Abstract
Machine learning is becoming the cornerstones of smart and autonomous systems. Machine learning algorithms can be categorized into supervised learning (classification) and unsupervised learning (clustering). Among many classification algorithms, the Support Vector Machine (SVM) classifier is one of the most commonly used machine learning algorithms. By incorporating convex optimization techniques into the SVM classifier, we can further enhance the accuracy and classification process of the SVM by finding the optimal solution. Many machine learning algorithms, including SVM classification, are compute-intensive and data-intensive, requiring significant processing power. Furthermore, many machine learning algorithms have found their way into portable and embedded devices, which have stringent requirements. In this research work, we introduce a novel, unique, and efficient Field Programmable Gate Array (FPGA)-based hardware accelerator for a convex optimization-based SVM classifier for embedded platforms, considering the constraints associated with these platforms and the requirements of the applications running on these devices. We incorporate suitable mathematical kernels and decomposition methods to systematically solve the convex optimization for machine learning applications with a large volume of data. Our proposed architectures are generic, parameterized, and scalable; hence, without changing internal architectures, our designs can be used to process different datasets with varying sizes, can be executed on different platforms, and can be utilized for various machine learning applications. We also introduce system-level architectures and techniques to facilitate real-time processing. Experiments are performed using two different benchmark datasets to evaluate the feasibility and efficiency of our hardware architecture, in terms of timing, speedup, area, and accuracy. Our embedded hardware design achieves up to 79 times speedup compared to its embedded software counterpart, and can also achieve up to 100% classification accuracy.
1. Introduction
With the advent of smart and autonomous systems, machine learning is becoming the cornerstone in creating these systems. Machine learning, a subset of artificial intelligence, enables a system to learn, identify patterns, and make decisions without being explicitly programmed, and with minimal or no human intervention [1,2]. The basic premise of machine learning is to create techniques (or algorithms) that can learn from the sample input data (known as the training data), by performing statistical analysis in order to make accurate predictions or decisions on the data [2,3]. In recent years, various fields including medical diagnosis in healthcare, credit risk analysis in finance, smart grids in energy, and smart cars in transportation, have been incorporating machine learning techniques [4]. According to a market research [5], the global market for machine learning was $1.4 billion in 2017 and is expected to reach $8.8 billion by 2022 with an annual growth rate of 43.6%. The aforementioned facts demonstrate that the machine learning market will continue to thrive as smart and autonomous systems emerge. Furthermore, as portable and embedded devices are gaining popularity, many machine learning applications are becoming common on these devices. For instance, with autonomous vehicles, real-time in situ data processing and analysis must be performed typically on embedded devices in order to make split-second decisions, to avoid catastrophic scenarios. Hence, novel and unique architectures, techniques, and methodologies are required to support and accelerate machine learning applications on highly constrained embedded devices.
Machine learning typically involves many important data mining tasks, and can be categorized into supervised learning (e.g., classification) and unsupervised learning (e.g., clustering) [6]. In this research work, we focus on supervised learning (specifically, classification) techniques. Classification techniques are employed by many machine learning applications in various domains including medical, finance, and transportation. There exist many classification algorithms; hence, selecting a suitable algorithm for a specific application (or dataset) would significantly impact the accuracy of the results as well as the overall system’s performance [6]. For instance, employing a linear classifier for non-linear separable data can diminish the accuracy and efficiency of the results, which in turn can hinder the subsequent analysis [6]. As a result, we investigate different classifiers and decide to focus on the Support Vector Machine (SVM) classification algorithm, since SVM has many advantages/traits that are deemed suitable for machine learning applications. For instance, SVM is more appropriate for classifying non-linear separable data [7], enables classification in multi-dimensional space [8], is capable of handling high dimensional data [9], and generates high accuracy results [8,9]. In addition, the accuracy and the classification process of the SVM can be further improved by incorporating non-linear optimization methods to find the optimal solutions [9]. Hence, we decide to integrate convex optimization (one of the best non-linear optimization methods) to the SVM classifier in order to clearly distinguish between the two separate classes by maximizing the margin width of the hyper-plane, which in turn leads to an optimal solution [9].
Most of the existing techniques/algorithms for machine learning, including SVM, are typically designed in high-level programming languages such as python and are executed on general-purpose computers such as desktops and servers. These processor-based (software-only) algorithms, in their current form, can not be executed directly on the embedded platforms/devices, since these devices have numerous constraints including stringent area and power, limited memory, increased speedup, and reduced cost and time-to-market requirements. Furthermore, today’s machine learning techniques/algorithms are becoming more compute- and data-intensive, requiring more processing power. For instance, the processing time for the training would increase exponentially with the number of input data samples. Additionally, for smart and autonomous systems, the data processing and analysis must be done in real-time in order to make split-second decisions. Consequently, to satisfy the constraints associated with the embedded devices, as well as the requirements of the machine learning applications, it is imperative to incorporate some applications-specific (or customized) hardware into embedded systems designs. In this regard, Field Programmable Gate Array (FPGA)-based hardware is one of the most promising avenues to deliver machine learning applications on highly constrained embedded platforms [10], not only because FPGAs provide a higher level of flexibility than ASICs (application-specific-integrated-circuits) and higher performance than software running on a processor, but also due to their many attractive traits including post-fabrication reprogrammability, dynamic partial reconfiguration capabilities, and reduced time-to-market. Our previous work demonstrated that FPGA’s aforementioned traits indeed facilitate the support and acceleration of many real-time compute/data-intensive applications (not only machine learning [2,11,12], but also data mining [13,14], control systems [15,16], and security [17,18]), especially on resource-constrained embedded devices.
Our main objective is to provide customized and optimized architectures and techniques to support and accelerate machine learning applications/algorithms on embedded platforms, considering the associated constraints of these platforms and the requirements of the applications running on these devices. In this research work, we focus on an FPGA-based hardware accelerator for a convex optimization-based (CO-based) support vector machine classifier for machine learning. By integrating the convex optimization to the SVM classifier, we can further enhance the efficiency and effectiveness of the machine learning applications [9].
Initially, we investigated some existing research work on hardware support for a CO-based SVM as well as for a general SVM algorithm. An FPGA-based implementation for SVM was introduced in [19] for telecommunication. In this case, only the SVM training phase was implemented on hardware. Some parts of the training phase, such as kernel and Q matrices, were also computed offline. In [20], a coprocessor was introduced only for the kernel matrix computation [20] of the SVM training. However, it is imperative to implement all the stages of both the training and testing phases on hardware, especially for real-time machine learning applications. In [21], a scalable FPGA-based architecture was proposed for the whole SVM algorithm. Although the proposed hardware classifier achieved a substantial speedup compared to its software counterpart, the design did not include techniques to solve the constraint quadratic formulation and to process and analyze the data in real-time. An FPGA-based accelerator was proposed for a different SVM algorithm known as least square SVM [22]. In this case, the SVM training was processed online utilizing a run-time reconfiguration framework and parallel processing architecture. This improved the speedup but with the penalty in area, which is not feasible especially for small-footprint embedded devices. In addition to the aforementioned works, we perform a detailed analysis of existing works on FPGA-based hardware architectures for CO-SVMs, which is presented in Section 5.5.
As mentioned above, there were several research works on hardware support for general SVM algorithms in the published literature. Most of these hardware architectures were not generic or parameterized. Additionally, these architectures were not designed with embedded devices in mind. None of these works proposed system-level architectures and associated techniques to facilitate the real-time processing of machine learning applications. From our extensive investigation (in Section 5.5) and to the best of our knowledge, we did not find any hardware support or any FPGA-based hardware accelerators, especially for the convex optimization-based SVM algorithm, in the published literature, which makes our research novel and unique.
In this paper, we make the following contributions:
- We introduce a novel, unique, customized, and optimized FPGA-based hardware accelerator specifically for the convex optimization (CO)-based SVM for machine learning with dynamic training capabilities. Our architectures are generic, parameterized, and scalable; hence, without changing the internal architectures, our designs can be used to process different datasets with varying sizes; can be executed on different embedded platforms; and can be utilized for linearly/non-linear separable, multi-dimensional datasets, making it suitable for various machine learning applications. We also introduce embedded software architecture for a CO-based SVM classifier.
- Our proposed design is also capable of solving a general convex optimization problem efficiently and effectively using a decomposition method. We provide three different options for mathematical kernels, which enable the user to select and utilize the most suitable option for a given machine learning application.
- We also introduce an efficient systems-level design in order to process the data efficiently and effectively and considering the real-time constraints. We design and integrate unique pre-fetching and burst transfer techniques to reduce the memory access latency and to facilitate real-time processing.
- We fully optimize our designs and techniques considering the requirements of the CO-based SVM algorithm and constraints associated with the embedded platforms. We perform experiments to evaluate the feasibility and efficiency of our designs using two different benchmark datasets. We analyze the timing, speedup, classification accuracy, and resource utilization for our embedded designs.
This paper is organized as follows: In Section 2, we discuss and present the SVM algorithm and the concepts of optimal hyper-plane, non-linear optimization methods, and convex optimization utilized in this research work. Our design approach, system-level architecture, and novel techniques to reduce the memory access latency are detailed in Section 3. In Section 4, our novel, unique, customized, and optimized FPGA-based embedded hardware accelerator for three stages of a CO-based SVM algorithm is introduced. Our embedded software design is also presented in Section 4. Experimental results and analysis in terms of timing, speedup, area, and accuracy are reported and discussed in Section 5. Analysis of existing works on FPGA-based hardware architectures for a CO-based SVM is also discussed and presented in Section 5. In Section 6, we summarize our work, conclude, and discuss future work.
2. Background: Convex Optimization and Support Vector Machines
The support vector machine (SVM), first introduced by Cortes and Vapnik in 1995, is one of the novel machine learning techniques based on the statistical learning theory [19]. The SVM was initially developed for classification tasks and was later extended to regression analysis [20]. As stated in [6,21], the SVM classifies the data points based on its location with respect to the hyper-plane. We investigate the published literature to get an insight into the concepts of the optimal hyper-plane, non-linear optimization methods for SVMs, and decomposition methods for solving the convex optimization for SVM.
2.1. Optimal Hyper-Plane
The SVM is a commonly used classification technique found in many different fields, such as digital channel equalization in signal processing, and protein structure prediction and cancer detection in medical diagnosis [7]. In this case, the concept is to formulate a hyper-plane with maximum margin width to distinguish between two classes [6,21]. It is a supervised classification method [6], which often involves a training set of {xi,yi}, where x is the set of input data samples/vectors, i is the total number of samples, and y is the output label of the binary classifier, used for identifying the class of the data sample. These are represented in Equation (1).
Each input vector x, constitutes different features of a dataset represented as, xi = {xi1,xi2,xi3, …, xin}, where, n is the number of features. The formula for the hyper-plane [6,21], and the decision function to determine the class, are represented by Equations (2) and (3), respectively.
where, w is the weight vector, and b is the bias value.
The margin width of a hyper-plane can be obtained by projecting a unit normal vector ŵ to the optimal hyper-plane, as in Equation (4) [6,21].
The SVM can typically be extended to a multi-class classification [22], in which the output label is represented as , where, m is the total number of classes. As stated in [21], the multi-class training process can be performed using a one-to-rest approach. With this approach, m different classes are trained independently, where one set of the input vectors forms the positive class, and the remaining input vectors form the negative class, in the hyper-plane. The classification process can further be carried out, similar to binary classification.
2.2. Non-Linear Optimization
In order to improve the overall accuracy of the classification tasks, the margin width of the hyper-plane can be maximized using a non-linear optimization method [9,23]. With these methods, an objective function subjected to boundary constraints of a hyper-plane can be derived from Equations (2) and (4) [9,23]. The formula for the objective function is presented in Equation (5), as follows:
Employing the duality theory and the Lagrange multipliers [21], we can efficiently calculate the local maxima or the local minima of the objective function in Equation (5) [9,21]. From Equation (5), the constrained optimization problem can be formulated similar to the non-linear programming/optimization [23]. This constrained optimization problem can be presented as the primal and dual form as in Equations (6) and (8) [21].
In order to obtain the minimum from Equation (6), let us consider the point where the gradient is zero [21]. The minimum value for the primal form can be obtained by applying the partial derivatives with respect to w and b to derive the following formulae in Equation (7).
Duality theory is useful for the constrained optimization problem since it provides a convenient way to improve the data classification by utilizing the non-linear optimization approach [9].
The primal form (in Equation (6)) can be solved using several methods [9], including the Newton method, least-squares algorithms, stochastic sub-gradient method, cutting plan algorithms, and interior point method. The dual form (in Equation (8)) can also be solved using the decomposition methods and interior point method [9]. The dual form has the advantage of utilizing mathematical kernels. Mathematical kernels are a set of algebraic transformation functions, which provide similarity information between data features [24]. To use the mathematical kernels, the dual form depends on the pair of samples, such as K(xi.xj) as in Equation (8). Utilizing the mathematical kernels, the SVM classification can be extended to non-linearly separable datasets [21].
After obtaining the local maxima or the local minima from Equations (6) and (8), the optimal value for α is evaluated to identify the support vectors [9,21]. The support vectors are the input vectors closest to the hyper-plane and have an α value greater than zero (α > 0) [9]. The dimensional coordinates of the support vectors determine the orientation of the hyper-plane. Any other vector may result in closer to zero α value, which indicates that the data points have less impact on the orientation of the hyper-plane, and are also situated further away from the hyper-plane [21].
2.3. Convex Optimization
Typically for large-scale applications, due to a high volume of data, the constrained optimization problem presented in Equation (8), must converge to the minimum value, in order to find the best fit for constructing an optimal hyper-plane [9]. As stated in [8], for convex optimization, all local minima are considered as the global minimum. In this case, due to the presence of noise in some datasets, the soft margin parameters such as l-norm error parameter (penalty parameter, C and slack variable (ξ)) must be considered for a better generalization of the primal form [9,21]. Since the aforementioned non-linear optimization/programming consists of an equality constraint, the dual form (in Equation (8)) can be reduced to the general form of the convex optimization problem by integrating soft margin parameters [23]. Hence, the overall objective function in Equation (8) can be modified to the following Equation (9).
Equation (9) is similar to the general form of convex optimization, as stated in [8], with the equality constraint and the box constraint, and can be written as Equation (10) below.
In convex optimization, the above objective function (in Equations (9) and (10)) should converge in such a way that it satisfies the Hessian matrix condition [9], which states that the contour of the convex plane should be continuously differentiable as in Equation (11). Then, the optimal solution can be found where the gradient value is zero [21].
In Equation (10), the P matrix is symmetric and positive semi-definite; and both the objective function and the constraint function are convex [8,23]. Especially, for large-scale applications, the resultant matrices are dense, thus, difficult to solve. Therefore, the decomposition methods are often used to break down the convex optimization process by finding the two working sets [25,26], as shown in Equation (12). Proper selection of the “working sets” impacts the performance of the convex optimization algorithm and its convergence properties [9,27]. In this case, the process of finding the two working sets to determine the support vectors in the training set is a compute-intensive and iterative process. Hence, by providing customized and optimized FPGA-based accelerators, we can dramatically enhance the speed-performance of these compute-intensive applications (or tasks).
The decomposition methods, such as sequential minimal optimization, are employed to solve the non-linear optimization applications/tasks by sequentially selecting the working set based on the proximal point with respect to the objective function (in Equation (10)) [26]. Considering the various methods employed to solve the non-linear optimization, the Sequential Minimal Optimization (SMO) is the most popular, due to its ability to handle large-scale datasets efficiently and effectively [25]. In this case, at each iteration, the input vector x (in Equation (10)) is divided into two working sets as , where, k is the current iteration count, xk is the current input vector, and is the previous input vector. Based on the specified starting point, the objective function (in Equation (10)) is solved to converge to a minimum value [26]. More information about Equations (9) and (10), the selection of the working sets, stopping criteria, and other methods to solve the non-linear optimization/programming can be found in [9,25].
For convex optimization (from Equation (10)), a feasible point of (0,0) is initially selected for the working sets, which is the first input data sample in Equation (1). Then based on the direction of the gradient descent, the next working set is selected [28,29]. Selecting the feasible point and the working set can impact the total time required to solve the convex quadratic optimization. As stated in [9], the solution for the convex optimization (in Equation (10)) is found, by selecting the suitable working set, which satisfies the following criteria in Equation (12).
In Equation (12), the R(α) and S(α) are the index sets used to characterize the descent direction and identify the optimality conditions. Based on the descent direction, the objective function (in Equation (9)) converges to the minimum α value, which is used to determine the orientation of the separating hyper-plane [9,21]. In this case, constructing the hyper-plane establishes an explicit distinction between the different classes of the data.
It has been illustrated [9,23] that the non-linear optimization methods for the SVM, employed to construct the hyper-plane, have significantly enhanced the overall speed-performance of the SVM classifier, and have also led to the high accuracy of the results. However, obtaining the minimum values for the objective function (in Equations (9) and (10)) is a compute-intensive task, which depends on the number of input samples. Hence, it is imperative to design, develop, and provide customized and optimized FPGA-based hardware accelerators to enhance the speedup of these compute-intensive tasks.
3. Design Approach and Development Platform
In our designs, both hardware and software versions of various operations/tasks are implemented using hierarchical platform-based and modular-based design approaches to facilitate component reuse at different levels of abstractions. As illustrated in Figure 1, at the highest level, our CO-based SVM algorithm comprises the training and testing tasks. During the training process, based on Equations (9) and (10), the objective function is formulated to define the hyper-plane, select a suitable mathematical kernel, and obtain the optimal solution. During the testing process, the decision function of the classifier is computed using the sign verification operation (in Equation (3)). The aforementioned intermediate operations to process the training and testing tasks involve vector addition/multiplication, matrix computations, and various other arithmetic operations, which are placed at the lowest level of our platform-based design hierarchy.
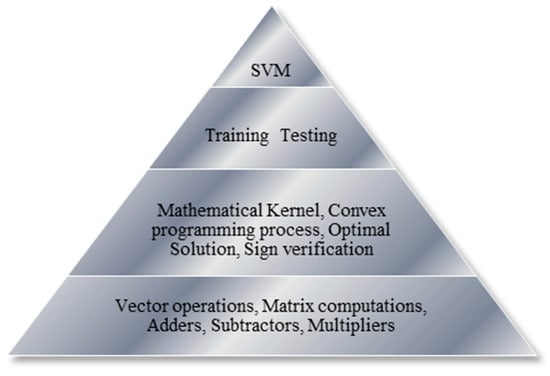
Figure 1.
Hierarchical and Modular-Based Design Approach.
During our early design phase, we investigate and utilize the integer units and also the double-precision floating-point units as our FPGA-based Intellectual Property (IP). From these experiments, the integer-based designs exhibit results with quite a low accuracy, whereas the double-precision floating-point designs occupy a larger area on the chip. These facts illustrate that the former might not be suitable for real-time applications, which typically require results with high accuracy; and the latter is not necessarily suitable for the embedded devices with stringent area requirements. Furthermore, for the high dimensional input vectors, the optimal solution for α could not be reached (in Equation (9)), utilizing the double-precision floating-point units, since these units not only create large on-chip logic resources requirements but also high latency requirements. Hence, we strive to approximate the results for the double-precision floating-point units, without compromising the accuracy of the results. As a result, we utilize the single-precision floating-point units, which create a tradeoff among the accuracy and area, and power. In this case, most of the lower-level operators are designed and implemented using the single-precision floating-point units in the Xilinx IP core library [30]. The same floating-point unit in [30] is configured and utilized for various fundamental operators (e.g., multipliers, adders, subtractors, dividers, etc.) of our proposed embedded hardware architectures presented in Section 4.2.
All our embedded hardware and software experiments are performed on the ML605 FPGA development platform [31,32,33] which utilizes a Xilinx Virtex-6 XC6VLX240T-FFH1156 device. This development platform consists of large on-chip logic resources (37,680 slices), 748 DSP48E1 slices, 512 MB DDR3-SDRAM (Double-Data-Rate Synchronous Dynamic Random Access Memory), and 2 MB on-chip BRAM (Block Random Access Memory) [34]. More details of the ML605 development platform and the specific Virtex-6 FPGA utilized can be found in [32,33].
The vector computations are designed in such a way by integrating the available DSP48E1 slices to enhance the speed performance. The large 512 MB off-chip memory resources are useful to store large datasets, typically found in many real-time machine learning applications.
All our customized embedded hardware modules are designed in mixed VHDL and Verilog, using Xilinx ISE 14.7 and XPS 14.7 design tools. They are executed on the aforementioned Virtex-6 FPGA running at 100 MHz (in real-time) to verify their correctness and performance. The results and the functionalities of the hardware designs are further verified using the ModelSim SE and Xilinx ISim tools. All our embedded software modules are written in C++ and executed on the 32-bit RISC MicroBlaze soft processor running at 100 MHz on the same FPGA. Xilinx XPS 14.7 and SDK 14.7 tools are used to design and verify the software modules. Unlike the hard processors, the MicroBlaze soft processor must be synthesized and mapped onto the configurable logic blocks of the FPGA. The MicroBlaze is also configured to use single-precision floating-point units for the software modules. The performance gain or the speedup is evaluated using the baseline software execution time over the improved hardware execution time. The hardware and software execution times are obtained from the AXI Timer [35]. Additional software experiments are performed on desktop computers to verify our results (i.e., to verify the correct functionality and operation, and accuracy) from the embedded hardware and embedded software designs.
The overall speedup is evaluated and reported using two different benchmark datasets obtained from the UCI machine learning repository [36]: Wisconsin breast cancer diagnostic dataset [37] and Ionosphere dataset for machine learning [38]. The Cancer dataset consists of 569 samples (or vectors), each having 30 features (or attributes) to describe the characteristics of the cancer cell nuclei. These cancer cells are obtained using a fine needle aspirate and the main features are obtained from a digitized image. The Ionosphere dataset comprises a set of phased-array data obtained from 16 high-frequency radars. It has 351 samples, each having 34 features to characterize the complex electromagnetic signals. The total sizes of these two datasets are 68,280 bytes and 47,736 bytes, respectively.
3.1. System-Level Architecture
Figure 2 demonstrates the system-level architecture for our embedded hardware and software designs. Since 2 MB on-chip BRAM, on Virtex-6 FPGA on ML605 board, is not sufficient to store the large amount of data commonly found in many machine learning applications, we integrate the 512 MB DDR3-SDRAM external memory into the system. In this case, DDR3-SDRAM and the DDR3-SDRAM memory controller run at 200 MHz, whereas the rest of the system is running at 100 MHz. As illustrated in Figure 2, we utilize the AXI (Advanced Extensible Interface) bus [39] to facilitate communication among the peripherals at the system level.
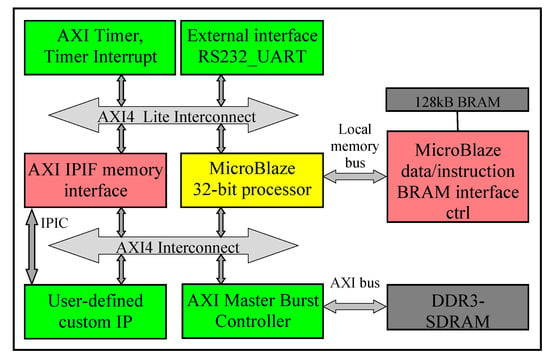
Figure 2.
System-Level Architecture.
During the initial software design phase, we configure the MicroBlaze processor to have the maximum available cache memory of 128 KB. However, this 128 KB of cache memory is not sufficient to execute our software code and to process the data, and our code starts hanging. Hence, we vary the heap and stack size and also increase the addressing of the cache memory to accommodate 256 KB. This indeed resolves our cache memory constraint issue, although the Xilinx XPS tool still reports the size of the cache memory as 128 KB.
As shown in Figure 2, our user-defined/designed custom Intellectual Property (IP) communicates with the DDR3-SDRAM and the MicroBlaze using the AXI bus through the AXI Intellectual Property Interface (IPIF) module, using a set of ports called the Intellectual Property Interconnects (IPIC) [39]. Typically, after the MicroBlaze processor sends a start signal to our user-defined IP via the AXI4-lite bus and the AXI IPIF interface, our user-defined IP starts processing, directly reads/writes data/results from/to the DDR3-SDRAM, and sends a finish signal to the MicroBlaze processor when the execution is completed.
For our hardware design, we provide an AXI4 burst/stream high-throughput interface for streaming the data from the DDR3-SDRAM to our user-defined IP for real-time processing. One of our design goals is to create our system-level architecture in such a way as to train and classify a continuous stream of data using the AXI4 burst/stream interface. In a real-world application scenario, this feature enables proving a direct connection between our user-defined hardware IP and a camera, (for instance, in an autonomous car), in order to process the input data on-the-fly. This enables our hardware IP to perform the training and classification processes dynamically to cater to the ever-changing environment. By streaming the data and processing the data directly, reduces the amount of memory storage required for the embedded designs.
3.2. Pre-Fetching Techniques and Top-Level Architecture
From our previous work [40,41], it was observed that a substantial amount of time was spent on accessing DDR3-SDRAM off-chip memory, which was a major performance bottleneck. Hence, in this work, we design and integrate a pre-fetching technique to address the memory access latency issue in our user-defined hardware IP.
As shown in Figure 2, the AXI4-lite and AXI4 interfaces act as glue logic for the whole system, including the MicroBlaze processor, internal peripherals, and the user-defined IP [39]. AXI4-lite is a single transaction memory-mapped bidirectional interface. In our design, the MicroBlaze sends/receives certain control signals, and also monitors the status of the user-defined IP via the AXI4-lite bus and via the slave registers (or software accessible registers). These 32-bit slave registers are also used to send the SVM specifications, such as kernel type, dimensions of the input vectors, penalty/slack variables, to the user-defined IP, along with the initial memory address of the DDR3-SDRAM to access the datasets.
Apart from the AXI4-lite, the user-defined IP reads/writes data/results from/to the DDR3-SDRAM via the AXI4 bus. AXI4 master burst is a high-performance memory-mapped interface capable of transferring burst size of up to 256 data beats, which are compatible with 16, 32, 64, and 128 data width with a single address transaction phase [42]. As stated in [39,42], with this data width, we can transfer up to 1 MB (2n − 1 bytes) per cycle on the IPIC command interface. Utilizing the AXI4 master burst capabilities, the memory access time is dramatically reduced for the SVM training and the SVM testing processes.
Our proposed pre-fetching technique is illustrated in Figure 3. During the pre-fetching mode, our user logic module (in Figure 3) determines the total number of bytes to be fetched utilizing the aforementioned SVM specifications provided by the MicroBlaze via the slave registers. Next, the address generator and the AXI master burst controller configure the control signals of the IPIC with a suitable data width, a burst length, and a number of beats per cycle. Then, the AXI master burst controller sends the aforementioned details about the data, as well as the “master read request” signal to the AXI interconnect core. Once the AXI master receives the “read request acknowledgment” signal from the AXI interconnect core, the user IP can start receiving the data from the DDR3-SDRAM, can store the data in the BRAM. Then the user logic can start the training process. In this case, the address generator is essential to index the correct values for matrix computations. After completing the training process, the AXI master burst controller is designed to automatically set up for the write operation, in order to store the weight vectors and the bias values in the DDR3-SDRAM, for subsequent computations/analysis.
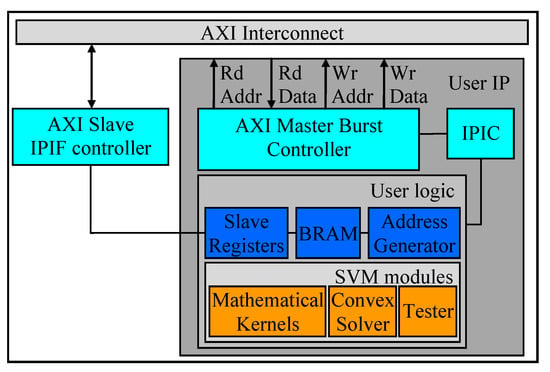
Figure 3.
Pre-fetching Techniques and Top-Level Architecture.
4. Embedded Architectures for the Convex Optimization-Based-SVM
In this section, we introduce novel, unique, and efficient embedded architectures (both hardware and software) for the convex optimization-based (CO-based) support vector machine (SVM) classification algorithm.
4.1. Embedded Software Design
Prior to our embedded software designs, we design and develop the software for our convex optimization-based SVM algorithm in C++ using the Microsoft Visual Studio development tools. This software design is executed on a desktop computer with an Intel i7 processor running at 2.3 GHz. Our results are compared and verified with the results from the open-source python code obtained from [43]. Both the C++ and python results are also used to verify our results (i.e., to verify the correct functionality and operation, and accuracy) from our embedded hardware and embedded software architectures.
In order to cater to the resource constraint nature of the embedded devices, we significantly modify the aforementioned C++ software architecture, initially developed for desktop computers. In this case, we create the codes leaner and simpler, in such a way to fit into the available program cache memory of the embedded microprocessor, i.e., MicroBlaze, without impacting the internal structure/flow and the functionalities of the overall algorithm.
For our embedded software designs, the MicroBlaze processor is configured to have the maximum available cache memory of 128 KB, from which 64 KB is used for the Instruction Cache and 64 KB is used for the Data Cache. The capacity of these data and instruction cache memories can be varied; however, the total can not exceed the maximum available cache memory.
During our embedded software design phase, we encounter several issues due to the stringent constraints of the embedded devices. One of the major issues is due to the limited memory resources. In this case, certain functionalities of the normal C++ programs, executed on desktop computers, can not be directly designed and implemented on the embedded devices. For instance, importing a pre-processor directive for vector computations from the desktop computer to the embedded devices resulted in memory limitations issues. To resolve this issue, initially, we vary the heap and stack sizes; then we design a compact function in software that is capable of performing the vector computation efficiently and effectively.
Our embedded software architecture for the convex optimization-based SVM algorithm comprises three stages. These three stages as well as the functional flow of our embedded software design are presented in Figure 4.
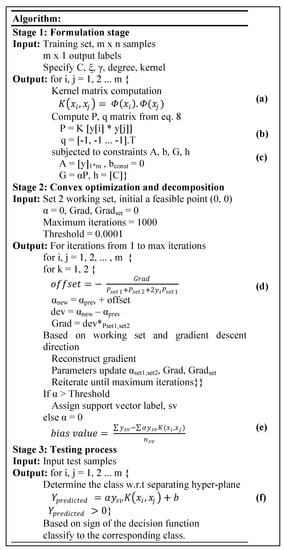
Figure 4.
Software and Functional Flow for the CO-Based SVM Algorithm.
4.2. Embedded Hardware Design
In this sub-section, we introduce our novel, customized, and optimized embedded hardware architecture for the convex optimization-based SVM algorithm. In this case, we examine and analyze the functional flow of the aforementioned algorithm. Subsequently, we partition this complex algorithm into three stages (SVM Module in Figure 3) to simplify the design process. The operations of these three consecutive stages are mathematical kernels, convex optimization (or convex solver), and testing (or tester). It should be noted that the solver is considered as the hardware IP to perform the optimization stage.
In this research work, we design and develop customized and optimized embedded hardware architectures for each stage as separate modules. The hardware designs for each stage comprise a datapath and a control path. The control path consists of finite state machines (FSMs) and manages the control signals of the datapath and the BRAMs. We also design a top-level module (i.e., SVM Module in Figure 3) to integrate the three modules for the three stages. The top-level module provides necessary communication/control among the three stages. The control path of the top-level module also consists of several FSMs, multiplexers, and tri-state buffers to control the timing, routing, and internal structures/functionalities of the designs.
The three stages of the CO-based SVM algorithm are executed non-sequentially to utilize the parallel processing nature of the FPGA-based hardware. Initially, the first stage, i.e., the mathematical kernel, is processed until a certain amount of results is obtained from this stage. Then the p and q matrices (in Equations (9) and (10)), in the second stage, i.e., the optimization (or the solver), is computed on the aforementioned kernel results, while the remainder of the mathematical kernel is proceeding. In this case, the execution time to compute the p and q matrices is typically less than the execution time to compute the mathematical kernel; and the former depends on the results of the latter. Hence, we design and develop a simple counter to create a time delay in order to wait until the kernel has processed at least 50 training data samples, i.e., closer to 10% of the total data size, before starting the second stage. For all the stages, the intermediate/final results are stored in the BRAM, and after all three stages are processed, the final results are written to the DDR3-SDRAM.
The internal architectures of these three stages of the convex optimization-based (CO-based) SVM algorithm are detailed in the following sub-sections. These internal architectures are customized and optimized in such a way by exploiting the inherent parallelism and pipeline nature of the CO-based SVM algorithm.
4.2.1. Stage 1: Mathematical Kernels
In the first stage of our embedded hardware design, we select and perform a suitable mathematical kernel. As stated in [44], linearly inseparable vectors in the input space can be transformed to linearly separable vectors in the feature space by mapping the data points to a higher dimensional space. This transformation can be performed with mathematical kernels (including linear, polynomial, and Gaussian kernels) since the bound does not depend on the dimensionality of the space for SVM [8]. This is an efficient way to obtain a well-defined separating hyper-plane [24]. Mathematical kernels are a set of algebraic transformation functions, which provides similarity information between data features [27]. In order to utilize the mathematical kernels, the mapping function (Φ(x)) in Equation (13) must satisfy the Mercer’s condition [24], which states that the inner product of the two input vectors must be defined for all the features, as represented in Equation (13) below.
In this research work, we decide to create customized and optimized embedded architectures for the linear, polynomial, and Gaussian radial basis function (RBF), since these are the three most popular mathematical kernels used for the SVM algorithm. The equations for these three mathematical kernels are as follows [6]:
The datapath for the polynomial kernel is illustrated in Figure 5 (corresponding to modules 2), whereas the datapath for the linear kernel is the dotted lines of Figure 5 (corresponding to modules 1). As shown, the datapath of the linear kernel consists of a multiplier, adder, and an accumulator register with the feedback loop to the adder, whereas the datapath for the polynomial kernel has a second adder and a power module.
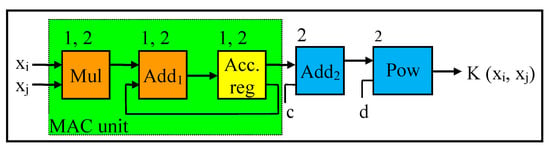
Figure 5.
Datapath for Liner and Polynomial Kernels.
The result of the linear kernel is the dot product operation of the two input data samples. In this case, initially, the first two elements of the two data samples are read from the BRAM, which is the first two elements of the first row, and the multiplication operation is performed, followed by the accumulation operation on each multiplier result. This process will continue until this multiplication and accumulation (MAC) operation is performed on the last two elements of the two data samples. Then, the final result of the MAC operation is forwarded and stored on the BRAM for subsequent analysis/computations. As depicted in Figure 5, the modules in the dotted line comprise the MAC operation.
For the datapath for the polynomial kernel, the inputs to the second adder are the final result from the linear kernel as well as the coefficient c. This addition result goes through the power module to perform the “power of d” on the addition results. In this case, we create a hardware module for the power function using a simple loop to iterate the multiplication based on the specified degree (d) value. Although this degree value is parameterized in our design, a commonly used quadratic kernel is performed by considering this degree value as two (2). The final result of this power module is also forwarded and stored on the BRAM for subsequent computations.
The datapath for the Gaussian kernel is demonstrated in Figure 6. The Gaussian radial basis function (RBF) kernel is the most popular among the aforementioned three mathematical kernels. The datapath comprises 3 MAC modules to perform part of Equation (16), which is (xi − xj)2, expanded into xi2 + xj2 − 2xi.xj, which requires three dot product operations. Next, the addition operation is performed on the results of the two square operations, while the result of the MAC3 operation is multiplied by two. Then the multiplier result is subtracted from the result of the addition operation. The subtractor result is multiplied by the parameter known as gamma (γ). As stated in [43], this gamma (γ) parameter determines the influence of a data sample on the separating hyper-plane. In this case, the gamma (γ) value typically varies from 10−3 to 10+3 as needed. In this research work, as detailed in Section 5, we vary the gamma (γ) value from 2 × 10−3 to 2 × 10+2 for both our embedded hardware and software designs. As shown in Figure 6, the result of the multiplier operation (with Mult2) goes through the exponent module to obtain the final result of the Gaussian RBF kernel. In this case, we create a parameterized exponent hardware module based on the Taylor series expansion, which is represented by Equation (17) [45]. Since the number of terms in the Taylor series becomes infinite, for both our embedded hardware and software designs, we limit the number of terms to be the first 5 terms/elements of the Taylor series, as shown in Equation (17), without compromising the accuracy of the final exponent result. Next, we create our exponent module in such a way to process each term/element of the Taylor series in sequence (i.e., to process one element at a time). Although not shown here, the datapath of the exponent module consists of a multiplier, divider, and adder, which are being reused to process each term of the Taylor series in an iterative fashion. The final result of this exponent module is forwarded and stored on the BRAM for subsequent computations.
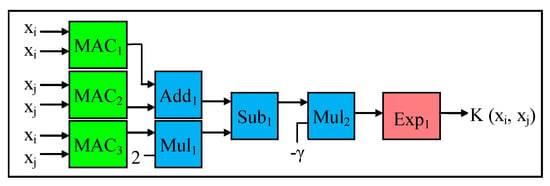
Figure 6.
Datapath for Gaussian Radial Basis Function (RBF) Kernel.
The output size of the matrix for the kernel computation depends on the number of input data samples (m). Thus, the size of the K(xi,xj) matrix is mXm. After processing 10% of the data for the kernel computation, the convex optimization process is initiated. The computation to track 10% of the data processing is implemented using a simple counter. The kernel computation is necessary to perform each iteration of the next stage, which is the convex optimization. Hence, initially, the results of the mathematical kernel are stored on the BRAM to ease the iterative process of convex optimization.
4.2.2. Stage 2: Convex Optimization
The optimization stage is the most complex operation among the three stages of the convex optimization-based (CO-based) SVM algorithm. In order to reduce the complexity, we divide stage 2 into three phases: parameter initialization, convex optimization, and bias value computation. In this stage, the dual form of the SVM (in Equation (8)) is utilized to formulate the general convex optimization as shown in Equations (9) and (10).
Parameter Initialization Phase
During the parameter initialization phase of stage 2, several parameters in Equations (9) and (10) are computed including the objective function parameters (i.e., P, q), constraint parameters (i.e., G, h, A, bconst), and other parameters (α, Grad, feasible point). In the convex optimization phase, the α value and the Grad value are evaluated for each iteration using the sequential minimal optimization (SMO) decomposition method (detailed in Section 2.3). Once the maximum number of iterations is reached, the bias value b is computed in the final bias value computation phase. This bias value is used to determine the intercept of the hyper-plane.
In this paper, for stage 2, we design a generic convex optimization solver utilizing the same naming convention for the parameters as the general form of convex optimization from Equation (10). In this case, the naming conventions b and bconst are the bias value (in Equation (2)) and the constraints value (in Equation (10)) respectively. The aforementioned objective function and constraint parameters are computed in Stage 2 (phase 1). These correspond to steps (b) and (c) in Figure 4.
Objective function parameters: The P parameter (in Equations (9) and (10)) is an mXm matrix, which is computed using the dot product of the output labels yi.yj. The y variable is the output label obtained from the dataset, and y represents the class of the data sample, as in Equation (1). In our design, during stage 1, y is typically pre-fetched to the BRAM with the input samples. During stage 2, the result of the dot product (yi.yj) is multiplied with the result of the kernel matrix K(xi,xj), in order to obtain the P matrix (in Equation (18)). In this case, as illustrated in Figure 7, the modules to compute the P matrix consist of a MAC unit and a multiplier (i.e., MAC1 Unit and Mul1, in Figure 7). The elements of the P matrix are stored in the BRAM. The elements of the P matrix in the BRAM are accessed using two separate address generators, in order to create two sets of matrices Pset1 and Pset2. As stated in [6], for big data analysis, the size of the P matrix increases in squared terms with the increasing number of input data samples; thus, performing the convex optimization using conventional methods such as interior-point methods become computationally challenging. To overcome this issue, a specific decomposition method in [46] is employed to perform convex optimization. With this decomposition method, at any instance of time during the optimization process, two working sets are selected. This decomposition method is detailed in the subsequent convex optimization phase. The q parameter (in Equations (9) and (10)) is an mX1 matrix. The q matrix is an array of ones as in Equation (19). For simplicity, during the design phase, each element of the q matrix is kept as constants of 1 s.
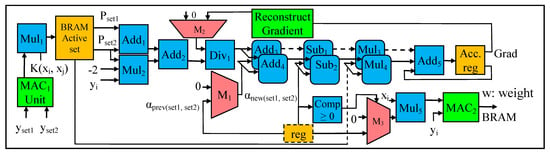
Figure 7.
Datapath for Convex Optimization.
Constraint parameters: In Equation (10), the objective function is subjected to the box constraint (which comprises G and h parameters), and the equality constraint (which consists of A and bconst). In this case, A parameter is a 1Xm matrix in Equation (10), which is the same as the output label y in Equation (9). Additionally, bconst parameter in Equation (10) is set to zero in Equation (9). In addition, the G parameter in Equation (10) is a constant value of 1. The maximum threshold of the box constraint is h parameter in Equation (10) (i.e., C parameter in Equation (9)). C is considered as the penalty parameter, which is a user-defined vector provided during the parameter specifications. The value of C impacts the overall speed-performance of the CO-based SVM algorithm. This impact is illustrated in Section 5.
Other parameters: α parameter in Equation (9), which corresponds to x in Equation (10), is crucial to identify the support vectors in the CO-based SVM algorithm. Since the coordinated dimensions of the support vectors determine the orientation of the hyper-plane, α values for all the input samples are initialized to zeros, prior to performing the convex optimization. The goal of performing the convex optimization is to compute the minimum value for α for each input sample. Based on the aforementioned threshold value (C), the input samples with α value greater than the threshold value will be considered as the support vectors. Furthermore, as discussed above, since the size of the P matrix increases exponentially with the increasing number of input samples, two working sets (or input samples) are selected and utilized to compute α in each iteration. In this case, for the initial starting point, a feasible set of (0, 0) is selected, which is the first input samples of the dataset; and the Grad (gradient) value is initialized to zero. The slope of the gradient to the minimum value typically corresponds to the gradient descent direction; hence, the gradient value is utilized to select the next working set.
Convex Optimization Phase
After the parameter initialization phase, the convex optimization phase is performed. During the convex optimization phase, five operations illustrated in steps (d) to (h) in Figure 4 are performed. The datapath for the convex optimization is shown in Figure 7, which consists of several adders, multipliers, subtractors, MAC modules, dividers, accumulator registers, comparators, and multiplexers. The convex optimization process (as in Equation (9)) involves finding the minimum α value. In this case, the α value is computed using the Add1 to Add4, Mul1, Div1, and M1 (multiplexer) modules, as illustrated in Figure 7. In order to find the minimum α value, additional comparators, multiplexers, and reconstruct gradient modules are utilized.
Initially, during the first iteration to find the minimum α value, the two input samples, Pset1 and Pset2 (stored in the BRAM) are accessed using the two separate address generators, simultaneously. Next, the result of the addition operation (with Add1 in Figure 7) of Pset1 and Pset2 are added (using Add2) to the result of the multiplier (i.e., Mul2), which corresponds to the step (f) in Figure 4. Then, the gradient value is divided (with Div1) by the result of the second addition operation (with Add2). Since the Grad (gradient) value is initialized to zero, the initial result of the division is also zero. In our design, the computation of the gradient parameter is modified using the reconstruct gradient module in Figure 7. The internal architecture of the reconstruct gradient module is demonstrated in Figure 8. During the first iteration, the value of α is zero. The output (or result) of the division operation (with Div1 in Figure 7) is considered as the offset value, which in turn is used to update the α value. Apart from the first iteration, where the initial α value is zero, from the second iteration onwards, the aforementioned offset value is added to the α value, computed in the former iteration. For instance, the offset value produced from the second iteration is added to the α value generated from the first iteration, and so on. In this case, two addition operations are performed in parallel (using Add3 and Add4), in order to generate new α values for each working set. The new α values are stored in the temporary register (i.e., reg in Figure 7), to be used for future iterations. Next, two subtraction operations are performed in parallel (using Sub1 and Sub2), to find the difference between the new α value and former α value, which provides the deviation between the successive iterations. Then, the new α values are multiplied in parallel (using Mul3 and Mul4), with Pset1 and Pset2, to find the gradient value (Grad). These Grad values are stored in the on-chip BRAM for subsequent iterations and operations. The aforementioned operations are illustrated in steps (d), (e), (f), and (g) in Figure 4.
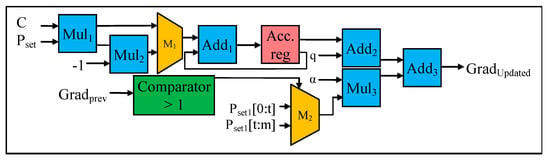
Figure 8.
Internal Architecture of Reconstruct Gradient Computation.
Finding the minimum value, for the objective function in Equation (9), is an iterative process. This process continues until it reaches the maximum number of iterations, typically defined by the user. Once the maximum iteration is reached, the α values are compared with the user-defined threshold value (C) of 10-3 using the Comp module (in Figure 7). In this case, the α values that are greater than the threshold value are considered as the support vectors, and the α values that are less than threshold values are discarded. These support vectors are important to determine the orientation of the hyper-plane. After obtaining the optimal solution for α (i.e., the minimum α value from Equation (9)), this optimal α value is forwarded via the multiplexer (M3) to a multiplier (Mul5) followed by a MAC module (MAC2) to compute the weight vectors (w) in Equation (7). These weight vectors and the α values are stored in the BRAM as well as in the DDR3-SDRAM for subsequent testing stage.
The internal architecture (or the datapath) of the reconstruct gradient computation is demonstrated in Figure 8. In case the optimization does not converge to a minimum α value during the iterative process of convex optimization (i.e., steps (d) to (h) in Stage 2, Figure 4), we utilize the reconstruct gradient module (in Figure 7) to adjust (or update) the value of the gradient parameter (Grad) and repeat the convex optimization process. Adjusting the values of the gradient parameter depends on the α values. As illustrated in Figure 8, the comparator module checks whether the gradient descent direction, of the current working set is positive or negative. In this case, for the Pset1, if the gradient descent direction is positive, then the remaining Pset1 values (from t to m) are assigned from the current number of active sets to the remaining number of data samples; and if the gradient descent direction is negative, then the working set is reset to select from the initial set (from 0 to t) as shown in step (h) in Figure 4. Next, the resulting Pset1 is selected via a multiplexer (Mux2), which is multiplied (with Mu13) with the α value. Then the addition operation is performed (using Add3) on the results from Add2 and the results from Mu13, in order to obtain the restructured Grad value. This updated Grad value is stored in the BRAM for subsequent iterations and analysis, i.e., the “Grad” signal shown in Figure 7.
Bias Value Computation Phase
The last phase of Stage 2 is the bias value computation phase. The bias value b in Equation (2) is also known as the offset value [6], which represents the intercept of the hyper-plane with respect to the origin. The bias value is computed with the following Equation (20) [9]:
Equation (20) is modified to include the sum of all the support vectors and then divided by the total number of support vectors to obtain the mean value as in Equation (21) [43]. As illustrated, Equation (21) provides the average values for all the support vectors, whereas Equation (20) identifies the max and min value for each class support vector.
The internal architecture (or datapath) of the bias value computation is demonstrated in Figure 9, which comprises a MAC module, adder, multiplier, subtractor, divider, and an accumulator register with a feedback loop to the adder. As shown in Figure 9, the results (elements) of the kernel matrix (K(xi,xj) obtained from Stage 1) are multiplied with the α value (obtained from Stage 2). The result of the Mul module is multiplied with the ysv value (which is the output label in Equation (1) corresponding to the support vector) and summed using the MAC module. The datapath of Mul and MAC corresponds to the second summation term in Equation (21). The output label of the support vector (ysv) is passed through the Add module and accumulator register (as in Figure 9), to obtain the first summation term in Equation (21). The result of the MAC is subtracted from the result of the accumulator register, in order to obtain the numerator in Equation (21). Finally, the result of the subtractor is divided by the total number of support vectors (nsv) to obtain the bias value. The final bias vale b is stored in the on-chip BRAM as well as in the DDR3-SDRAM for subsequent analysis.
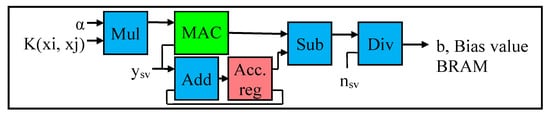
Figure 9.
Internal Architecture of Bias Value Computation.
4.2.3. Stage 3: Testing
Stage 3, which is our final stage, is the testing process. Typically for classification, the input dataset is divided into two samples: training and testing. During Stages 1 and 2, training is performed using the training set, whereas, in Stage 3, testing is performed using the testing set. For the testing process, the testing data samples are classified into -1 (minus one) class or +1 (plus one) class, based on the sign value of the function f(x) in Equation (3). In this case, if the output of Equation (3) is less than zero (f(x) < 0), then the test vectors are assigned to -1 class; and if the output of Equation (3) is greater than zero (f(x) > 0), then the test vectors are assigned to +1 class. We utilize the following formula (derived from [9]), which is known as the decision function (Equation (22)), to design and develop our testing (or classification) stage of the CO-based SVM algorithm.
The datapath of the testing process, which performs the final SVM classification, is illustrated in Figure 10. As mentioned before, this datapath is designed and developed based on Equation (22). In this stage, as in Equation (22), it is necessary to perform the sign verification for the testing (classification) process. As depicted in Figure 10, the datapath for the testing (classification) comprises a multiplier, a MAC module, an adder, a multiplexer, and a comparator. Initially, the test vectors (from the testing sample) are pre-fetched to the BRAM from the DDR3-SDRAM and forwarded to the kernel block in a pipelined fashion. As detailed in Stage 1 (Section 4.2.1), the kernel module maps these test vectors to the feature space. The two aforementioned steps are done in stage 1. In Stage 3 of our design, we reuse the pre-fetch and kernel modules from Stage 1 to reduce the total area occupied by our hardware design. The resource utilization is detailed in Section 5.
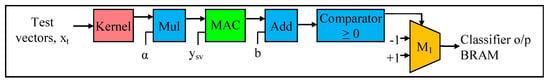
Figure 10.
Datapath of Testing Process.
In stage 3, as shown in Figure 10, the result of the kernel computation, which is the kernel matrix, is initially multiplied by the α value (obtained from Stage 2) using the Mul module. Then the result of the Mul module is forwarded to the MAC module to multiply with the output label of the support vector (ysv), and then perform the summation operation corresponding to Equation (22). The α value and the support vector parameters (ysv) are computed during the convex optimization process, as detailed in Section 4.2.2. Next, the bias value (b) (obtained in Stage 2 (in Equation (21)) is added (using the Add module) to the final summation result of the MAC module. Then the result of the adder is forwarded to the comparator to determine whether the adder result is greater than or equal to zero. Based on the results of the comparator, the test data samples are assigned to +1 class or -1 class, via the multiplexer.
5. Experimental Results and Analysis
We perform experiments to evaluate the feasibility and efficiency of our proposed embedded designs, for the convex optimization (CO)-based SVM algorithm, in terms of the speed-performance (speedup), accuracy, as well as scalability to handle different datasets with varying sizes. We measure the classification accuracy [43] and the speed-performance utilizing the following Equations (23) and (24), respectively. The scalability metric is to demonstrate our embedded designs’ capability to handle different datasets with varying data sizes, and a varying number of attributes, and other varying parameters that are commonly found in many datasets of machine learning applications.
During the initial design phase, we compare our results with the results from an open-source python code [43], in order to verify the correctness and functionality and operation, and accuracy of our proposed embedded designs. The execution times for the embedded designs are obtained in clock cycles and converted to seconds. Our proposed embedded hardware architectures (both the hardware and software) are executed on Virtex-6 FPGA (running at 100 MHz) and our proposed embedded software architectures are executed on the MicroBlaze embedded processor (running at 100 MHz) on the same FPGA, whereas the python code is executed on the desktop computer, with Intel-i7 processor, running at 2.3 GHz.
In this research work, the experiments are carried out to evaluate our embedded designs on two different benchmark datasets: the Ionosphere dataset [38] and the Wisconsin breast cancer diagnostic dataset [37] for machine learning applications. These datasets are stored in the DDR3-SDRAM and formatted accordingly to distinguish between the input features and the output labels. The data size is measured by considering the number of input vectors/samples (n) and the number of dimensions/features (m) in each vector. For our experiments, data sizes are varied to examine its impact on accuracy, speedup, and scalability.
For all our experiments, we partition the datasets into two sets: training and testing. The test set is considered as a percentage of the dataset to investigate the classification accuracy. In this case, the training set is varied from 10% to 90%, with an increment of 10%. Our experimental results on execution times, speedup, and classification accuracy for embedded hardware and embedded software designs are presented in Tables 2–7.
Apart from varying the data sizes for the testing and training, the number of iterations (to find the minima) is also varied for the training process, in order to examine the ability and the speedup of the convex optimization process to find these minima values.
5.1. Analysis of Resource Utilization
In order to examine the feasibility and area efficiency of our embedded hardware architectures, cost analysis on space (resource utilization) is carried out. In this case, after the implementation process, we obtain the significant resource utilization parameters, including the number of occupied slices, number of BRAMs, and the number of DSP48E1 slices, whereas the number of occupied slices typically consist of the slice registers and slice LUTs. These resource utilization statistics for our proposed embedded hardware design are presented in Table 1. As illustrated, for our embedded hardware design, the total number of occupied slices, number of DSP48E1, and number of BRAMs are 5216, 110, and 118 respectively. Considering the total number of logic slices (37,680 slices) in Virtex-6 FPGA, our hardware design occupies only 12.7% of the chip area. These resource utilization values (presented in Table 1) are compatible with our previously proposed FPGA-based hardware accelerators for applications with similar computation complexity (e.g., data mining/analytics) on an embedded device [13,40].

Table 1.
Resource Utilization for Embedded Hardware.
During our initial design phase, we explore the feasibility and tradeoff of utilizing the registers versus BRAMs to store the intermediate results. From this investigation, it is observed that utilization of BRAMs leads to a substantial reduction of the total number of occupied slices on the chip compared to that of the registers. Furthermore, BRAMs are imperative to hold the intermediate minima values during the sequential minimal optimization (SMO) process. For certain operations (such as vector computations), we utilize the DSP48E1 slices for single-precision floating-point computations. This design decision also leads to more area-efficient and lower clock latency for the floating-point operations compared to ones using the pure logic-based options.
Utilizing the on-chip BRAMs (and in few cases, using the registers) to hold the intermediate results, substantially reduces the execution time for numerous matrix computations inherent in the CO-based SVM algorithm, thus enhancing the overall speed-performance of this algorithm as illustrated in Section 5.4.
The pre-fetching techniques introduced to reduce the memory access latency and the on-chip BRAM to hold the data/results, indeed add more space (i.e., extra resources) to the overall design of the CO-based SVM algorithm. Thus, it is important to consider the speed-space tradeoffs, when designing certain algorithms/techniques, such as CO-based SVM, for machine learning applications, which typically require processing a large volume of data, especially on embedded platforms with their stringent area constraints.
5.2. Analysis of Classification Accuracy
We perform experiments to evaluate the classification accuracy of our proposed embedded designs for the CO-based SVM algorithm. In this case, the classification accuracy for the CO-based SVM algorithm is obtained with the varying data sizes for the maximum number of iteration of 1000. The classification accuracy is measured using Equation (23).
In order to measure the classification accuracy, we partition the datasets into two sets: training and testing. The training set is varied from 10% to 90%, with an increment of 10% to investigate the classification accuracy. Furthermore, for the linear, polynomial, and Gaussian radial basis function (RBF) in Stage 1, we investigate and select the following specifications: penalty parameter (C) to 1; degree of the polynomial (d) to 2, with a coefficient of 1; and γ to 0.0001.
The aforementioned parameters are varied to find a good fit for constructing the hyper-plane. Varying these parameters can potentially lead to under-fitting and over-fitting problems [43]. The under-fitting occurs when the SVM generalizes the main features of the data; whereas the over-fitting occurs when the SVM learns that it is sensitive to the noise [9,43]. As a result, for our experiments, we partition the datasets and utilize the cross-validation method [9] to select suitable constants. The cross-validation methods enable us to train the SVM by partitioning the dataset and also enable us to adjust the aforementioned parameters to obtain the best accuracy results. In this case, in order to avoid the under-fitting and over-fitting issues, the SVM is trained and tested with different parameters (C, degree, gamma, number of iterations) to construct a better data classifier.
The classification accuracy results (for both the embedded hardware and embedded software designs) for the overall CO-based SVM algorithm using Cancer and Ionosphere benchmark datasets are presented in Table 2, Table 3, Table 4, Table 5, Table 6 and Table 7, respectively. Three sets of accuracy results (in percentage) are obtained separately, when using three different mathematical kernels for Stage 1, i.e., linear (in Table 2 and Table 5), polynomial (in Table 3 and Table 6), and Gaussian RBF (in Table 4 and Table 7). The accuracy results are presented in column 7 of these tables.

Table 2.
Embedded Hardware and Embedded Software: Execution Time, Speedup, Accuracy using Cancer Benchmark Dataset for Linear Kernel, with C = 1, d = 2, γ = 0.0001.

Table 3.
Embedded Hardware and Embedded Software: Execution Time, Speedup, Accuracy using Cancer Benchmark Dataset for Polynomial Kernel, with C = 1, d = 2, γ = 0.0001.

Table 4.
Embedded Hardware and Embedded Software: Execution Time, Speedup, Accuracy using Cancer Benchmark Dataset for Gaussian RBF Kernel, with C = 1, d = 2, γ = 0.0001.

Table 5.
Embedded Hardware and Embedded Software: Execution Time, Speedup, Accuracy using Ionosphere Benchmark Dataset for Linear Kernel, with C = 1, d = 2, γ = 0.0001.

Table 6.
Embedded Hardware and Embedded Software: Execution Time, Speedup, Accuracy using Ionosphere Benchmark Dataset for Polynomial Kernel, with C = 1, d = 2, γ = 0.0001.

Table 7.
Embedded Hardware and Embedded Software: Execution Time, Speedup, Accuracy using Ionosphere Benchmark Dataset for Gaussian RBF Kernel, with C = 1, d = 2, γ = 0.0001.
5.2.1. Analysis of Classification Accuracy with Varying Data Sizes
From Table 2, Table 3, Table 4, Table 5, Table 6 and Table 7, it is observed that the classification accuracy varies with the different datasets as well as with varying percentages of training sets. At a glance, the classification accuracy seems to increase with the increasing percentage of the training set for both datasets. For instance, classification accuracy increases: from 83–93% (in Table 2) and from 66–100% (in Table 5) with the linear kernel; from 81–96% (in Table 3) and 66–100% (in Table 6) with the polynomial kernel; and 74–90% (in Table 4) and 60–98% (in Table 7) with the Gaussian RBF kernel. From Table 5, Table 6 and Table 7, the Ionosphere datasets have 100% classification accuracy, when the percentage of the training set is 90% of the dataset with linear and polynomial kernels. From Table 2, Table 3 and Table 4, the Cancer benchmark dataset achieves the best classification accuracy of 96% with the polynomial kernel, when the percentage of the training set is 90% of the dataset. It should be noted that the classification accuracy results are the same for our embedded hardware designs as well as for our embedded software designs.
Apart from our embedded hardware and software designs, the classification accuracy experiments are also performed on the python code running on the desktop computer. The accuracy graphs of the python code design results when using the linear, polynomial, and Gaussian RBF mathematical kernels are presented in Figure 11 and Figure 12 for the Cancer and Ionosphere benchmark datasets (with the maximum number of iteration of 1000), respectively. The accuracy results of our designs when using the linear, polynomial, and Gaussian RBF mathematical kernels (from Table 2, Table 3, Table 4, Table 5, Table 6 and Table 7) are also presented in Figure 11 and Figure 12 for the Cancer and Ionosphere benchmark datasets, respectively.
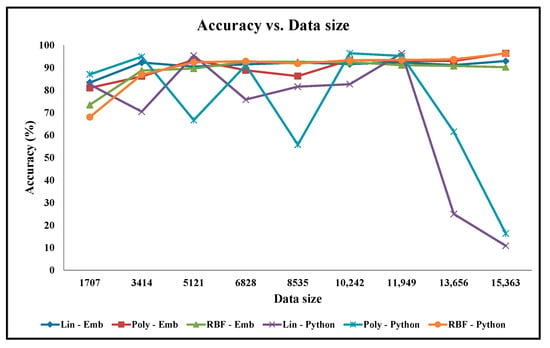
Figure 11.
Graph of Classification Accuracy vs. Data Size for Cancer Benchmark Dataset.
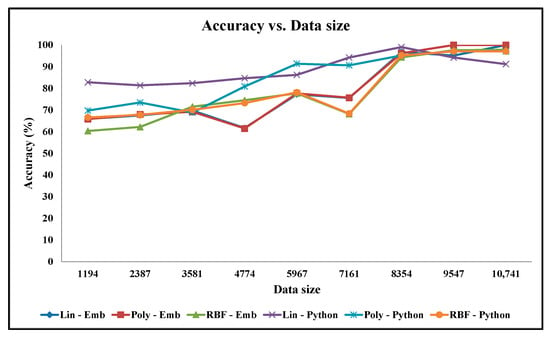
Figure 12.
Graph of Classification Accuracy vs. Data Size for Ionosphere Benchmark Dataset.
From Figure 11, it is observed that the classification accuracy of the python codes does not increase with the increasing percentage of the training set for the Cancer benchmark dataset. In this case, the classification accuracy results of the python code seem highly inconsistent compared to the classification accuracy results of our embedded designs. Furthermore, the classification accuracy results seem inconsistent especially among the two python codes: one with the linear kernel and one with the polynomial kernel. Conversely, classification accuracy results for our embedded designs are almost overlapping for all three kernels. Visually, as depicted in Figure 12, the classification accuracy results of the python code seem to increase with the increasing percentage of the training set for the Ionosphere benchmark dataset. In this case, the classification accuracy results seem slightly inconsistent especially among the two python codes: one with the linear kernel and the other with the polynomial kernel; whereas the classification accuracy results for our embedded designs are almost overlapping for all three kernels. As illustrated, the classification accuracy results of the python code, with the Gaussian RBF kernel, are quite similar to that of our embedded designs for both the benchmark datasets.
The inconsistencies from the python codes (in Figure 11) might be due to the over-fitting problems when executed with a constant number of iterations (i.e., 1000 iterations). From our experiments, it is observed that these python codes converge to a minimum value with a free-running number of iterations. In this case, the number of iterations is quite high, for instance, the number of iterations is 18,000 for 10% of the training dataset, and 13,592,546 for 60% of the training dataset. Although these inconsistencies may require further investigations on the python codes on desktop computers, it is beyond the scope of this paper as well as and our research on hardware accelerators for machine learning applications on embedded devices.
From these results, it is evident that classification accuracy varies with different datasets, with varying data sizes, as well as with different classification techniques. As detailed in Section 1, selecting a suitable classifier for a specific dataset is not a trivial task. By employing the cross-validation method, we can vary and select the most appropriate parameters that can indeed facilitate this task, which in turn will lead to better classification results.
5.2.2. Analysis of Classification Accuracy with Varying Number of Iterations
The aforementioned classification accuracy results are obtained with varying data sizes and with a constant number of iterations. We perform additional experiments to analyze the classification accuracy results with a varying number of iterations and with constant data size. In this case, we select the training set data size of 50% for both the Ionosphere and Cancer datasets. In this case, we vary the maximum number of iterations from 100 to 2500 with an increment of 500, to find the minima value. The classification accuracy results of our designs as well as of the python codes when using the linear, polynomial, and RBF mathematical kernels are shown in Figure 13 and Figure 14, for the Cancer and Ionosphere benchmark datasets, respectively.
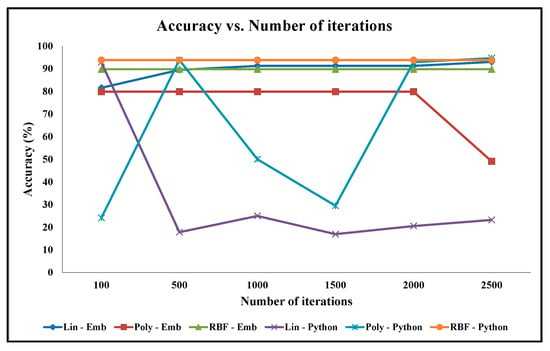
Figure 13.
Graph of Classification Accuracy vs. Number of Iterations for Cancer Benchmark Dataset.
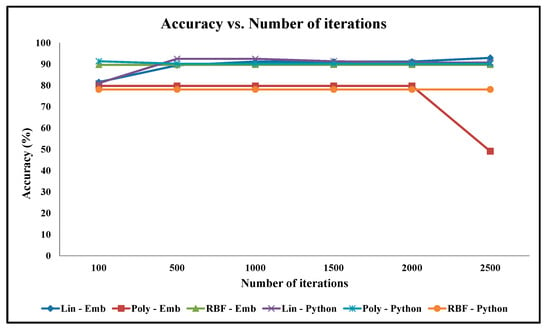
Figure 14.
Graph of Classification Accuracy vs. Number of Iterations for Ionosphere Benchmark Dataset.
As illustrated in Figure 13, the classification accuracy results of the python code seem highly inconsistent compared to the classification accuracy results of our embedded designs, for a varying number of iterations for the Cancer dataset. Furthermore, the classification accuracy results seem highly inconsistent especially among the two python codes: one with the linear kernel and the other with the polynomial kernel for the Cancer dataset. Visually, as depicted in Figure 13, the classification accuracy results of our embedded designs are almost the same; for instance, for our CO-based SVM with the linear kernel, the accuracy varies from 83% to 92%, with the maximum number of iterations varying from 100 to 2500, respectively, whereas with the Gaussian RBF kernel, the accuracy remains the same at 90% regardless of the number of iterations. For our CO-based SVM with the polynomial kernel also, the accuracy remains the same at 80%, with the maximum number of iteration varying from 100 to 2000. However, the accuracy decreases from 80% to 50%, with the maximum number of iterations varying from 2000 to 2500, due to the over-fitting problem. As illustrated, the classification accuracy results with the Gaussian RBK kernel are not impacted dramatically by the varying number of iterations and are almost the same for the python code as well as for our embedded designs.
For the Ionosphere datasets, it is observed that the convex optimization process converges to the minima with less number of iterations, for instance in 100 iterations in some cases. As a result, the impact of the number of iterations on accuracy is insignificant, as shown in Figure 14.
5.3. Analysis of Execution Time
As detailed in Section 4.1, in order to evaluate the speed-performance of our embedded hardware designs, we design and implement the embedded software for the CO-based SVM algorithm. The embedded software design is executed on the MicroBlaze soft processor on the same ML605 development platform. The execution times for both the embedded hardware and embedded software designs are obtained using the AXI Timer running at 100 MHz on the ML605 board. These execution times are measured in real-time, while our embedded designs are actually running on the chip. In this case, we design the AXI Timer in cascade mode to measure the accurate execution time for all three stages of the CO-based SVM algorithm. This is mainly because in certain scenarios, especially for large datasets, the execution time exceeds the allowable timer counter value of the AXI Timer. In order to resolve the counter overflow issue, the AXI timer is designed utilizing two timers in cascade mode.
The execution times for both our embedded hardware and embedded software designs are obtained with the varying data sizes for the maximum number of iterations of 1000. The execution times for the overall CO-based SVM algorithm using the Cancer and Ionosphere benchmark datasets are presented in Table 2, Table 3, Table 4, Table 5, Table 6 and Table 7, respectively. Similar to the classification accuracy results, three sets of execution times for embedded software and embedded hardware designs are obtained separately, when using three different mathematical kernels for Stage 1, i.e., linear (in Table 2 and Table 5), polynomial (in Table 3 and Table 6), and RBF (in Table 4 and Table 7). The execution time for each set (for both the embedded hardware and software) is measured 10 times and the average is presented in columns 4 and 5 of these tables, respectively.
The execution times for the embedded hardware and embedded software designs for the CO-based SVM algorithm using the linear, polynomial, and Gaussian RBF kernels are illustrated in Figure 15 and Figure 16, respectively, for the Cancer benchmark datasets. As illustrated, the execution times increase almost exponentially with the increasing data sizes, for both the embedded hardware and embedded software designs. Somewhat similar results are obtained when using the Ionosphere dataset. The CO-based SVM with linear kernel takes less execution time compared to that of the polynomial kernel. As illustrated in Figure 15 and Figure 16, the execution times are the highest for CO-based SVM with the polynomial kernels, whereas the execution times are the lowest for CO-based SVM with the linear kernels for the Cancer dataset.
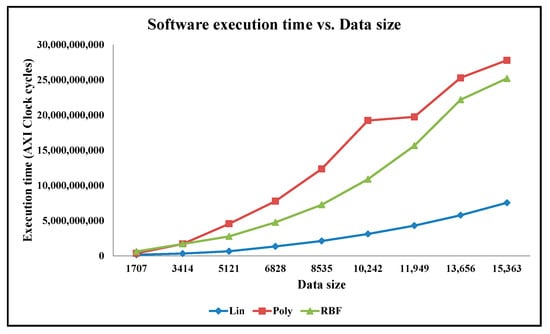
Figure 15.
Embedded Software for CO-Based SVM: Execution Times vs. Data Size for Cancer Benchmark Dataset.
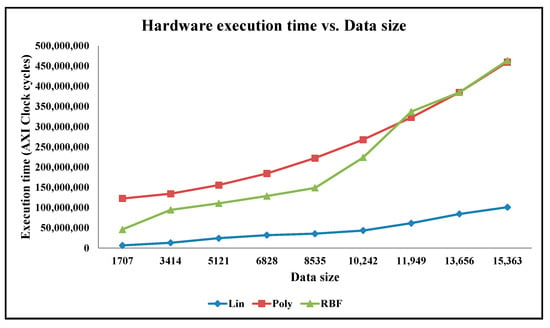
Figure 16.
Embedded Hardware for CO-Based SVM: Execution Times vs. Data Size for Cancer Benchmark Dataset.
Analysis of Execution Times with Varying Number of Iterations
The aforementioned execution times are obtained with varying data sizes and with a constant number of iterations. Similar to accuracy analysis, we perform additional experiments to analyze the execution times with a varying number of iterations and with constant data size. In this case, we vary the maximum number of iterations from 500 to 3000 with an increment of 500, to find the minima value. The execution times for both the embedded hardware and embedded software are also obtained with the varying number of iterations for both the benchmark datasets with the training set data size of 50%. The embedded hardware execution times for Cancer and Ionosphere datasets are presented in Figure 17 and Figure 18, respectively. Visually, as shown in Figure 17, for our embedded hardware designs, the execution times increase almost linearly with the increasing number of iterations, for all three kernels, for the Cancer benchmark datasets. For the Ionosphere benchmark dataset, as depicted in Figure 18, the embedded hardware execution times increase almost linearly with the increasing number of iterations, for the linear and polynomial kernels; whereas for the RBF kernel, the embedded hardware execution times remain the same with the increasing number of iterations.
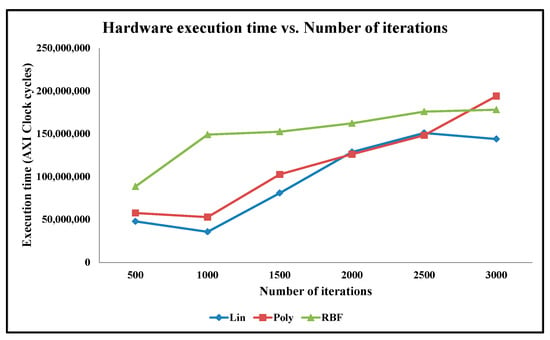
Figure 17.
Embedded Hardware for CO-Based SVM: Execution Times vs. Number of Iterations for Cancer Benchmark Dataset.
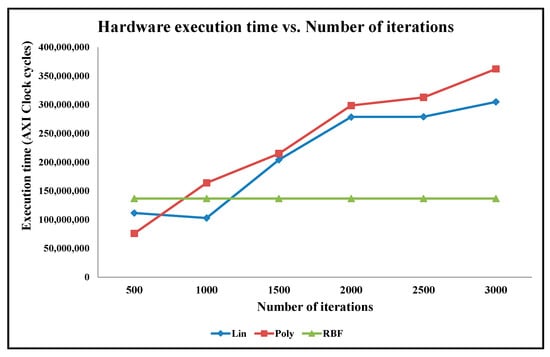
Figure 18.
Embedded Hardware for CO-Based SVM: Execution Times vs. Number of Iterations for Ionosphere Benchmark Dataset.
For our embedded designs, we utilize the maximum number of iterations (in our case, 1000 iterations) as our threshold point to find the minima value, instead of implementing a specific stopping criterion. Hence, our convex optimization solver has to reach the maximum number of iterations, in order to complete the execution of the CO-based SVM algorithm. Conversely, a stopping criterion terminates the execution of the CO-based SVM algorithm, when the objective function converges to the minima value, which may or may not reduce (or increase) the total execution time.
5.4. Analysis of Speedup
The performance gain (or speedup), resulting from the embedded hardware design over embedded software running on MicroBlaze, for the CO-based SVM algorithm using three different mathematical kernels, is presented in column 6 in Table 2, Table 3, Table 4, Table 5, Table 6 and Table 7. The speedup is measured using Equation (24). Figure 19 and Figure 20 demonstrate the speedup versus the data sizes (percentage of the training set) for our embedded hardware design for the CO-based SVM with the linear, polynomial, and Gaussian RBF kernels for the Cancer and Ionosphere benchmark datasets, respectively. At a glance, as shown in Figure 19 and Figure 20, the speedup typically increases as the percentage of training set increases for both datasets.
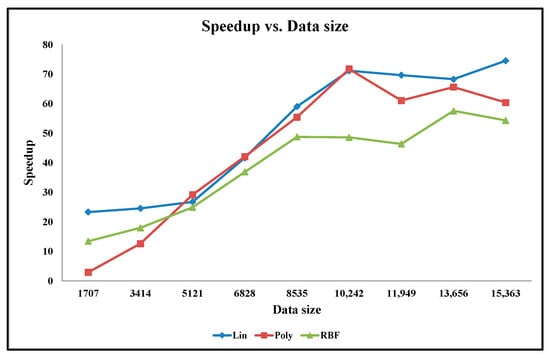
Figure 19.
Embedded Hardware vs. Embedded Software for CO-Based SVM: Speedup vs. Data Size using Cancer Benchmark Dataset.
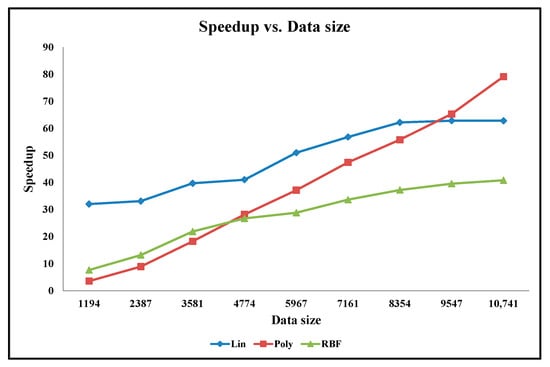
Figure 20.
Embedded Hardware vs. Embedded Software for CO-Based SVM: Speedup vs. Data Size using Ionosphere Benchmark Dataset.
For the Ionosphere benchmark dataset, as in Figure 20 (and from Table 5, Table 6 and Table 7), for the one with the polynomial kernel, the speedup increases linearly (from 4 times to 79 times faster than the software counterparts) when the percentage of training set increases from 10% to 90%; for the one with the linear kernel, the speedup increases almost linearly (from 32 times to 62 times faster than the software counterparts) when the percentage of training set increases from 10% to 70% and the speedup remains the same (62 times faster) when the percentage of training set increases from 70% to 90%; for the one with the RBF kernel, the speedup also increases almost linearly (from 8 times to 41 times faster than the software counterparts) when the percentage of the training set increases from 10% to 90%.
For the Cancer benchmark dataset, as in Figure 19 (and from Table 2, Table 3 and Table 4), for the one with the polynomial kernel, the speedup increases linearly (from 3 times to 72 times faster than the software counterparts) when the percentage of training set increases from 10% to 60% and the speedup drops slightly when the percentage of training set increases from 70% to 90%; for the one with the linear kernel, the speedup increases linearly (from 23 times to 75 times faster than the software counterparts) when the percentage of training set increases from 10% to 60% and from 70% to 90%; for the one with the RBF, the speedup increases linearly (from 14 times to 58 times faster than the software counterparts) when the percentage of training set increases from 10% to 60% and from 80% to 90%.
As mentioned earlier, additional software experiments are performed on a desktop computer; thus, we also compare our embedded hardware designs running at 100 MHz on Virtex-6 FPGA with the baseline python software design on the Intel i7 processor running at 2.3 GHz. In this case, our embedded hardware design achieves 3.1 times speedup compared to the python design for the Cancer dataset with the linear kernel; and our embedded hardware design achieves 34.8 times speedup compared to the python design for the Ionosphere dataset with the polynomial kernel. These results further justify the efficiency of our proposed embedded hardware designs.
In summary: It is observed that for the CO-based SVM algorithm, as the number of samples (i.e., vectors) increases, the accuracy and total speedup also increase. In this case, when the CO-based SVM classifier has more samples to learn, it could lead to identifying complex patterns, and also generating a better separating hyper-plane. Furthermore, as the size of the matrices is increasing, as well as the complexity of the computations/operations is increasing, customized and optimized designs might be the best avenue to accelerate and enhance various performance metrics of the CO-based SVM algorithms, compared to the conventional computing platforms such as general-purpose processors.
5.5. Analysis of Existing Works on FPGA-Based Hardware Architectures for CO-Based SVM
We performed an extensive investigation on the existing works on FPGA-based hardware architectures for CO-based SVM algorithms in the published literature. Since we could not find any related work specifically for the CO-based SVM, we extended our investigations to the existing works on FPGA-based hardware for a general SVM. Our investigation revealed that there are many papers on FPGA-based hardware for a SVM; however, we decided to select, discuss, and present some papers that are most recent and/or closely related to our proposed hardware architectures and techniques for creating a CO-based SVM. Hence, it should be noted that this is not an exhaustive analysis of the existing works on FPGA-based hardware architectures for SVMs. Detailed analysis of other existing works can be found in some survey papers such as [47,48].
An FPGA-based parallel processing hardware architecture was proposed for a SVM using stochastic gradient descent (SGD) as the training method, in [49]. The authors demonstrated the scalability of the SGD approach for a SVM in terms of fixed-point vs. single-precision floating-point computations. The hardware design was generated using the Xilinx System Generator design tool and executed on the Xilinx ML605 board with Virtex-6 FPGA. The authors did not discuss the advantages/disadvantages of utilizing system generator tools versus ISE tools, since the former might not generate the most efficient hardware designs. As a result, the occupied area reported seems very high for the hardware architecture, considering that the authors did not provide a system-level design. Furthermore, no testing phase was proposed in the hardware. In this case, the synthesis results were obtained and reported, in terms of area, time, and throughput; however, the classification accuracy results were not reported. From the results, it is evident that parallelization led to the increase in the occupied area, thus confirming that higher speedup due to parallelization, comes with the penalty of a larger occupied area on the chip. The proposed design could not execute datasets with more than 4 features/attributes, which is indeed a limitation when executing a large volume of data with many attributes. Conversely, our proposed design can execute datasets with varying sizes and with any number of features/attributes.
In [50], an energy-efficient embedded binarized SVM architecture was proposed and implemented on an FPGA. The computation kernels were designed in C/C++ and transformed into HDL using Xilinx HLS (high-level synthesis) tools. The proposed hardware design was executed on Xilinx Virtex-6/7 FPGAs. The results were obtained and reported, in terms of area, speedup, power, and classification accuracy. The FPGA’s performance matric results (especially speedup and power) were compared with that of the CPU and GPU. From the results, it is evident that FPGA and GPU achieved significant speedup compared to the CPU. However, the power consumption of the GPU was significantly higher than that of the FPGA. These results illustrate that FPGA-based hardware architecture for SVM can achieve better performance-per-watt, thus are suitable for embedded devices with stringent power requirements. The authors only proposed the hardware design for the testing phase; however, in order to support real-time machine learning applications, it is imperative to provide hardware designs for both the training and the testing phases.
An FPGA-based hardware accelerator was proposed for approximate SVM in [51], utilizing two approximation techniques, including precision scaling and loop perforation. The hardware was designed using the Xilinx Vivado HLS tool and executed on the Xilinx Zynq7 ZC706 board. The results were obtained and reported, in terms of area, speedup, and classification accuracy. From the results, it is evident that the approximate computing led to higher speedup, but with the penalty of larger occupied area (or resource utilization) on the chip, and lower classification accuracy. In some cases, the significant accuracy loss did not compensate with a significant increase in speedup. Since the machine learning algorithms/techniques are being incorporated into many accuracy-critical applications, it is important to consider certain classification accuracy thresholds for approximating. Furthermore, as the authors indicated, the utilization of HLS led to more resource-intensive hardware architectures. It is not clear how the speedup values were obtained, since the authors did not discuss or present the experimental setup to obtain the execution times for the approximate SVM, in order to compute speedup. Additionally, numerical precision plays a major role in training the SVM using optimization; hence, it would be interesting to see how the precision scaling would impact the training phase since the proposed approximate SVM hardware accelerator is only for the testing phase.
In [52], an FPGA-based hardware design was proposed for a SVM classifier. In this case, three variable-size SVM models were implemented using different optimization techniques. The proposed hardware was designed using the Xilinx Vivado HLS tool and executed on the Xilinx Zynq7 ZC702 board. The results were obtained and reported in terms of area, speedup, power, and classification accuracy. As illustrated in [53], the HLS tools utilized to transform the designs written in C/C+ to HDL, do not necessarily produce the most efficient hardware, which would be an issue when realizing these designs on resource-constrained embedded devices. Additionally, in this paper, the training phase was done offline on software; hence, the support vectors were pre-computed and forwarded to the proposed hardware design, which is created only for the testing phase. This could be an issue for real-time machine learning applications.
An FPGA-based parallel processing architecture was proposed in [54] for the training phase of SVM using Sequential Minimal Optimization (SMO). The proposed hardware design was executed on Xilinx Virtex-6/7/Ultra-scale FPGAs. The synthesis results were obtained and reported in terms of area, throughput, and speedup; however, the classification accuracy results were not reported. In this case, the authors utilized the hardware-friendly kernel (HFK) for SVM training, which leads to a reduction in the precision of the floating-point operations. This in turn reduces the classification accuracy, which depends on the numerical precision. Although a marginal loss in accuracy is acceptable for testing, utilizing HFKs for training would result in an inefficient construction of a hyper-plane during training. Furthermore, the maximum operating frequencies of the designs seem quite low, considering the FPGAs utilized can run at 100 MHz or more. From the results and analysis, it is evident that the reported times, speedups, and throughputs, were estimated theoretically. In addition, the authors did not discuss and present any details regarding the hardware designs for exponent computations, memory management, and decision boundary conditions. Additionally, no testing phase was proposed in hardware.
In [53], an FPGA-based hardware-software co-design was proposed to accelerate the SVM algorithm by utilizing a two-level approach: first to optimize the global structure of the SVM, and; second, to refine it through the design exploration. The proposed architecture was designed using the Xilinx Vivado HLS tool and executed on the Xilinx Zynq Zedboard. The results were obtained and reported in terms of area, latency, and speedup; however, the classification accuracy results were not reported. As the authors indicated, for high values of SVM parameters, the resource utilization (i.e., occupied area) increased significantly, which would be an issue for embedded devices with stringent area requirements. In this paper, the authors extensively discuss and analyze the advantages/disadvantages of utilizing the HLS tools to transform the designs written in C/C+ to HDL, thus providing insight into the HLS inefficiencies, which would be very useful when creating optimized hardware architectures in order to improve certain performance metrics, including the latency. Additionally, no training phase is proposed in hardware.
An FPGA-based coarse-grained reconfigurable hardware architecture was proposed in [55] for various machine learning (ML) algorithms, including SVM, decision trees, and artificial neural networks. The hardware was designed using the Xilinx Vivado tool and executed on the Xilinx Virtex-7 FPGA. The results were obtained and reported in terms of area, and speedup; however, the classification accuracy results were not reported. The authors’ intention was to provide a single universal solution for the aforementioned ML algorithms. In this case, in order to change from one ML algorithm to another, the authors claim that the reconfigurable processing nodes (RPNs) of the proposed architecture can be reconfigured individually; however, no details are provided on how this can be done. This requires partial reconfiguration of the FPGA; thus, adding significant complexity to the design process, which has not been addressed or discussed in the paper. In addition, with the experiments and analysis, only one ML algorithm (or instance) was implemented at a time. In order to implement a different ML algorithm, the RPNs need to be reconfigured, which incurs a reconfiguration time overhead. This reconfiguration time overhead is not considered in the reported speedup. Furthermore, the proposed hardware architecture is only for the testing phase.
A scalable FPGA-based architecture was proposed in [56] to accelerate the SVM classification. The hardware was designed in VHDL and executed on the Altera Stratix III EP3SE260 board. The results were obtained and reported in terms of speedup; however, the occupied area was not reported. It is imperative to report resource utilization (or area) details in order to distinguish the speed-space tradeoffs, especially for small-footprint embedded devices. Additionally, no detailed discussion and analysis was provided on the classification accuracy results. Furthermore, in this paper, the authors only proposed the hardware design for the testing phase. Hence, the support vectors were pre-computed and stored in the on-chip memory for subsequent processing during the testing phase. The same authors proposed a design flow for the SVM training phase in [57]; however, the experimental results and analysis do not show any evidence of an actual hardware design and implementation.
From this investigation, it is evident that most of the existing works proposed hardware architectures either for testing or for training, but not for both. Furthermore, most of these proposed hardware architectures were not generic or parameterized. Additionally, most of these architectures were not designed with embedded devices in mind. None of these works proposed system-level architectures and associated techniques to facilitate the real-time processing of machine learning applications. Consequently, the existing works did not report the corresponding system-level area and did not consider the associated memory access latency while reporting timing/speedup. As a result, we could not make any direct performance comparisons with the existing works on FPGA-based hardware architectures in the published literature. In summary, from this investigation, and to the best of our knowledge, we could not find any similar work to ours in the published literature that provides FPGA-based hardware accelerators for a CO-based SVM, especially on embedded devices, nor could we find any similar work that proposed system-level architectures, which is imperative for the machine learning applications in real-world scenarios.
6. Conclusions and Future Work
In this paper, we introduced a novel, customized, and optimized FPGA-based hardware accelerator for convex optimization (CO)-based support vector machines (SVM) on embedded platforms. Our embedded architectures are generic, parameterized, and scalable. Thus, without changing the internal hardware architectures, our embedded designs can be used for different datasets with varying sizes, can be executed on different embedded platforms, including the platforms with recent FPGAs such as Virtex-7 chips; and can be utilized for linear/non-linear separable, multi-dimensional datasets, making it suitable for various machine learning applications such as medical testing for cancer diagnosis, data analysis for quality control, image classifications, and speech recognition [58], while satisfying the associated constraints of the embedded devices. By providing generic and independent IPs (intellectual properties) for each stage, these independent IPs can be utilized for any machine learning application or other applications that require a SVM algorithm, and are not limited to a specific application in a specific field.
Our proposed design can be reconfigured to select a suitable mathematical kernel (out of three, i.e., linear, polynomial, and Gaussian radial basis function), based on the requirements of a given machine learning application. Our proposed designs were created and optimized considering the associated constraints/requirements of embedded devices/applications. We also introduced efficient system-level architecture for our FPGA-based hardware accelerator in order to process the data efficiently and effectively. With this system-level design, we designed and integrated unique pre-fetching techniques to reduce the memory access latency and to facilitate real-time data analysis and processing.
From our investigation on the existing works and to the best of our knowledge, we could not find any similar work in the published literature that provides FPGA-based architectures, especially for a CO-based SVM on embedded devices, nor could we find any similar work that proposed system-level architectures, which is imperative for machine learning applications for real-world scenarios.
Our embedded hardware accelerator for a CO-based SVM executed up to 79 times faster than its software counterpart running on the embedded microprocessors. This significant performance improvement was due to several hardware optimization techniques incorporated into our embedded architectures, including creating customized and optimized architectures by exploiting inherent parallelism and pipeline nature of the computations/tasks; designing computations/tasks to overlap with memory access; burst transfer and pre-fetching techniques. Our embedded designs (both hardware and software) achieved up to 100% classification accuracy and better performance (i.e., with a constant number of iterations to find the minima for the convex optimization, requiring much less number of iterations) compared to the python code on desktop computers. From the results and analysis of our proposed hardware accelerator, it was also observed that the accuracy results and the speedup results increased with the increasing data size. These performance metrics are crucial especially for real-time machine learning applications, which typically require processing a large volume of data. We also introduced lean and compact embedded software architecture for CO-based SVM, which was designed to fit into the available resources of the embedded microprocessor on-chip. These experimental results are encouraging and indeed show great potential in utilizing FPGA-based systems to support and accelerate machine learning applications, specifically on resource-constrained embedded platforms. The compact size of our proposed architectures, as well as the ability of our embedded designs to dynamically train from the unstructured datasets, further enhance the potential of implementing machine learning on embedded devices.
Currently, we are exploring the most recently proposed optimization techniques for SVMs [9] and deep neural mapping SVMs. As future work, we will investigate artificial and deep neural networks for machine learning applications in order to create hardware accelerators for these networks. We are also planning to provide dynamic reconfigurable hardware accelerators [13,59] for machine learning applications to integrate smart and adaptive traits into our designs. The experimental results presented in this paper were based on floating-point arithmetic. As future work, we will investigate and utilize suitable fixed-point arithmetic for our fundamental operators and will analyze the tradeoffs in terms of area, speedup, and accuracy. Additionally, as future work, we will investigate and utilize our proposed embedded architectures for the CO-based SVM for various applications/fields including fault diagnosis [60].
Author Contributions
Conceptualization, S.R.; Formal analysis, S.R. and D.G.P.; Investigation, S.R.; Methodology, S.R.; Project administration, D.G.P.; Supervision, D.G.P.; Validation, S.R.; Writing—original draft, S.R.; Writing—review & editing, D.G.P. Both authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Simplilearn Inc. Machine Learning: What It Is and Why It Matter. February 2019. Available online: https://www.simplilearn.com/what-is-machine-learning-and-why-it-matters-article (accessed on 1 August 2020).
- Mohsin, M.A.; Perera, D.G. An FPGA-Based Hardware Accelerator for K-Nearest Neighbor Classification for Machine Learning on Mobile Devices. In Proceedings of the 9th International Symposium on Highly-Efficient Accelerators and Reconfigurable Technologies, Toronto, ON, Canada, 6–7 June 2018; pp. 1–7. [Google Scholar]
- Smola, A.; Vishwanathan, S.V.N. Introduction to Machine Learning; Cambridge University Press: Cambridge, UK, 2008. [Google Scholar]
- Marr, B. Top 10 AI and Machine Learning Use Cases Everyone Should Know About. September 2016. Available online: https://www.forbes.com/sites/bernardmarr/2016/09/30/what-are-the-top-10-use-cases-for-machine-learning-and-ai/#57083d5994c9 (accessed on 1 August 2020).
- BCC Market Research. Machine Learning: Global Markets to 2022. May 2018. Available online: https://www.bccresearch.com/market-research/information-technology/machine-learning-global-markets.html (accessed on 1 August 2020).
- Bishop, C.M. Pattern Recognition and Machine Learning; Springer: New York, NY, USA, 2006. [Google Scholar]
- Wang, L. Support Vector Machines: Theory and Applications; Springer: Berlin/Heidelberg, Germany; New York, NY, USA, 2005. [Google Scholar]
- Campbell, C.; Ying, Y. Learning with Support Vector Machines; Morgan and Clay-pool Publishers: San Rafael, CA, USA, 2010; Volume 10. [Google Scholar]
- Piccialli, V.; Sciandrone, M. Nonlinear optimization and support vector machines. 4OR 2018, 16, 111–149. [Google Scholar] [CrossRef]
- Perera, D.G.; Li, K.F. Analysis of Single-Chip Hardware Support for Mobile and Embedded Applications. In Proceedings of the IEEE Pacific Rim Conference on Communication, Computers, and Signal Processing, Victoria, BC, Canada, 27–29 August 2013; pp. 369–376. [Google Scholar]
- Miro, J.P. FPGA-Based Accelerators for Convolutional Neural Networks on Embedded Devices. Master’s Thesis, Department of Electrical & Computer Engineering, University of Colorado Colorado Springs, Colorado Springs, CO, USA, May 2020. [Google Scholar]
- Ramadurgam, S. Optimized Embedded Architectures and Techniques for Machine Learning Algorithms for On-Chip AI Acceleration. Ph.D. Thesis, Department of Electrical & Computer Engineering, University of Colorado Colorado Springs, Colorado Springs, CO, USA, May 2021. [Google Scholar]
- Perera, D.G.; Li, K.F. FPGA-Based Reconfigurable Hardware for Compute Intensive Data Mining Applications. In Proceedings of the 6th IEEE International Conference on P2P, Parallel, Grid, Cloud, and Internet Computing, Catalonia, Spain, 26–28 October 2011; pp. 100–108. [Google Scholar]
- Raghavan, R.; Perera, D.G. A Fast and Scalable FPGA-Based Parallel Processing Architecture for K-Means Clustering for Big Data Analysis. In Proceedings of the IEEE Pacific Rim International Conference on Communications, Victoria, BC, Canada, 21–23 August 2017; pp. 1–8. [Google Scholar]
- Madsen, A.K.; Perera, D.G. Efficient embedded architectures for fast-charge model predictive controller for battery cell management in electric vehicles. EURASIP J. Embed. Syst. 2018, 2018, 2. [Google Scholar] [CrossRef]
- Madsen, A.K.; Trimboli, M.S.; Perera, D.G. An Optimized FPGA-Based Hardware Accelerator for Physics-Based EKF for Battery Cell Management. In Proceedings of the 2020 IEEE International Symposium on Circuits and Systems (ISCAS), Seville, Spain, 17–20 May 2020; pp. 1–5. [Google Scholar]
- Alkamil, A.; Perera, D.G. Efficient FPGA-Based Reconfigurable Accelerators for SIMON Cryptographic Algorithm on Embedded Platforms. In Proceedings of the IEEE International Conferences on Reconfigurable Computing and FPGAs, Cancun, Mexico, 9–11 December 2019. [Google Scholar]
- Alkamil, A.; Perera, D.G. Towards Dynamic and Partial Reconfigurable Hardware Architectures for Crypto-graphic Algorithms on Embedded Devices. IEEE Access 2020, 8, 221720–221742. [Google Scholar] [CrossRef]
- Anguita, D.; Boni, A.; Ridella, S. A digital architecture for support vector machines: Theory, algorithm, and fpga implementation. IEEE Trans. Neural Networks 2003, 14, 993–1009. [Google Scholar] [CrossRef] [PubMed]
- Venkateshan, S.; Patel, A.; Varghese, K. Hybrid Working Set Algorithm for SVM Learning with a Kernel Coprocessor on FPGA. IEEE Trans. Very Large Scale Integr. (VLSI) Syst. 2015, 23, 2221–2232. [Google Scholar] [CrossRef]
- Papadonikolakis, M.; Bouganis, C. Novel Cascade FPGA Accelerator for Support Vector Machines Classification. IEEE Trans. Very Large Scale Integr. (VLSI) Syst. 2012, 23, 1040–1052. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Peng, Y.; Zhao, G.; Peng, X. Accelerating on-line training of LS-SVM with run-time reconfiguration. In Proceedings of the 2011 International Conference on Field-Programmable Technology, New Delhi, India, 12–14 December 2011; pp. 1–6. [Google Scholar]
- Vapnik, V.N. The Nature of Statistical Learning Theory, 2nd ed.; Springer: New York, NY, USA, 2000. [Google Scholar]
- Vapnik, V.; Golowich, S.E.; Smola, A. Support Vector Method for Function Approximation, Regression Estimation and Signal Processing. In Proceedings of the 9th International Conference on Neural Information Processing Systems, Denver, CO, USA, 2–5 December 1996; pp. 281–287. [Google Scholar]
- Hsu, C.-W.; Lin, C.-J. A comparison of methods for multiclass support vector machines. IEEE Trans. Neural Networks 2002, 13, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Deng, N.; Tian, Y.; Zhang, C. Support Vector Machines: Optimization Based Theory, Algorithms, and Exten-Sions; Chapman & Hall, CRC: London, UK, 2012. [Google Scholar]
- Schölkopf, B.; Smola, A.J. Learning with Kernels: Support Vector Machines, Regularization, Optimization, and Beyond; MIT Press: Cambridge, MA, USA, 2002. [Google Scholar]
- Boyd, S.P.; Vandenberghe, L. Convex optimization; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2004. [Google Scholar]
- Chang, C.-C.; Hsu, C.-W.; Lin, C.-J. The analysis of decomposition methods for support vector machines. IEEE Trans. Neural Networks 2000, 11, 1003–1008. [Google Scholar] [CrossRef] [PubMed]
- Xilinx, Inc. DS-816 LogiCORE IP Floating-Point Operator v6.0. 18 January 2012. Available online: https://www.xilinx.com/support/documentation/ip_documentation/floating_point/v6_0/ds816_floating_point.pdf (accessed on 1 May 2021).
- Xilinx Inc. Virtex-6 Family Overview—DS150 (v2.5). August 2015. Available online: https://www.xilinx.com/support/documentation/data_sheets/ds150.pdf (accessed on 1 May 2021).
- Xilinx, Inc. ML605 Hardware User Guide, UG534 (v1.5). February 2011. Available online: www.xilinx.com/support/documentation/boards_and_kits/ug534.pdf (accessed on 23 May 2021).
- Xilinx, Inc. Available online: https://www.xilinx.com/products/boards-and-kits/ek-v6-ml605-g.html#hardware (accessed on 23 May 2021).
- Xilinx Inc. Virtex-6 FPGA Memory Resources—User Guide UG363 (v1.8). February 2014. Available online: https://www.xilinx.com/support/documentation/user_guides/ug363.pdf (accessed on 1 May 2021).
- Xilinx Inc. AXI Timer v2.0, LogiCORE IP Product Guide. October 2016. Available online: https://www.xilinx.com/support/documentation/ip_documentation/axi_timer/v2_0/pg079-axi-timer.pdf (accessed on 1 May 2021).
- Dua, D.; Graff, C. UCI—Machine Learning Repository. University of California, Irvine, School of Information and Computer Sciences. 2017. Available online: http://archive.ics.uci.edu/ml (accessed on 1 August 2020).
- Wolberg, W.H.; Street, W.N.; Mangasarian, O.L. Breast Cancer Wisconsin (Diagnostic). September 1995. Available online: https://archive.ics.uci.edu/ml/datasets/Breast+Cancer+Wisconsin+(Diagnostic) (accessed on 1 August 2020).
- Sigillito, V. Ionosphere Data Set Machine Learning Repository. 1989. Available online: https://archive.ics.uci.edu/ml/datasets/ionosphere (accessed on 1 August 2020).
- Xilinx Inc. AXI Reference Guide—UG761 (v13.1). March 2011. Available online: https://www.xilinx.com/support/documentation/ip_documentation/ug761_axi_reference_guide.pdf (accessed on 1 May 2021).
- Shahrouzi, S.N.; Perera, D.G. Optimized hardware accelerators for data mining applications on embedded platforms: Case study principal component analysis. Microprocess. Microsyst. 2019, 65, 79–96. [Google Scholar] [CrossRef]
- Perera, D.G.; Li, K.F. Embedded Hardware Solution for Principal Component Analysis. In Proceedings of the IEEE Pacific Rim International Conference on Communication, Computers, and Signal Processing, Victoria, BC, Canada, 23–26 August 2011; pp. 730–735. [Google Scholar]
- Xilinx Inc. LogiCORE IP AXI Master Burst DS844 (v1.00.a). June 2011. Available online: https://www.xilinx.com/support/documentation/ip_documentation/axi_master_burst/v1_00_a/ds844_axi_master_burst (accessed on 1 May 2021).
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Schölkopf, B.; Burges, C.J.C.; Smola, A.J. Advances in Kernel Methods: Support Vector Learning; MIT Press: Cambridge, MA, USA, 1999. [Google Scholar]
- Nilsson, P.; Shaik, A.U.R.; Gangarajaiah, R.; Hertz, E.; Nilsson, P. Hardware implementation of the exponential function using Taylor series. In 2014 NORCHIP; IEEE: Piscataway, NJ, USA, 2014; pp. 1–4. [Google Scholar]
- Platt, J. Sequential Minimal Optimization: A Fast Algorithm for Training Support Vector Machines; Microsoft: Washington, DC, USA, 1998; Available online: https://web.iitd.ac.in/~sumeet/tr-98-14.pdf (accessed on 1 August 2020).
- Afifi, S.M.; GholamHosseini, H.; Poopak, S. Hardware implementations of SVM on FPGA: A state-of-the-art review of current practice. Int. J. Innov. Sci. Eng. Technol. 2015, 2, 732–752. [Google Scholar]
- Afifi, S.; Hosseini, H.G.; Sinha, R. FPGA Implementations of SVM Classifiers: A Review. SN Comput. Sci. 2020, 1, 1–17. [Google Scholar] [CrossRef]
- Lopes, F.F.; Ferreira, J.C.; Fernandes, M.A.C. Parallel Implementation on FPGA of Support Vector Machines Using Stochastic Gradient Descent. J. Electron. 2019, 8, 631. [Google Scholar] [CrossRef]
- Elgawi, O.; Mutawa, A.M.; Ahmad, A. Energy-Efficient Embedded Inference of SVMs on FPGA. In Proceedings of the 2019 IEEE Computer Society Annual Symposium on VLSI (ISVLSI), Miami, FL, USA, 15–17 July 2019; pp. 164–168. [Google Scholar]
- Koliogeorgi, K.; Zervakis, G.; Anagnostos, D.; Zompakis, N.; Siozios, K. Optimizing SVM Classifier Through Approximate and High Level Synthesis Techniques. In Proceedings of the 2019 8th International Conference on Modern Circuits and Systems Technologies (MOCAST), Thessaloniki, Greece, 13–15 May 2019; pp. 1–4. [Google Scholar]
- Afifi, S.; Gholamhosseini, H.; Sinha, R. A system on chip for melanoma detection using FPGA-based SVM classifier. Elsevier J. Microprocess. Microsyst. (MICPRO) 2019, 65, 57–68. [Google Scholar] [CrossRef]
- Tsoutsouras, V.; Koliogeorgi, K.; Xydis, S.; Soudris, D. An Exploration Framework for Efficient High-Level Synthesis of Support Vector Machines: Case Study on ECG Arrhythmia Detection for Xilinx Zynq SoC. J. Signal Process. Syst. 2017, 88, 127–147. [Google Scholar] [CrossRef]
- Noronha, D.H.; Torquato, M.F.; Fernandes, M.A.C. A Parallel Implementation of Sequential Minimal Optimization on FPGA. Elsevier J. Microprocess. Microsyst. (MICPRO) 2019, 69, 138–151. [Google Scholar] [CrossRef]
- Vranjković, V.; Struharik, R. Coarse-grained reconfigurable hardware accelerator of machine learning classifiers. In Proceedings of the IEEE International Conference on Systems Signals and Image Processing (IWSSIP’16), Bratislava, Slovakia, 23–25 May 2016; pp. 1–5. [Google Scholar]
- Papadonikolakis, M.; Bouganis, C.-S. A novel FPGA-based SVM classifier. In Proceedings of the 2010 International Conference on Field-Programmable Technology, Beijing, China, 8–10 December 2010; pp. 283–286. [Google Scholar]
- Papadonikolakis, M.; Bouganis, C.-S. A Heterogeneous FPGA Architecture for Support Vector Machine Training. In Proceedings of the 2010 18th IEEE Annual International Symposium on Field-Programmable Custom Computing Machines, Charlotte, NC, USA, 2–4 May 2010; pp. 211–214. [Google Scholar]
- Available online: https://data-flair.training/blogs/applications-of-svm/ (accessed on 25 May 2021).
- Shahrouzi, S.N.; Perera, D.G. Dynamic partial reconfigurable hardware architecture for principal component analysis on mobile and embedded devices. EURASIP J. Embed. Syst. 2017, 2017, 212. [Google Scholar] [CrossRef]
- Glowacz, A. Ventilation Diagnosis of Angle Grinder Using Thermal Imaging. Sensors 2021, 21, 2853. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).