del Hospital Clínico de Valencia” (INCLIVA), from a medical study about the effects of DNA methylation in patients with Diabetes Mellitus 2 (DM2). The samples used in this test were extracted from four different patient groups, and they are denoted as A, B, C, and D. We used only groups A and D for this test to compare the results with the same test shown in [19]. Group A denotes those patients who do not suffer from DM2 and do not show resistance to insulin treatment. Group D denotes those patients suffering from DM2 and showing resistance to insulin.
The hardware platform used as the web server consists of a computer based on the Intel® Xeon® CPU E5620 @2.4 GHz processor, with 8 cores and 2 threads per core, and 24 Gb RAM memory, as well as a GeForce RTX 2070 graphic device, with 2304 NVidia cores and 8 GB GDDR6 RAM memory. The procedure steps implemented in the GPU have been carried out using the CUDA V10.2.89 library. The client browsers to communicate with the server were Mozilla Firefox and Google Chrome, and they were installed in personal computers. The first test was executed with the client computer located in the same university campus network, and the second test was executed from a personal computer out of the campus network, but in the same metropolitan area.
The main parameter values for the first test (the first dataset) were coverage ratios one and five, 0.25 and 0.3 as threshold values and DWT levels five, six and seven. Combining these values, 12 different experiments were carried out. Table 1 shows the evaluation results. This table shows the number of experiments in the column labeled as “exp”, the coverage of each experiment in the column labeled as “cov”, the threshold value in the column labeled as “thr”, and the DWT level in the column labeled as “dwt level”. The rest of the columns shows the evaluation results: the number of DMRs found are shown in the column labeled as “num DMRs”, the processing time in the web server is shown in the column labeled as “process”, the response time provided to the user webpage is shown in the column labeled as “response”, and the communication delay (in milliseconds), i.e., the difference between the response time and the server processing time for each experiment, are shown in the column labeled “comm delay”.

Table 1.
Evaluation results for very low coverage value of chromosome 21 (BsseqDataset).
The number of DMRs detected (values in column “num DRMs”) is high due to the very low coverage ratios used. Nevertheless, it can be seen that for a coverage ratio of 5 the number of detected DMRs is about one third of the number of DMRs detected with a coverage ratio of 1, where there are a high number of samples with a single occurrence of a read aligned in that DNA location. If we compare the number of DMRs found for different dwt levels, we can see how the number of DMRs found decreases as the level of DWT increases. This is due to the compression of the signal as the transformation level is increased. All the times shown in the rest of the columns are directly related to the number of DMRs found, as it could be expected. Thus, the response times are shorter for a coverage ratio of 5, where a maximum response time of 1182 s is needed. Although these times cannot be rated as interactive, they are efficient times if we take into account that we are performing an automatic detection of all the DMRs in a chromosome. Also, it is worth mention that for a coverage ratio of 1 the fraction of the response time devoted to computation (column labeled as “process”) is between 0.64 and 0.75 for the different levels of DWT, while this fraction is about 0.85 for a coverage ratio of 5.
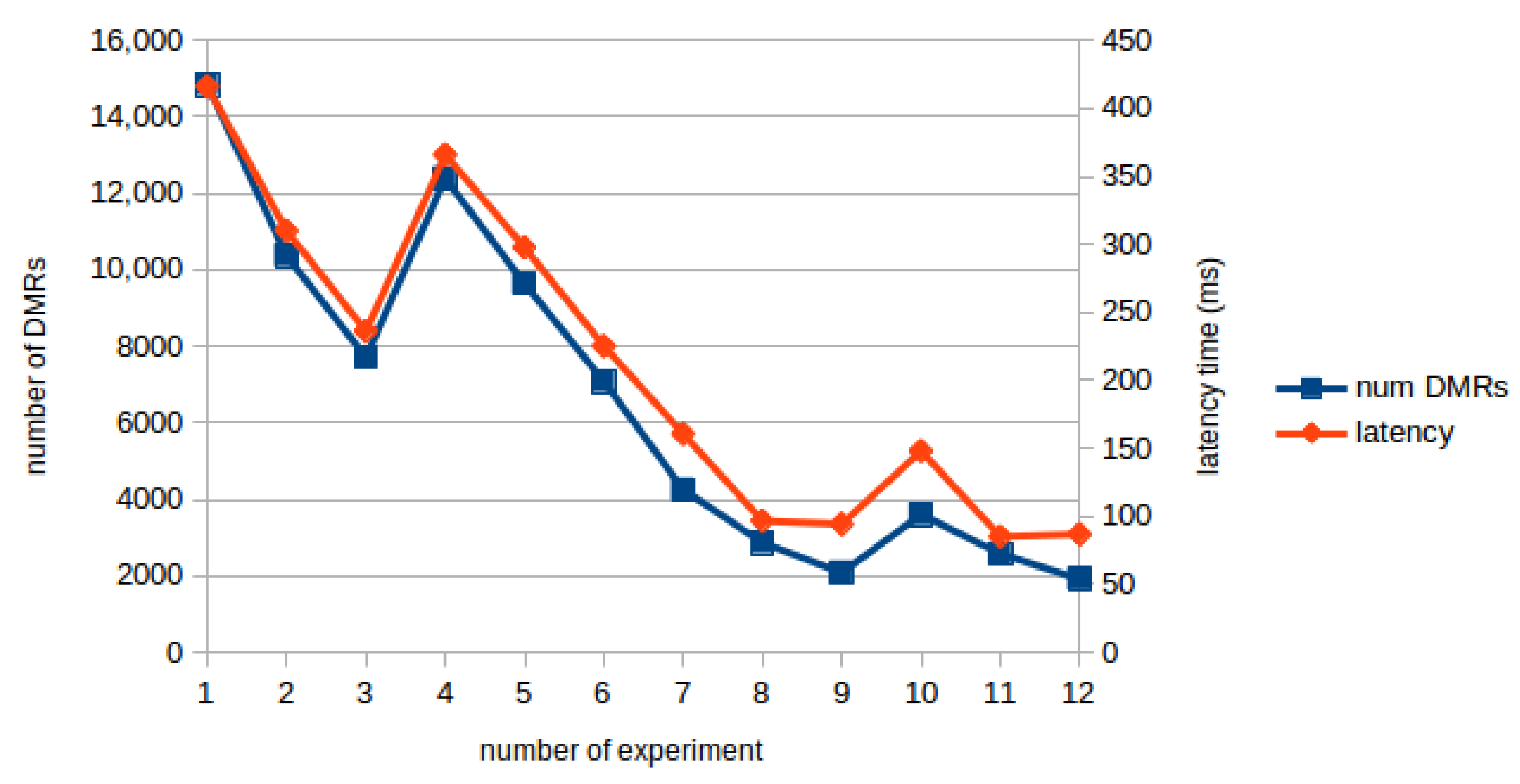
Figure 8 shows the communication delay in milliseconds and the number of DMRs found in each experiment. This figure shows a clear correlation between the two variables, showing that the communication delay corresponds to the transmission of the results from the server to the client. In fact, the number of DMRs identified is the main responsible for the increase of communication delay. Nevertheless, even in cases of many DMRs the communication delay time is no longer than half a second.
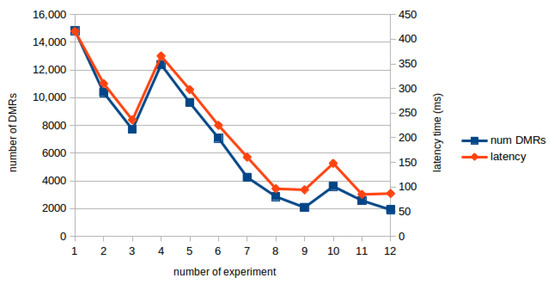
Figure 8.
Communication delay and number of DMRs correlation.
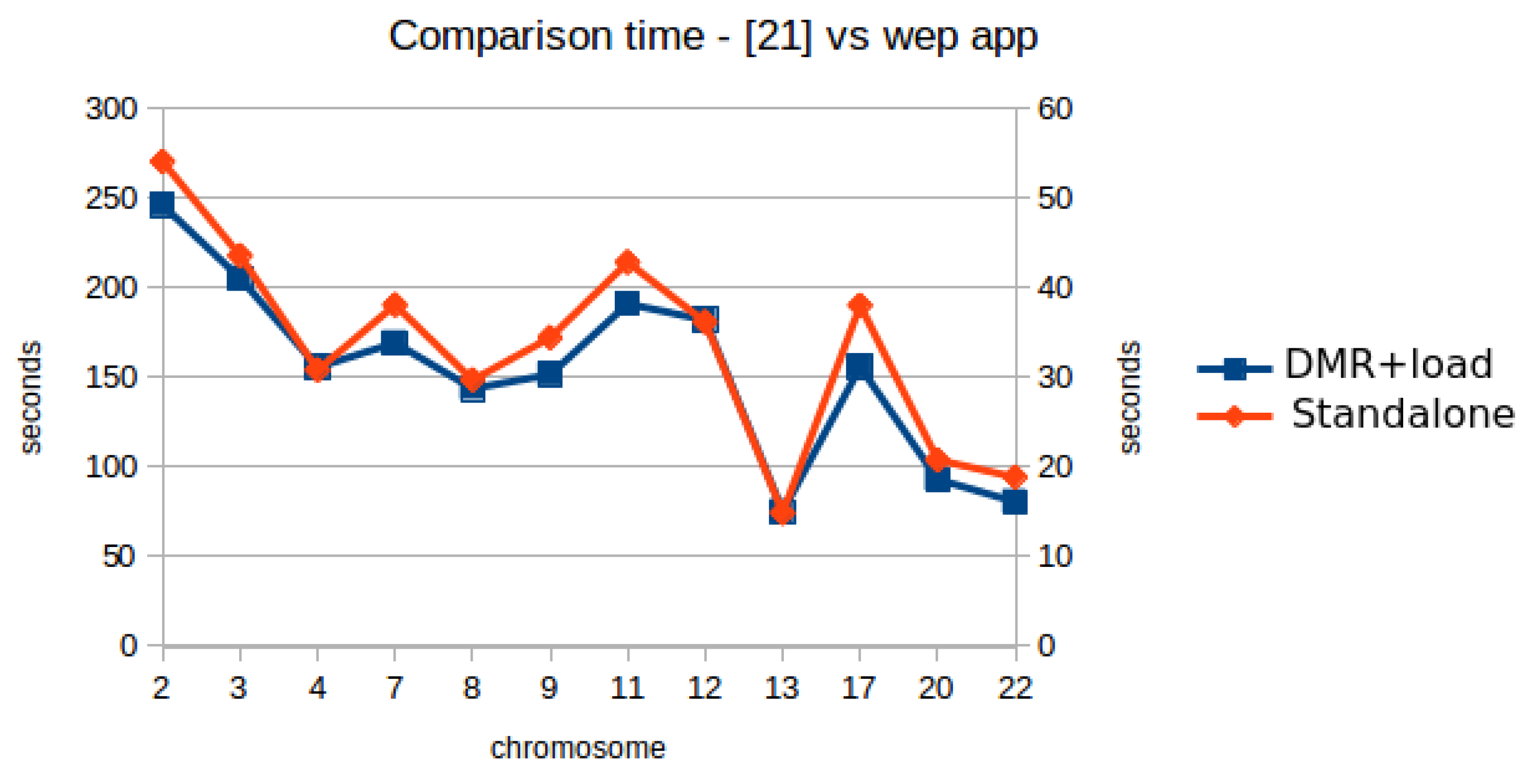
The second test analyzes the differential methylation levels between two groups of samples coming from the INCLIVA database, A and D. The selected chromosomes were indicated by the INCLIVA staff as the most likely chromosomes to show DMRs on genes of interest. In this case, the parameter values were set as follows: a DMR threshold of 0.35, a DWT level of 6 and a minimum coverage of 50. Table 2 shows the evaluation results. The leftmost column, labeled as “chrom”, shows the chromosome number. The column labeled as “DMR server” shows the time (in milliseconds) required by the server for identifying DMRs. The column labeled as “DMR client” shows the time between the request from the user and the reception of response by the user, i.e., the response times perceived by the user. The column labeled as “comm delay” shows the communication delay between the processing time in the server and the response time by the user. The column labeled as “num DMRs” shows the number of DMRs found. The column labeled as “load files” shows the time dedicated to load the files selected by the user from the HDD to the RAM memory. The column labeled as “T1 DMR + load” shows the total time of “DMR client” plus the time needed to load the files. The column labeled as “T2 [19] Table 13 A–D” shows the processing time of A–D datasets analysis in Table 13 at [19], to compare the performance of the web application version with the standalone version of the tool. For comparison purposes, the last column, labeled as “speedup T2/T1”, shows the speedup between the total analyzing time in the web application (T1) and the processing time of the standalone version [19] (T2) for the same group of samples.

Table 2.
Response times for detecting all the DMRs in 12 chromosomes of real dataset (milliseconds).
Table 2 shows that the longest response time (around 23 s) corresponds to the detection of DMRs in chromosome 2. These response times show that the web application can efficiently provide the user with the list of automatically detected DRMs. Moreover, the column labeled “speedup” shows that the web application pre-processes the data more efficiently and achieves an average speedup of 5.54 in the total time of analysis, including the time for loading the files, with respect to the analysis carried out in the standalone version [19]. This significant speedup is due to some improvements in the web-based version. On the one hand, we have improved HPG-Hmapper tool [56] to filter the methylation map by the coverage ratio requested by the user, generating a new mix of forward and reverse strands files. With these new features, the size of the input files for the web application is significantly reduced in regard to the analysis carried out in [19], and the mixing process of the forward and reverse strands is carried out offline, i.e., before the process of loading the files in the HPG-Dhunter server. In this way, the time to load the files to the web server is significantly reduced. On the other hand, we have added several improvements in the HPG-Dhunter algorithm which performs a better use of GPU memory and RAM memory, improving the computation times. First, the backend of the web-based application does not use OpenGl for the visualization of data in any monitor. Instead, the visualization takes place in the client computer. As a result, the GPU is exclusively dedicated to DMR detection, and much bigger data chunks can be stored in the GPU memory and processed in parallel, thus decreasing the total required time. Second, all the methylated locations in the selected samples are stored in a matrix when loading the input files. Instead of computing each time the number of methylated locations in the considered DNA segment in the process of DMR detection, this huge matrix is checked, significantly reducing the computing time for DMR detection. These two improvements significantly reduce the response time of the web-based application.
Figure 9 shows a comparison between the performance evaluation in [19] and the same test done with the web application (user waiting time for DMRs detection plus time required for loading files, the values in the column labeled as “T1 DMR + load” in Table 2). The scale on the right side of Figure 9 (Y-axis) correspond to the values shown in Table 2. The scale on the left side of Figure 9 correspond to the values for the same metric shown by the standalone tool [19]. It can be seen a very similar shape in the two curves, due to the type of files for each chromosome. However, a significant speedup is achieved by the web-based application (around 5×, since the values in the scale of the left side are five times higher than the ones on the right side). Therefore, we can state that the web-based application improves the user experience regarding the standalone version. Table 2 also shows the communication delay in milliseconds and the number of DMRs identified. The behavior of these variables is the same of the first dataset, where the communication delay has a direct relation with the number of DMRs identified.
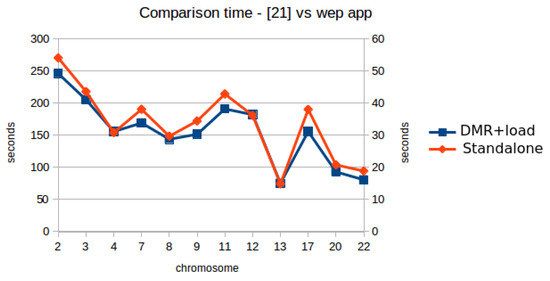
Figure 9.
Comparison between the performance evaluation in [19] and the same test with the web-based application.
Finally, we have measured the response time provided to the user by the web application for the visualization of a DMR, i.e., once the DMR identification is done, we have measured the time required for receiving from the server all the necessary data for drawing in the client computer the DMR the user wants to visualize. We have measured this response time for visualization under two different configurations: the first case is when most of the response time is due to the processing time in the server and the network connection does not add significant delay. In this case, we have located the client computer in the same network (a 100 Mbps Ethernet network) where the server is located. In the second configuration, the client computed is placed in a different LAN, with a domestic internet connection, although geographically located in the same metropolitan area where the server is located. In both configurations we have taken 30 measurements, to have statistically significant values. The average time measured in the first configuration is 350 milliseconds. Regarding some references in the field [57,58], this response time can be considered to be an immediate response-time. The average value obtained in the second configuration is 533 milliseconds. These results show that the web application can provide immediate response times also for the visualization of the results, and the delay in drawing the required DMR in the client computer will depend on the network connections.
5. Conclusions
In this paper, we have proposed the implementation of HPG-Dhunter [19] as a web-based application for identifying and visualizing DMRs in the DNA coming from different samples. The tool has been installed on a server, together with a file server and an authentication server. The adaptation of our tool to the new web environment has helped to improve some sections of the software. Furthermore, the web-based version improves the efficiency in the identification of DMRs regarding the standalone version, because in the web-based application the GPU is fully available for computation tasks exclusively, and the visualization of the DMRs found takes place in the client computer. These improvements result in an average speedup of 5.54 regarding the standalone version. The data transmission through a LAN is included in the total response time, showing that if the network connection does not add significant delay, then the response time offered by the server is better than the one offered by the standalone version. Also, the response time offered by the web-based application to DMR visualization requests coming from the client computer can be considered to be immediate. Thus, the proposed web-based application avoids the need for user skills or knowledge about GPU installation, setting up and maintenance, which prevented this tool from being used by biomedical researchers with little or no knowledge about this hardware and related software packages.
As a future work to be done, we plan to use an array of GPU devices to support massive multiuser access to the web application.
Author Contributions
L.F. and R.O. have programmed the necessary changes in the software. M.P. has designed the tests batteries, and J.M.O. has leaded and supervised the writting of this paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Spanish MCIU and EU ERDF programs under grant RTI2018-098156-B-C55.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| CGH | Comparative Genomic Hybridization |
| CRISPR | Clustered Regularly Interspaced Palindromic Repeats |
| DMR | Differentially Methylated Region |
| DNA | DeoxyriboNucleic Acid |
| DWT | Discrete Wavelet Transform |
| GO | Gene Ontology |
| GPU | Graphics Processing Unit |
| SOA | Service Oriented Architecture |
| PCR | Polymerase Chain Reaction |
References
- Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef]
- Jones, P.A.; Takai, D. The Role of DNA Methylation in Mammalian Epigenetics. Science 2001, 293, 1068–1070. [Google Scholar] [CrossRef]
- Wu, H.; Tao, J.; Sun, Y.E. Regulation and function of mammalian DNA methylation patterns: A genomic perspective. Brief. Funct. Genom. 2012, 11, 240–250. [Google Scholar] [CrossRef]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2013, 13, 484–492. [Google Scholar] [CrossRef]
- Laird, P.W. Principles and challenges of genome-wide DNA methylation analysis. Nat. Rev. Genet. 2010, 11, 191–203. [Google Scholar] [CrossRef]
- Krueger, F.; Andrews, S.R. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011, 27, 1571–1572. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.Y.; Cokus, S.; Pellegrini, M. BS Seeker: Precise mapping for bisulfite sequencing. BMC Bioinform. 2010, 11, 203. [Google Scholar] [CrossRef]
- Harris, E.Y.; Ponts, N.; Le Roch, K.G.; Lonardi, S. BRAT-BW: Efficient and accurate mapping of bisulfite-treated reads. Bioinformatics 2012, 28, 1795–1796. [Google Scholar] [CrossRef] [PubMed]
- Olanda, R.; Pérez, M.; Orduña, J.M.; Tárraga, J.; Dopazo, J. A new parallel pipeline for DNA methylation analysis of long reads datasets. BMC Bioinform. 2017, 18, 161. [Google Scholar] [CrossRef]
- Hansen, K.D.; Langmead, B.; Irizarry, R.A. BSmooth: From whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol. 2012, 13, R83. [Google Scholar] [CrossRef]
- Hebestreit, K.; Dugas, M.; Klein, H.U. Detection of significantly differentially methylated regions in targeted bisulfite sequencing data. Bioinformatics 2013, 29, 1647–1653. [Google Scholar] [CrossRef]
- Sun, D.; Xi, Y.; Rodriguez, B.; Park, H.J.; Tong, P.; Meong, M.; Goodell, M.A.; Li, W. MOABS: Model based analysis of bisulfite sequencing data. Genome Biol. 2014, 15, R38. [Google Scholar] [CrossRef]
- Wu, H.; Xu, T.; Feng, H.; Chen, L.; Li, B.; Yao, B.; Qin, Z.; Jin, P.; Conneely, K.N. Detection of differentially methylated regions from whole-genome bisulfite sequencing data without replicates. Nucleic Acids Res. 2015, 43, e141. [Google Scholar] [CrossRef]
- Shafi, A.; Mitrea, C.; Nguyen, T.; Draghici, S. A survey of the approaches for identifying differential methylation using bisulfite sequencing data. Brief. Bioinform. 2018, 19, 737–753. [Google Scholar] [CrossRef]
- Xie, C.; Leung, Y.K.; Chen, A.; Long, D.X.; Hoyo, C.; Ho, S.M. Differential methylation values in differential methylation analysis. Bioinformatics 2018, 35, 1094–1097. [Google Scholar] [CrossRef]
- Catoni, M.; Tsang, J.M.; Greco, A.P.; Zabet, N. DMRcaller: A versatile R/Bioconductor package for detection and visualization of differentially methylated regions in CpG and non-CpG contexts. Nucleic Acids Res. 2018, 46, e114. [Google Scholar] [CrossRef]
- Blume, A. methylKit, DNA methylation analysis from high-throughput bisulfite sequencing data. Genome Biol. 2018. [Google Scholar] [CrossRef]
- Fernández, L.; Pérez, M.; Orduña, J.M. Visualization of DNA methylation results through a GPU-based parallelization of the wavelet transform. J. Supercomput. 2019, 75, 1496–1509. [Google Scholar] [CrossRef]
- Fernández, L.; Pérez, M.; Olanda, R.; Orduña, J.M.; Marquez-Molins, J. HPG-DHunter: An ultrafast, friendly tool for DMR detection and visualization. BMC Bioinform. 2020, 21, 287. [Google Scholar] [CrossRef]
- Wang, Z.; Li, X.; Jiang, Y.; Shao, Q.; Liu, Q.; Chen, B.; Huang, D. swDMR: A Sliding Window Approach to Identify Differentially Methylated Regions Based on Whole Genome Bisulfite Sequencing. PLoS ONE 2015, 10, e0132866. [Google Scholar] [CrossRef]
- Liu, H.; Liu, X.; Zhang, S.; Lv, J.; Li, S.; Shang, S.; Jia, S.; Wei, Y.; Wang, F.; Su, J.; et al. Systematic identification and annotation of human methylation marks based on bisulfite sequencing methylomes reveals distinct roles of cell type-specific hypomethylation in the regulation of cell identity genes. Nucleic Acids Res. 2016, 44, gkv1332. [Google Scholar] [CrossRef]
- Müller, F.; Scherer, M.; Assenov, Y.; Lutsik, P.; örn Walter, J.; Lengauer, T.; Bock, C. RnBeads 2.0: Comprehensive analysis of DNA methylation data. Genome Biol. 2019, 20, 55. [Google Scholar] [CrossRef]
- Kuo, H.C.; Lin, P.Y.; Chung, T.C.; Chao, C.M.; Lai, L.C.; Tsai, M.H.; Chuang, E.Y. DBCAT: Database of CpG Islands and Analytical Tools for Identifying Comprehensive Methylation Profiles in Cancer Cells. J. Comput. Biol. 2011, 18, 113–1017. [Google Scholar] [CrossRef]
- Illumina DRAGEN Bio-IT Platform. Available online: https://www.illumina.com/products/by-type/informatics-products/dragen-bio-it-platform.html (accessed on 30 April 2021).
- Qlucore Omics Explorer—Analysis of DNA Methylation Data. Available online: https://qlucore.com/dnamethylation (accessed on 30 April 2021).
- Al-Shahrour, F.; Minguez, P.; Vaquerizas, J.M.; Conde, L.; Dopazo, J. BABELOMICS: A suite of web tools for functional annotation and analysis of groups of genes in high-throughput experiments. Nucleic Acids Res. 2005, 33, W460–W464. [Google Scholar] [CrossRef]
- Yang, J.H.; Kim, B.W.; Nam, S.J.; Kwon, Y.; Lee, D.H.; Lee, J.Y.; Hwang, C.S.; Lee, Y.H.; Hwang, S.H.; Park, I.C.; et al. MetaCore: An application-specific programmable DSP development system. IEEE Trans. Very Large Scale Integr. Syst. 2000, 8, 173–183. [Google Scholar] [CrossRef]
- Blankenberg, D.; Kuster, G.V.; Coraor, N.; Ananda, G.; Lazarus, R.; Mangan, M.; Nekrutenko, A.; Taylor, J. Galaxy: A web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. 2010, 89, 19.10.1–19.10.21. [Google Scholar] [CrossRef]
- Antonov, A.V.; Schmidt, T.; Wang, Y.; Mewes, H.W. ProfCom: A web tool for profiling the complex functionality of gene groups identified from high-throughput data. Nucleic Acids Res. 2008, 36, 347. [Google Scholar] [CrossRef]
- Zheng, Q.; Wang, X.J. GOEAST: A web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res. 2008, 36, W358–W363. [Google Scholar] [CrossRef]
- Rainer, J.; Sanchez-Cabo, F.; Stocker, G.; Sturn, A.; Trajanoski, Z. CARMAweb: Comprehensive R- and bioconductor-based web service for microarray data analysis. Nucleic Acids Res. 2006, 34, W498–W503. [Google Scholar] [CrossRef]
- Carver, T.; Thomson, N.; Bleasby, A.; Berriman, M.; Parkhill, J. DNAPlotter: Circular and linear interactive genome visualization. Bioinformatics 2009, 25, 119–120. [Google Scholar] [CrossRef]
- Derrien, T.; André, C.; Galibert, F.; Hitte, C. AutoGRAPH: An interactive web server for automating and visualizing comparative genome maps. Bioinformatics 2007, 23, 498–499. [Google Scholar] [CrossRef][Green Version]
- Adler, P.; üri Reimand, J.; Jänes, J.; Kolde, R.; Peterson, H.; Vilo, J. KEGGanim: Pathway animations for high-throughput data. Bioinformatics 2008, 24, 588–590. [Google Scholar] [CrossRef]
- Lai, W.; Choudhary, V.; Park, P.J. CGHweb: A tool for comparing DNA copy number segmentations from multiple algorithms. Bioinformatics 2008, 24, 1014–1015. [Google Scholar] [CrossRef][Green Version]
- Mitra, A.; Kesarwani, A.K.; Pal, D.; Nagaraja, V. WebGeSTer DB—A transcription terminator database. Nucleic Acids Res. 2011, 39, 129. [Google Scholar] [CrossRef]
- Humann, J.L.; Lee, T.; Ficklin, S.; Main, D. Structural and Functional Annotation of Eukaryotic Genomes with GenSAS. In Gene Prediction: Methods and Protocols; Springer: New York, NY, USA, 2019; pp. 29–51. [Google Scholar] [CrossRef]
- Ferrè, F.; Clote, P. BTW: A web server for Boltzmann time warping of gene expression time series. Nucleic Acids Res. 2006, 34, 482. [Google Scholar] [CrossRef] [PubMed]
- Rousseau, C.; Gonnet, M.; Romancer, M.L.; Nicolas, J. CRISPI: A CRISPR interactive database. Bioinformatics 2009, 25, 3317–3318. [Google Scholar] [CrossRef]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.M. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef]
- Bao, Y.; Bolotov, P.; Dernovoy, D.; Kiryutin, B.; Tatusova, T. FLAN: A web server for influenza virus genome annotation. Nucleic Acids Res. 2007, 35, 280. [Google Scholar] [CrossRef] [PubMed]
- Cantarel, B.; Korf, I.; Robb, S.; Parra, G.; Ross, E.; Moore, B.; Holt, C.; Alvarado, A.S.; Yandell, M. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008, 18, 188–196. [Google Scholar] [CrossRef]
- Bhave, S.V.; Hornbaker, C.; Phang, T.L.; Saba, L.; Lapadat, R.; Kechris, K.; Gaydos, J.; McGoldrick, D.; Dolbey, A.; Leach, S.; et al. The PhenoGen informatics website: Tools for analyses of complex traits. BMC Genet. 2007, 8, 59. [Google Scholar] [CrossRef]
- Sulyman, S. Client-Server Model. IOSR J. Comput. Eng. 2014, 16, 57–71. [Google Scholar] [CrossRef]
- Zeilinger, H.; Anees, T. SOA Model for High Availability of Services. In Proceedings of the IARIA Conference on Communication Theory, Reliability, and Quality of Service, Venice, Italy, 21–26 April 2013. [Google Scholar]
- Perkins, B.; Hammer, J.; Reid, J. XML and JSON; John Wiley & Sons: Hoboken, NJ, USA, 2018; pp. 673–696. [Google Scholar] [CrossRef]
- Mason, P. JavaScript. In SAS Stored Processes; Apress: Berkeley, CA, USA, 2020; pp. 65–80. [Google Scholar] [CrossRef]
- Bühler, P.; Schlaich, P.; Sinner, D. Html5. In HTML5 und CSS3. Bibliothek der Mediengestaltung; Springer Vieweg: Berlin/Heidelberg, Germany, 2017; pp. 1–42. [Google Scholar] [CrossRef]
- Olsson, M. CSS Properties. In CSS3 Quick Syntax Reference; Apress: Berkeley, CA, USA, 2019; pp. 53–56. [Google Scholar] [CrossRef]
- bin Uzayr, S.; Cloud, N.; Ambler, T. JavaScript Frameworks for Modern Web Development: The Essential Frameworks, Libraries, and Tools to Learn Right Now; Apress: Berkeley, CA, USA, 2019; pp. 523–539. [Google Scholar] [CrossRef]
- Biswal, M. React Lifecycle Methods. Available online: https://www.researchgate.net/publication/337731350_React_lifecycle_methods (accessed on 30 April 2021).
- Bucea-Manea-Tonis, R. Angular JS—The Newest Technology in Creating Web Applications. Ann. Spiru Haret Univ. Econ. Ser. 2016, 16, 103. [Google Scholar] [CrossRef]
- Neto, M.; Andrade, S.; Novais, R. Cross-Platform Multimedia Application Development: For Mobile, Web, Embedded and IoT with Qt/QML; Association for Computing Machinery: Gramado, Brazil, 2017; pp. 23–26. [Google Scholar] [CrossRef]
- Siboldi, M. Node JS Performance Testing. Available online: https://digitalcommons.calpoly.edu/laessp/35 (accessed on 30 April 2021).
- Moroney, L. Using Authentication in Firebase. In The Definitive Guide to Firebase; Apress: Berkeley, CA, USA, 2017; pp. 25–50. [Google Scholar] [CrossRef]
- González, C.; Pérez, M.; Orduña, J.M.; Chaves, J.; García, A.B. HPG-HMapper: A DNA hydroxymethylation analysis tool. Int. J. High Perform. Comput. Appl. 2020, 34, 57–65. [Google Scholar] [CrossRef]
- Dabrowski, J.; Munson, E.V. 40 years of searching for the best computer system response time. Interact. Comput. 2011, 23, 555–564. Available online: https://academic.oup.com/iwc/article-pdf/23/5/555/1900920/iwc23-0555.pdf (accessed on 30 April 2021). [CrossRef]
- Card, S.; Robertson, G.; Mackinlay, J. The Information Visualizer, an Information Workspace. In Proceedings of the SIGCHI Conference on Human Factors in Computing Systems, New Orleans, LA, USA, 27 April–2 May 1991; pp. 181–186. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).