A Self-Spatial Adaptive Weighting Based U-Net for Image Segmentation
Abstract
1. Introduction
2. Related Work
3. Our Approach
3.1. Image Segmentation Problem
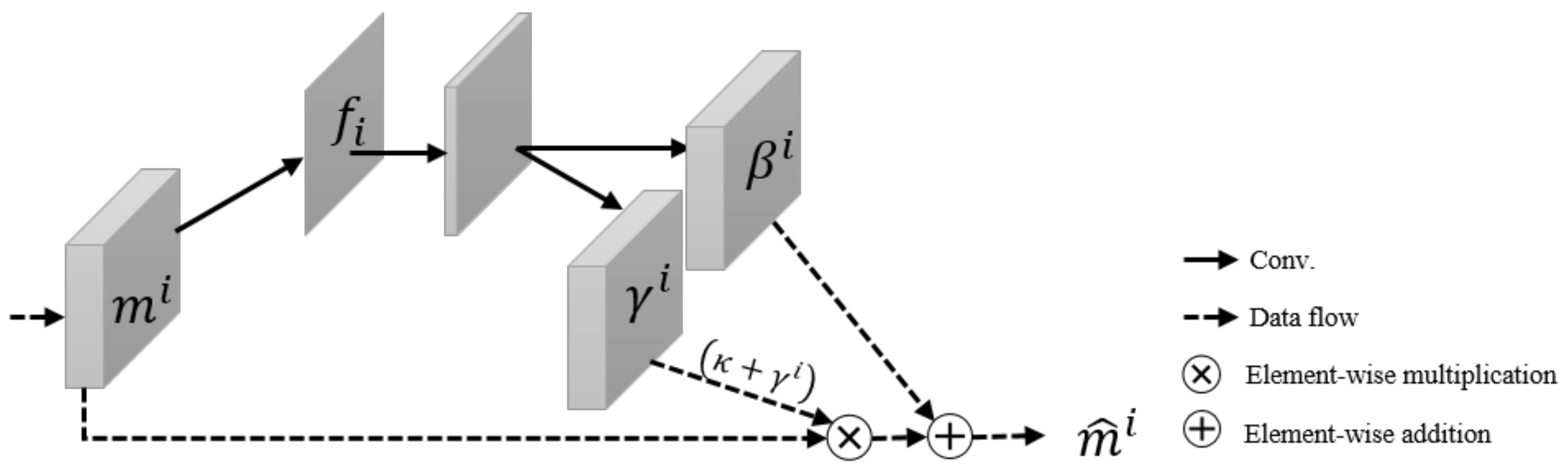
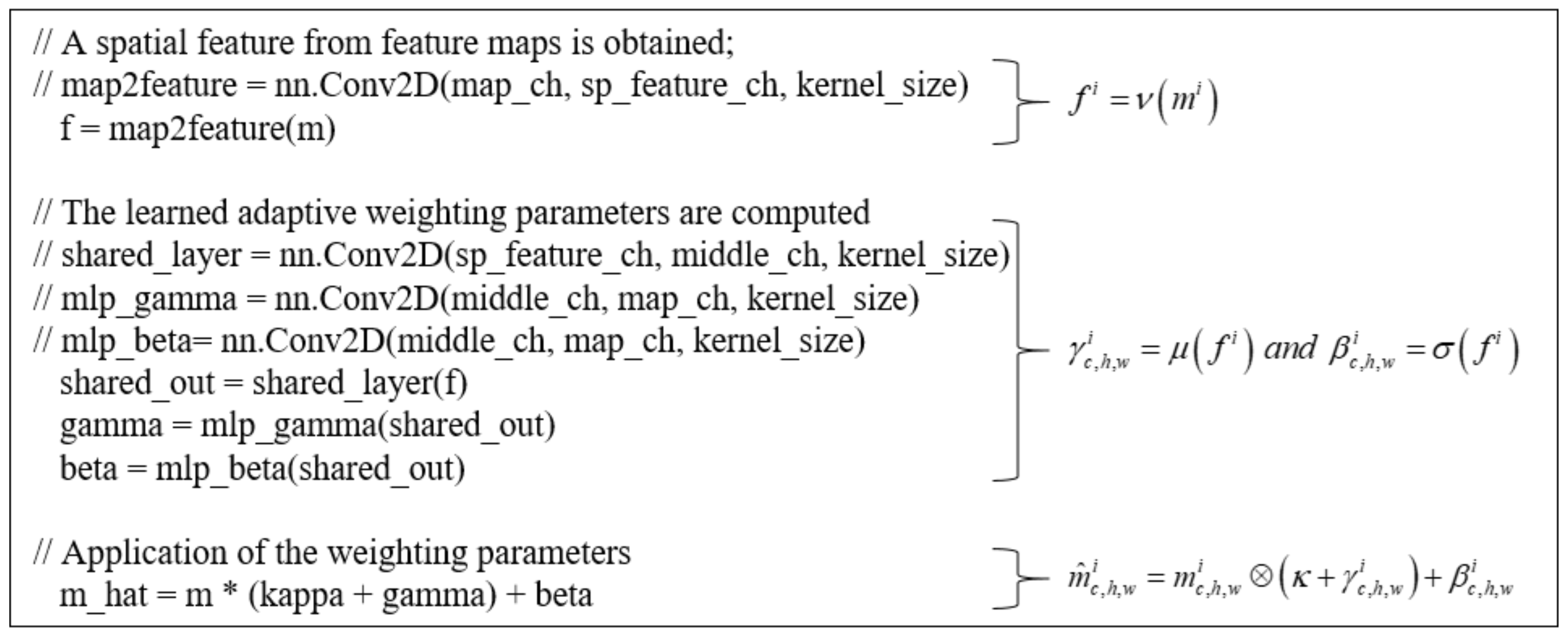
3.2. Self-Spatial Adaptive Weighting
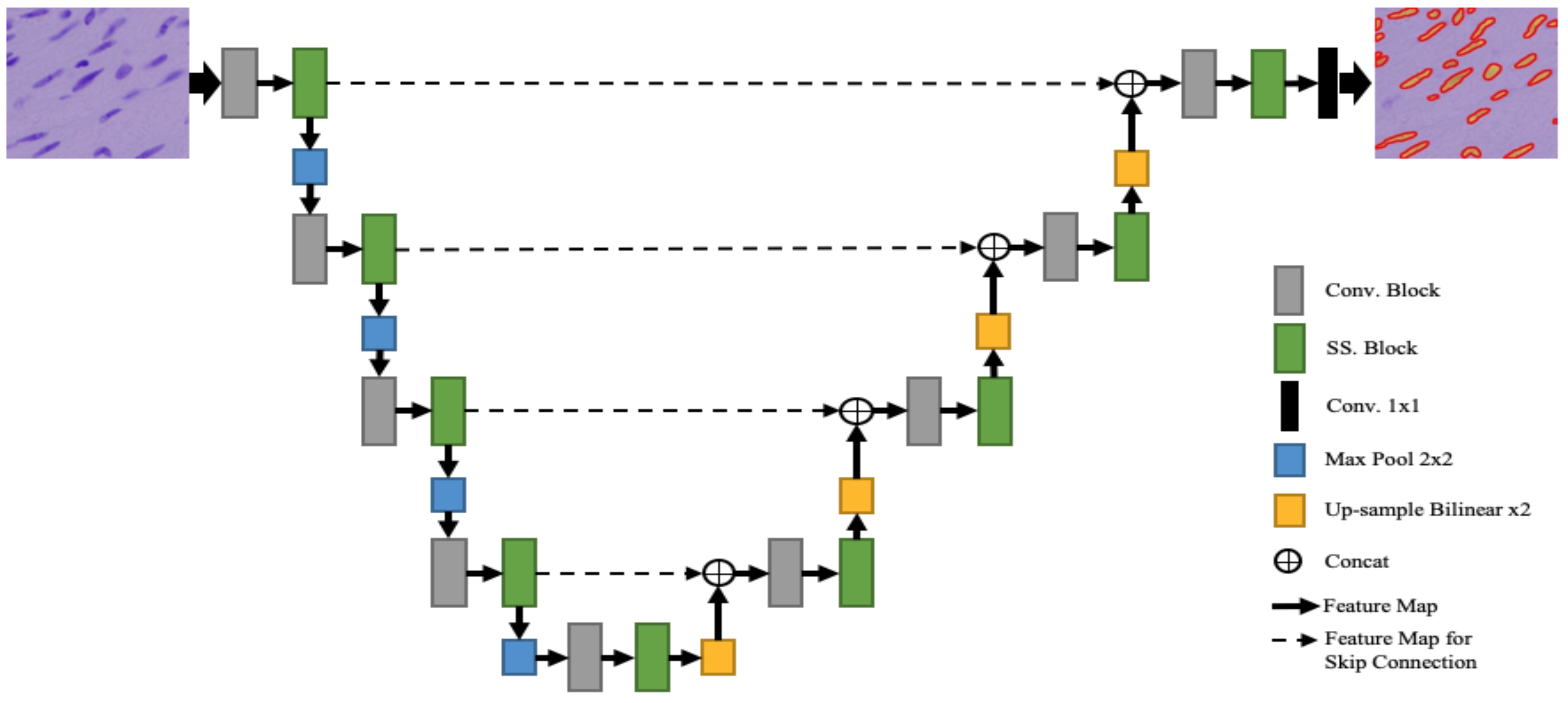
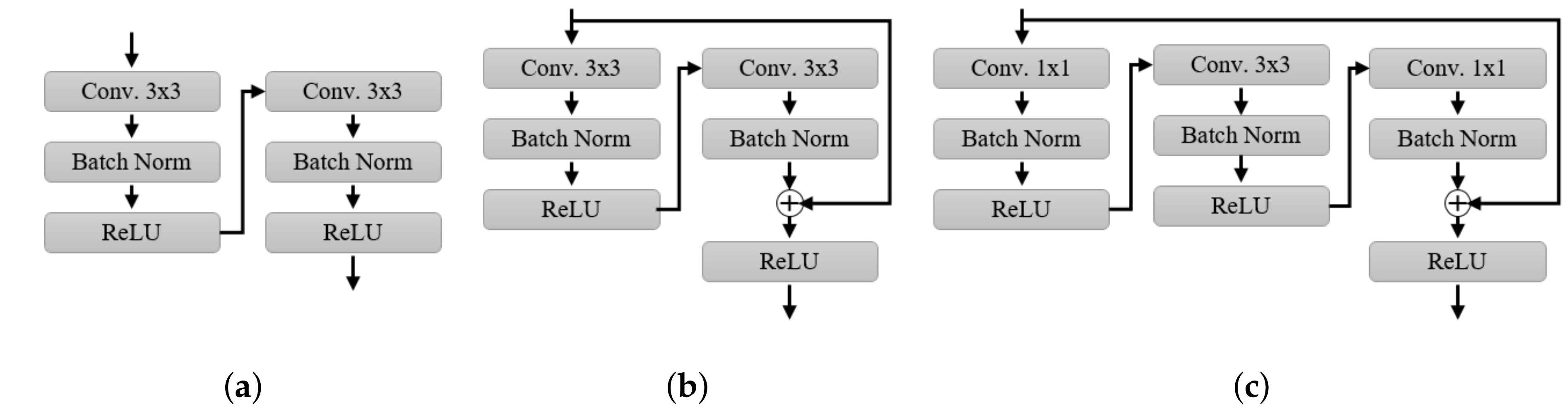
3.3. Self-Spatial Adaptive Weighting-Based U-Net Structure for Image Segmentation
4. Experimental Results
4.1. Datasets
4.2. Training Setup
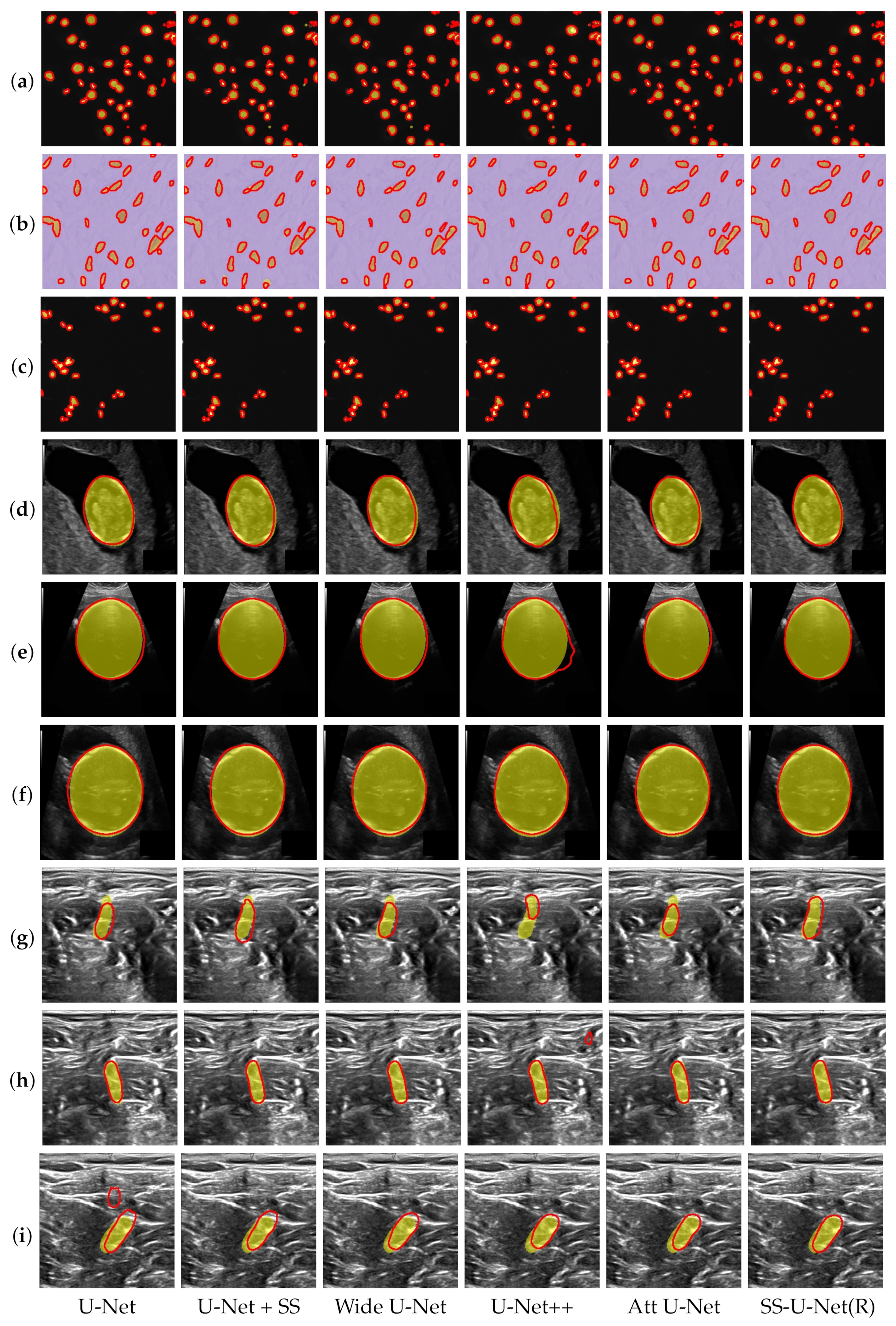
4.3. Performance Comparison
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-Excitation Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar]
- Gao, M.; Yu, R.; Li, A.; Morariu, V.I.; Davis, L.S. Dynamic zoom-in network for fast object detection in large images. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 21–26. [Google Scholar]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid scene parsing network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2881–2890. [Google Scholar]
- Zhao, H.; Zhang, Y.; Liu, S.; Shi, J.; Change Loy, C.; Lin, D.; Jia, J. Psanet: Point-wise spatial attention network for scene parsing. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 267–283. [Google Scholar]
- Donahue, J.; Anne Hendricks, L.; Guadarrama, S.; Rohrbach, M.; Venugopalan, S.; Saenko, K.; Darrell, T. Long-term recurrent convolutional networks for visual recognition and description. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 2625–2634. [Google Scholar]
- Luvizon, D.C.; Picard, D.; Tabia, H. 2d/3d pose estimation and action recognition using multitask deep learning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; pp. 5137–5146. [Google Scholar]
- Xu, K.; Ba, J.; Kiros, R.; Cho, K.; Courville, A.; Salakhudinov, R.; Zemel, R.; Bengio, Y. Show, attend and tell: Neural image caption generation with visual attention. In Proceedings of the International Conference on Machine Learning, Lille, France, 6–11 July 2015; pp. 2048–2057. [Google Scholar]
- Dai, B.; Lin, D. Contrastive Learning for Image Captioning. In Proceedings of the Advances in Neural Information Processing Systems (NeurIPS), Long Beach, CA, USA, 4–9 December 2017; pp. 898–907. [Google Scholar]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. Unet++: Redesigning skip connections to exploit multiscale features in image segmentation. IEEE Trans. Med Imaging 2019, 39, 1856–1867. [Google Scholar] [CrossRef] [PubMed]
- Cho, C.S.; Lee, S. Low-complexity topological derivative-based segmentation. IEEE Trans. Image Process. 2014, 24, 734–741. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Li, X.; Chen, H.; Qi, X.; Dou, Q.; Fu, C.W.; Heng, P.A. H-DenseUNet: Hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans. Med Imaging 2018, 37, 2663–2674. [Google Scholar] [CrossRef]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Lu, J.; Xiong, C.; Parikh, D.; Socher, R. Knowing when to look: Adaptive attention via a visual sentinel for image captioning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 375–383. [Google Scholar]
- Li, W.; Liu, K.; Zhang, L.; Cheng, F. Object detection based on an adaptive attention mechanism. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Chen, X.; Wang, J. Object-contextual representations for semantic segmentation. arXiv 2019, arXiv:1909.11065. [Google Scholar]
- Wang, J.; Sun, K.; Cheng, T.; Jiang, B.; Deng, C.; Zhao, Y.; Liu, D.; Mu, Y.; Tan, M.; Wang, X.; et al. Deep high-resolution representation learning for visual recognition. arXiv 2020, arXiv:1908.07919. [Google Scholar] [CrossRef] [PubMed]
- Tao, A.; Sapra, K.; Catanzaro, B. Hierarchical Multi-Scale Attention for Semantic Segmentation. arXiv 2020, arXiv:2005.10821. [Google Scholar]
- Chan, T.F.; Vese, L.A. Active contours without edges. IEEE Trans. Image Process. 2001, 10, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Huang, R.; Ding, Z.; Gatenby, J.C.; Metaxas, D.N.; Gore, J.C. A level set method for image segmentation in the presence of intensity inhomogeneities with application to MRI. IEEE Trans. Image Process. 2011, 20, 2007–2016. [Google Scholar] [PubMed]
- Isola, P.; Zhu, J.Y.; Zhou, T.; Efros, A.A. Image-to-image translation with conditional adversarial networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1125–1134. [Google Scholar]
- Wang, T.C.; Liu, M.Y.; Zhu, J.Y.; Liu, G.; Tao, A.; Kautz, J.; Catanzaro, B. Video-to-video synthesis. arXiv 2018, arXiv:1808.06601. [Google Scholar]
- Wang, T.C.; Liu, M.Y.; Zhu, J.Y.; Tao, A.; Kautz, J.; Catanzaro, B. High-resolution image synthesis and semantic manipulation with conditional gans. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 8798–8807. [Google Scholar]
- Park, T.; Liu, M.Y.; Wang, T.C.; Zhu, J.Y. Semantic image synthesis with spatially-adaptive normalization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 2337–2346. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Sobhaninia, Z.; Rafiei, S.; Emami, A.; Karimi, N.; Najarian, K.; Samavi, S.; Soroushmehr, S.R. Fetal ultrasound image segmentation for measuring biometric parameters using multi-task deep learning. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 6545–6548. [Google Scholar]
- Zou, K.H.; Warfield, S.K.; Bharatha, A.; Tempany, C.M.; Kaus, M.R.; Haker, S.J.; Wells III, W.M.; Jolesz, F.A.; Kikinis, R. Statistical validation of image segmentation quality based on a spatial overlap index1: Scientific reports. Acad. Radiol. 2004, 11, 178–189. [Google Scholar] [CrossRef]
- Caicedo, J.C.; Goodman, A.; Karhohs, K.W.; Cimini, B.A.; Ackerman, J.; Haghighi, M.; Heng, C.; Becker, T.; Doan, M.; McQuin, C.; et al. Nucleus segmentation across imaging experiments: The 2018 Data Science Bowl. Nat. Methods 2019, 16, 1247–1253. [Google Scholar] [CrossRef] [PubMed]
- Ultrasound Nerve Segmentation Kaggle. 2016. Available online: https://www.kaggle.com/c/ultrasound-nerve-segmentation (accessed on 2 December 2020).
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. Pytorch: An imperative style, high-performance deep learning library. In Proceedings of the Advances in Neural Information Processing Systems (NeurIPS), Vancouver, BC, Canada, 8–14 December 2019; pp. 8026–8037. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980v9. [Google Scholar]

| Data Set | # of Images | Input Size | Modality | Provider |
|---|---|---|---|---|
| Cell Nuclei [33] | 670 | 320 × 256 | Microscopy | Data Science Bowl competition |
| Fetal Head [31] | 999 | 800 × 540 | Ultrasound | HC18 Grand Challenge |
| Nerve [34] | 5635 | 580 × 420 | Ultrasound | Ultrasound Nerve competition Kaggle |
| Method | Model Size Param (MB) | Cell Nuclei | Fetal Head | Nerve | |||
|---|---|---|---|---|---|---|---|
| IoU | DICE | IoU | DICE | IoU | DICE | ||
| U-Net(B) | 20.01 | 86.24 | 91.59 | 94.12 | 96.49 | 65.55 | 76.72 |
| SS-U-Net(B) | 21.29 | 86.38 | 91.65 | 95.26 | 97.24 | 68.56 | 79.61 |
| U-Net(V) | 29.96 | 86.05 | 91.34 | 95.26 | 97.24 | 68.32 | 79.20 |
| SS-U-Net(V) | 31.24 | 86.36 | 91.68 | 95.24 | 97.26 | 68.84 | 79.90 |
| U-Net(R) | 31.62 | 85.91 | 91.38 | 95.22 | 97.23 | 68.01 | 79.17 |
| SS-U-Net(R) | 32.89 | 86.58 | 91.84 | 95.48 | 97.41 | 69.16 | 80.14 |
| Segmentation Method | Model Size Param (MB) | Cell Nuclei | Fetal Head | Nerve | |||
|---|---|---|---|---|---|---|---|
| IoU | DICE | IoU | DICE | IoU | DICE | ||
| U-Net [14] | 32.95 | 86.09 | 91.39 | 95.31 | 97.29 | 68.35 | 79.34 |
| U-Net + SS | 34.22 | 86.14 | 91.63 | 95.37 | 97.33 | 68.90 | 79.73 |
| Wide U-Net [11] | 34.85 | 86.10 | 91.51 | 95.13 | 97.21 | 68.94 | 79.75 |
| U-Net++ [11] | 34.96 | 85.83 | 91.58 | 95.41 | 97.30 | 67.87 | 79.85 |
| Att U-Net [18] | 33.63 | 85.83 | 91.49 | 95.36 | 97.35 | 68.53 | 79.31 |
| U-Net(R) + SS(our) | 32.89 | 86.58 | 91.84 | 95.48 | 97.41 | 69.14 | 80.14 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, C.; Lee, Y.H.; Park, J.; Lee, S. A Self-Spatial Adaptive Weighting Based U-Net for Image Segmentation. Electronics 2021, 10, 348. https://doi.org/10.3390/electronics10030348
Cho C, Lee YH, Park J, Lee S. A Self-Spatial Adaptive Weighting Based U-Net for Image Segmentation. Electronics. 2021; 10(3):348. https://doi.org/10.3390/electronics10030348
Chicago/Turabian StyleCho, Choongsang, Young Han Lee, Jongyoul Park, and Sangkeun Lee. 2021. "A Self-Spatial Adaptive Weighting Based U-Net for Image Segmentation" Electronics 10, no. 3: 348. https://doi.org/10.3390/electronics10030348
APA StyleCho, C., Lee, Y. H., Park, J., & Lee, S. (2021). A Self-Spatial Adaptive Weighting Based U-Net for Image Segmentation. Electronics, 10(3), 348. https://doi.org/10.3390/electronics10030348






