Abstract
Skin aging is a complex, multifaceted process influenced by both intrinsic and extrinsic factors. Understanding the molecular mechanisms underlying skin aging is crucial for developing effective anti-aging strategies. Dermal stem cells play a pivotal role in maintaining skin homeostasis, but their functionality is compromised with aging. This study investigated the impact of aging on dermal stem cells and explored the potential of natural extracts in modulating their biological characteristics. Using bulk RNA barcoding and sequencing (BRB-seq), we identified differentially expressed genes (DEGs) between young and aged dermal stem cells, revealing alterations in cellular processes, including cell proliferation, ECM synthesis, and RNA splicing. We also demonstrated that a natural extract, comprising callus cells and Alpine rose leaf extracts, influenced RNA splicing in aged dermal stem cells, leading to improved dermal structure and integrity in vitro. Our findings suggest that natural extracts may exert their effects through senolytic activity and the modulation of RNA splicing, a process crucial to gene expression and cellular function. This study underscores the potential of integrating high-throughput transcriptomics in understanding skin aging, presenting new avenues for the development of innovative, sustainable, and effective anti-aging strategies.
1. Introduction
Skin aging is a multifaceted process influenced by both intrinsic and extrinsic factors, leading to changes in skin elasticity, texture, and resilience [1]. Understanding the molecular mechanisms of skin aging is notably critical for the development of effective anti-aging strategies. Stem cells, particularly dermal stem cells, play a pivotal role in maintaining skin homeostasis and regeneration [2]. However, these cells are not unaffected by the aging process. Aging-related changes in stem cell functionality, including diminished proliferative capacity and altered differentiation potential, can significantly contribute to tissue aging [3]. The cellular mechanisms underlying stem cell aging are of great interest, as enhancing stem cell regenerative capacity is a favorable strategy to promote healthy aging. Several molecular pathways affect skin stem cells at the cellular level, including alterations in DNA repair and stability, telomere shortening, oxidative stress, mitochondrial dysfunction, and altered hormonal signaling [4,5,6]. As such, these pathways may further be interdependent, delivering cues that affect the skin stem cell niche in a complex manner. For instance, estrogen signaling is reported to help maintain telomere length [7], while oxidative stress and the resulting reactive oxygen species result in DNA damage and telomere shortening [8]. Several reports have found that maintaining adult stem cells required for skin maintenance can prevent premature skin aging [9,10,11]. Therefore, exploring the complex impact of aging on skin stem cells is essential to comprehending and combating skin aging at a cellular level.
Plant extracts have shown significant potential in stem cell modulation, attributed in part to their high content of beneficial molecules such as flavonoids, polyphenols, and isoflavones [12]. For example, flavonoids identified in an Alpine rose leaf extract (Rhododendron ferrugineum) have been identified as probable senolytic molecules [13]. A limitation of natural plant-derived materials is the sustainable cultivation and sourcing [14]. In this context, plant callus cells offer a sustainable alternative to obtain bio-active material from whole plant extracts. Grown under controlled conditions, a consistent profile of bioactive compounds can be achieved, further eliminating the risk of contamination by environmental pollutants and pathogens [15]. Callus cell extracts have further demonstrated promising potential for skin cell treatments, as they have been found to promote antioxidant activity, enhance collagen formation, and stimulate cell migration and proliferation [16,17]. However, the effects of plant callus extracts on dermal stem cells remain largely unexplored.
RNA sequencing (RNA-seq) offers numerous benefits in the field of molecular biology and clinical diagnostics [18]. Particularly bulk RNA-seq, which is performed on pooled cell populations, has been proven useful in gaining novel insights into the biological mechanisms underlying pathologies, such as very early onset inflammatory bowel disease [19], glioblastoma [20], and systemic sclerosis [21]. Despite being a powerful tool, the limitations, including the extensive time and cost of the technology, have driven researchers to find viable alternatives to address these constraints. One such approach is bulk RNA barcoding and sequencing (BRB-seq), a cost-effective method for high-throughput transcriptomics, which combines cost-effective single-cell RNA-seq efficiency with the performance of the bulk RNA-seq procedure [22]. Employing this technology facilitates the generation of libraries that parallel the benchmark TruSeq in terms of gene expression quantification and quality, even applied to low-quality RNA samples [22,23]. As such, BRB-seq represents a valuable tool to explore the molecular mechanisms present in both physiology and pathology.
Our study primarily aims to investigate the impact of aging on dermal stem cells by exploring the gene expression changes in both young and aged cells. Given the reported potential of plant extracts, particularly plant callus extracts, in skin cell treatments and the detailed insight gained from BRB-seq analysis, we identify a significant research opportunity. Our study, therefore, will further investigate the impact of a natural extract, comprised of grape (Vitis vinifera Linné) and apple (Malus Domestica Borkh) callus cells, and Alpine rose leaf extract (Rhododendron ferrugineum) on the biological and molecular attributes of aged dermal stem cells. By employing BRB-seq, we aim to generate a comprehensive gene expression profile of young and aged dermal stem cells, thereby elucidating the cellular mechanisms occurring during aging, and explore whether they can be influenced by the callus cell extract.
2. Materials and Methods
2.1. Preparation of Extract
The extract evaluated in this study was prepared with a mixture of callus cells obtained from grape callus cells (Vitis vinifera Linné) and a specific apple variety of Malus Domestica (Malus Domestica Borkh), the Uttwiler Spätlauber, according to an established and patented in-house protocol following the principle of wound-induced callus formation [24]. The mixture was further enriched with an Alpine rose leaf extract (Rhododendron ferrugineum). The final extract, SenoCellTecTM (hereafter referred to as “SCT extract”), was provided by Mibelle Biochemistry, Buchs, Switzerland.
2.2. Validation of Xeno-Free Culture Medium
To validate the use of a xeno-free media for the primary culture of human dermal stem cells (DSCs), human dermal fibroblasts were cultured in DSC media supplemented with 2% human serum (Curio Biotech, Visp, Switzerland, reference CB-DSC-GM) for a total of 7 days. Control cells were cultured in CB-DSC-GM supplemented with 2% fetal bovine serum (FBS). Brightfield images of the cultured cells, were taken after 7 days to compare cell morphology (Nikon ECLIPSE Ts2-FL, Tokyo, Japan).
2.3. CCK-8 Cell Viability Assay
For the determination of cell viability, cell proliferation and cytotoxicity assays using the Cell Counting kit-8 (CCK-8) (Cat. N.: HY-K0301, MCE (Med Chem Express, Monmouth Junction, NJ, USA)) were performed according to the kit’s instructions. Briefly, 8000 cells/cm2 were seeded into 96-well plates and preincubated for 24 h in a humidified incubator (37 °C, 5% CO2). Then, various concentrations of the SCT extract were added into the appropriate wells, and plates were incubated for 48 h (37 °C, 5% CO2). After this period, cell media were replaced with standard media, and 10 μL of CCK-8 solution was added into each well of the plates. After a further 2 h of incubation (37 °C, 5% CO2), the absorbance was measured at 450 nm using a microplate reader.
2.4. Dermal Stem Cell (DSC) Isolation and Treatment
DSCs were isolated from human skin explants by a standard methodology. Human skin samples from the abdomen of Caucasian females from two age groups (“young” group, 18–30 years old and “aged” group, >70 years old, n = 3 per group) were acquired from a tissue collection site in France with license number AC-2017 3030 from French Ministry of Higher Education and Research according to French laws including ethics regulations. The tissues had been collected during plastic surgery after informed consent had been obtained. Tissues were washed and treated with dispase II enzyme (Sigma Aldrich, Taufkirchen, Germany), overnight at 40 °C. After, the epidermis was peeled, the microvessels from the upper part of dermis were extruded mechanically, and the remaining tissue was washed several times. The tissue was then chopped into tiny pieces with a scalpel blade and digested with collagenase I (Thermo Fischer Scientific, Waltham, MA, USA) till fully digested by shaking every hour. Isolated cells were filtered using a cell strainer (Milian, Switzerland) and cultured in validated (supplementary data) xeno-free dermal stem cell growth medium (CB-DSC-GM, Curio Biotech, Visp, Switzerland) containing 2% human serum (Type AB—heat inactivated, P30-2501-HI, PAN-Biotech, Aidenbach, Germany). Cells were grown to a confluency of 60% in growth medium (CB-DSC-GM), and then the media were exchanged with fresh medium containing 0.5% human serum and the cells treated for 2 days either in presence or absence of 0.4% SCT extract. After 2 days of treatment, cells were detached, lysed in RNA lysis buffer (Bio-Rad Laboratories, Hercules, CA, USA), and RNA extraction was performed according to the manufacturer’s protocol. Untreated DSCs of young donors and aged donors were cultured as a control until confluency, without extract treatment, for 2 days according to the above-described procedure.
2.5. Flow Cytometry Analysis
The populations of stem-cell marker-expressing cells were determined on a Cytoflex S flow cytometer (Beckman Coulter, Brea, CA, USA). Cells were fluorescently labeled with anti-human monoclonal antibodies (BioLegend, San Diego, CA, USA) against CD29 (Cat. No. # 303015), CD49f (# 313607), CD133 (# 397905), CD34 (# 343503), CD31 (# NB100-65336AF488), CD73 (# 344003), CD90 (# 328107), and CD105 (# 323203). Unstained cells and appropriate IgG controls were used in parallel as controls. The protocol used was as follows: Cell suspensions were first fixed in 4% formaldehyde for 10 min, washed in a wash buffer (WB: 0.25% Tween-20 in DPBS + 5% FBS), and spun down. The cell pellets were resuspended in the presence of the labeled monoclonal antibody diluted in WB and incubated for 1 h on ice in the dark. Then the samples were washed twice with cold WB, and the cell pellets were resuspended in cold PBS and subjected to flow cytometry analysis. A total of 105 cells were measured at medium speed and gated to exclude doublets. Data acquisition s was performed with CytExpert Acquisition Software (version 2.5, Beckman Coulter, Brea, CA, USA) and analysis was performed using FloJoTM software (version 10.8.1, FlowJo, Ashland, OR, USA).
2.6. RNA Extraction and Quantification
Total RNA was extracted from the samples using the Aurum Total RNA Mini Kit (Bio-Rad Laboratories, Hercules, CA, USA) according to the manufacturer’s instructions. Briefly, adherent cells were lysed in lysis buffer, then one volume of 70% ethanol was added, and the resulting mix was directly added into each spin column and further processed following several sequential washes with high and low stringency buffers included in the kit. DNase I treatment was performed to eliminate eventual residual genomic DNA. Finally, RNA was eluted in 10 mM Tris-Cl pH 8.0. The resulting total RNA was quantified via NanoDrop 2000 apparatus (Thermo Fisher Scientific, Waltham, MA, USA).
2.7. Bulk RNA Barcoding (BRB) and Sequencing
For BRB mRNA sequencing, library preparation was performed using MERCURIUS BRB Sequencing Library preparation Kit (Alithea Genomics, Epalinges, Switzerland) and were subjected to Illumina sequencing with 5 million reads/sample, as previously described [22]. Briefly, NanoDrop quantified RNA was further quality controlled for RNA integrity on BioAnalyzer (Agilent technologies, Santa Clara, CA, USA). RNA was reverse transcribed directly in the reverse transcription (RT) reaction with barcoded oligo-dT primers. The resulting cDNA was pooled in a single tube prior to cDNA second strand synthesis. The Illumina-compatible Unique Dual Indexed (UDI) library preparation was performed, and the libraries were sequenced on the Illumina NovaSeq (Illumina, San Diego, CA, USA) instrument. The FASTQ files were sample-demultiplexed and aligned to the reference genome using STARsolo version 2.7.9a [25]. The resulting count matrices were used for downstream gene expression analysis. For the identification of differentially expressed genes (DEGs), the data were analyzed using the DESeq2 R package, and functional enrichment tests were performed for finding associated biological pathways. First, the raw count data obtained from BRB-seq experiments were pre-processed to remove low-quality reads and filter out lowly expressed genes. The data were then normalized using DESeq2 to account for differences in library size and composition. The variance of the data was estimated using a negative binomial distribution to model the read counts. LRT hypothesis testing was performed to identify DEGs based on adjusted p-values and logFC (fold change). The detected DEGs were further analyzed to identify Gene Ontology terms and KEGG pathways associated with them using functional enrichment tests. Significant enrichment was indicated by a minimum gene overlap of 3, p-value cut-off of 0.01, and minimum enrichment factor of 1.5.
2.8. Dermis Culture and Treatment
DSCs from young and aged donors were seeded in 3D cell culture inserts with a seeding density of 200,000 cells/insert in growth medium (CB-DSC-GM), serum-free, overnight. The following day, the culture medium was exchanged with fresh medium containing 0.5% human serum and treated in the presence or absence of 0.4% SCT extract for 10 days, with media change and treatment renewal every alternate day. After exposure, 3D models were harvested at day 10, fixed and processed for histological staining.
2.9. Histology and Immunohistochemistry
Inserts were fixed in 4% PFA for 15 min, then PFA was changed for 70% ethanol. The inserts were then processed for paraffin embedding. Paraffin-embedded sections (5 mm) were stained with H&E (Hematoxylin and Eosin) for gross morphology. Bright field histological microscopy (Nikon TS2-FL, Tokio, Japan, magnification 20×) images were acquired, and the thickness of 3D dermis formed in the presence and absence of SCT extract was quantified using an open-source software, ImageJ (Version 20, NIH, Bethesda, MD, USA).
2.10. Statistical Analysis
Statistical comparisons were performed using GraphPad Prism 8 software (GraphPad, Boston, MA, USA), and all data are expressed as mean ± SD of at least 3 independent experiments. Comparisons among different groups were made using one-way ANOVA or Student’s t-test with homoscedasticity. p values < 0.05 were considered significant.
3. Results
3.1. Characterization of Dermal Stem Cells by Fluorescence-Activated Cell Sorting (FACS)
To ensure optimal culture conditions for human stem cells, including DSCs, ideally defined culture conditions are applied. This includes the removal of non-human animal-derived components, which helps to eliminate issues associated with batch variation and pathogen transmission [26]. Therefore, isolated DSCs were cultured, and the experiments performed in xeno-free culture media supplemented with human serum. The primary cell culture in the defined xeno-free media was validated prior to the experiment’s start by assessing cell viability and morphology (Supplemental Figure S1) and provides, to the best of our knowledge, the first evidence of successful xeno-free human DSC culture. The isolated cells were characterized by FACS analysis to confirm their identity, with CD29, CD73, and CD90 as positive markers, whereas CD31 and CD34 were chosen as negative markers [27,28]. CD29 is a commonly used skin stem cell marker [29,30], which identifies dermal mesenchymal stem cells. Further, CD73 and CD90 are a favorable combination to label dermal stem cells [31,32], as they can distinguish different subsets of human dermal stem cells, including dermal stromal cells [33]. After cell isolation, FACS analysis of three independent donors demonstrated clearly negative signals for both CD31 and CD34, confirming the cells are not of a hematopoietic or endothelial origin (Supplemental Figure S2). In contrast, the isolated cell populations from all three donors had clear positive signals for the dermal stem cell markers CD29, CD73, and CD90. The combination of these three markers confirms that the cells, as isolated by the protocol used for this study, are indeed a dermal stem cell population.
3.2. Differential Gene Expression in Young and Aged Human DSCs
The BRB sequencing of DSCs isolated from young and aged donor tissues identified a total of 7409 genes, of which 75 were differentially expressed genes (DEGs). Compared to young DSCs, 20 genes were upregulated, and 55 genes were significantly downregulated in the aged DSC samples (Figure 1a). The volcano plot highlights the most significant up- and downregulated genes in aged donors, compared to young DSCs (Figure 1b). The full names and log2 fold changes of the top 10 up- and downregulated genes in aged cells are further depicted in Table 1. Compared to young DSCs, old cells had significantly upregulated genes associated with the inhibition of cell proliferation, such as PHLDA2 and SFRP1 [34,35]. Furthermore, protective factors such as glutathione peroxidase 3 (GPX3) and clusterin (CLU) were identified to be significantly downregulated [36,37]. Further downregulated genes include DKK1, CCN5, PTGIS, and SCARA5, which are involved in processes affecting cellular aging, such as cell proliferation [38], immune cell infiltration [39], and protein clearance [40].
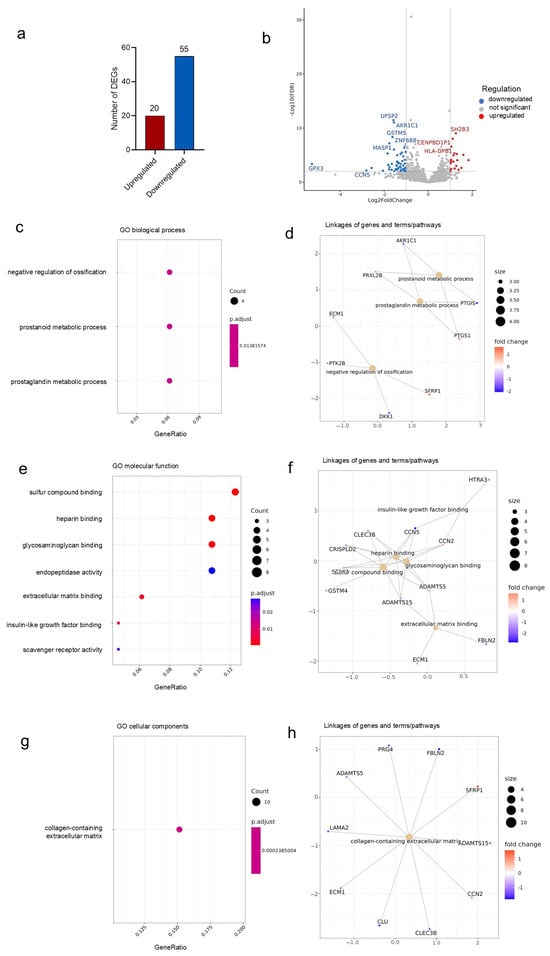
Figure 1.
Differential gene expression in young and aged human DSCs. BRB sequencing of young and aged DSCs identified 75 differentially expressed genes (DEGs), with 20 upregulated, and 55 significantly downregulated in the aged DSCs compared to young DSCs (a). A volcano plot (b) was used to visualize the DEGs identified. Gene Ontology analysis performed reveals changes in biological process (c), molecular function (e), and cellular components (g). Linkages of genes, terms, and pathways for the respective GO categories are graphed in (d,f,h), respectively. Colors correspond to significant fold change expression. Red, high expression; blue, low expression.

Table 1.
Top 10 DEGs up- and downregulated in aged DSCs.
Gene Ontology (GO) functional enrichment analysis was further performed for the DEGs identified by RNA-seq and grouped according to biological process (Figure 1c), molecular function (Figure 1e), and cellular components (Figure 1g). The analysis revealed altered processes (and linked genes) in the aged DSCs, such as the negative regulation of ossification (downregulated ECM1, DKK1, and PTK2B and upregulated SFRP), and prostanoid and prostaglandin metabolic processes (downregulated AKR1C1 and PTGIS and upregulated PTGS1 and PRXL2B) (Figure 1d). When analyzing the molecular functions, an altered regulation of various cell binding processes (sulfur compound, heparin, glycosaminoglycan, IGF, and ECM) was identified, which was linked to cell-communication genes including CCN2 (upregulated), CCN5 (downregulated), and the extracellular matrix (ECM)-modulating factors ADAMTS5, ADAMTS15, FBLN2, and ECM1 (downregulated) (Figure 1f). The GO further identified the collagen-containing ECM to be a cellular component significantly altered in aged DSCs (Figure 1h). In line with this, the DEGs associated with the ECM include LAMA2, ECM1, ADAMTS5, ADAMTS15, FBLN2, CLU, CLEC3B, and PRG4 (downregulated) and CCN2 and SFRP1 (upregulated).
3.3. Natural Extract Affects RNA Splicing in Aged Cells
RNA-seq can yield detailed insights to the cellular transcriptome and has been frequently used to identify effects of botanical actives on biological processes in vitro [41,42]. To identify possible effects of the SCT extract on aged DSCs, cells treated with the extract were compared to their untreated aged and cultured controls. In total, 8006 genes were identified, wherein, interestingly, only seven DEGs were found to be downregulated with the treatment in comparison to untreated aged controls (Figure 2a). Further, no significantly upregulated genes were detected in this analysis. The heat map of the RNA-seq analysis shows the expression pattern of the downregulated DEG in both technical and biological replicates, highlighting the strong differential expression between untreated and SCT-treated aged DSCs (Figure 2b). The seven DEGs identified were PNISR, RSRP1, MEG3, PNN, LUC7L3, AHSA2P, and GOLGA4 (Figure 2b). The full names and log2 fold changes are highlighted in Table 2.
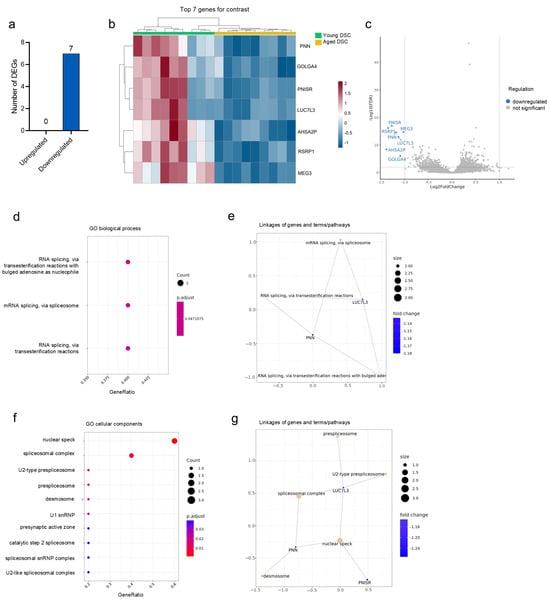
Figure 2.
Altered gene expression in aged DSCs treated with SCT Extract. BRB sequencing of aged DSCs treated with or without SCT identified 7 significantly downregulated genes in the treated DSCs (a). The heat map highlights the expression pattern of the downregulated DEGs in the different groups (b), and the volcano plot (c) visualizes the DEGs identified. Gene Ontology analysis performed reveals changes in biological process (d) and cellular components (f). Linkages of genes, terms, and pathways for the respective GO categories are graphed in (e,g) respectively. Colors correspond to significant fold change expression. Red, high expression; blue, low expression.

Table 2.
DEGs downregulated in aged DSCs treated with SCT extract.
Gene Ontology functional enrichment analysis identified molecular targets and pathways, which suggest that RNA splicing could underlie the biological activity of the SCT extract. Four of the seven DEGs identified, namely, PNN, PNISR, RSRP1, and LUC7L3, are associated with RNA binding activity and RNA splicing via spliceosome or transesterification reactions (Figure 2d) [43]. This was also reflected in the biological processes (Figure 2e) and cellular components identified to be involved, which included the nuclear speck, prespliceosome, and spliceosomal complex (Figure 2f). Further, GO analysis identified desmosomes as critical cell components modulated with the SCT treatment. This cellular component was in turn linked to PNN, which was originally identified as a desmosome-associated protein [44].
3.4. Improved Dermal Structure with Extract Treatment
To assess the functional outcome of the treatment, primary DSCs of aged tissues were cultured in the presence of SCT extract to form a stratified 3D structure and compared to untreated young and aged DSC controls. By adjusting the culture conditions and scaffold, the formation of organoids could be avoided [45], and the cells formed a stratified dermal structure. Compared to young DSCs, which formed a dense stratified multilayer (Figure 3a), the aged DSCs failed to form a comparable structure (Figure 3b). Morphologically, the structures appeared less dense and, upon the visualization and quantification of the dermis thickness (Figure 3d), it was observed that the dermal structures formed by aged DSCs were significantly thinner compared to those formed by young DSCs. Intriguingly, when aged DSCs were cultured in the presence of SCT extract, the structural integrity was rescued, and the DSC formed a dermal structure almost equivalent to the young DSCs (Figure 3c). This highlights the potential of the extract to stimulate aged cells, presumably by modifying RNA splicing machinery. As a result, dermal stem cell function was enhanced, which subsequently led to improved dermal integrity in vitro.
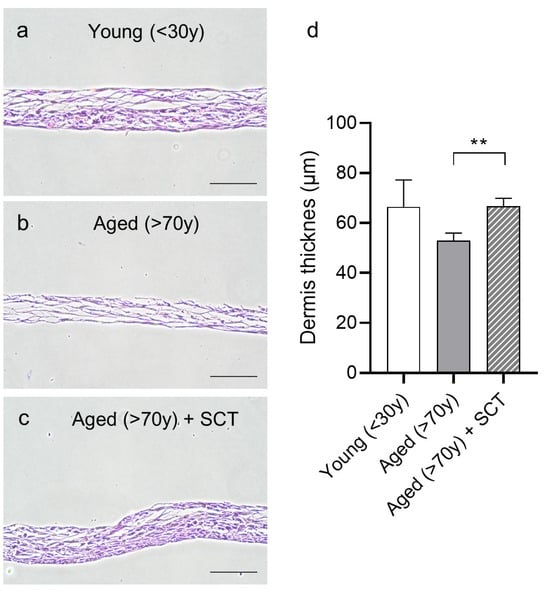
Figure 3.
Effects of SCT treatment on 3D dermis reconstruction. Representative images of 3D reconstructed dermis of cells isolated from young (<30 y) donors (a), aged donors (>70 y), (b) and aged donors treated with SCT extract (c) after 10 days of culture. Quantification and graphical representation of dermis thickness (µm) (d). Scale bar = 100 μm. Students t-test, mean ± SD, n = 3, ** p < 0.01 compared to aged (>70 y) untreated DSCs.
4. Discussion
The process of skin aging is a complex and inevitable phenomenon that has garnered increasing interest in recent years. Understanding the molecular mechanisms underlying skin aging is crucial for the development of effective anti-aging therapies and tissue rejuvenation strategies. The skin’s ECM plays a fundamental role in the aging process, both intrinsic and extrinsic [46]. Among the known ECM modulators, the family of CCN matricellular proteins plays a critical role in various physiological processes in the skin, such as cell proliferation, migration, adhesion, and ECM synthesis [47]. However, some may also contribute to pathological conditions such as fibrotic disease [48]. In this study, a differential regulation of two CCNs was identified in aged DSCs, with an upregulation of CCN2 and a downregulation of CCN5, respectively. The expression of different CCN proteins is indeed spatially and temporally regulated [49] and can further be modulated by environmental factors such as UV irradiation. In healthy human volunteers, the UV irradiation of sun-protected buttock skin demonstrated a significant increase in CCN2, whilst CCN5 was significantly reduced 24 h post irradiation [50]. Furthermore, in human fibroblasts, TGF-β-induced fibrosis led to the upregulated expression of CCN2 whilst CCN5, in contrast, was downregulated [48]. In regard to skin aging, studies report both the increased [51] and reduced [52,53] expression of CCN2 in human dermal fibroblasts. The increased expression of CCN2 was found to induce cellular senescence in human foreskin fibroblasts via p53 and P16INK4a [51]. In contrast, an age-related downregulation of CCN2 was observed in aged human skin, which was found to mediate collagen loss [52,53]. These differing observations may in part be allocated to the different culture conditions and exposures used. Elevated levels of CCN2 are further commonly observed in inflammatory and fibrotic skin conditions [54,55]. In contrast, CCN5 has been described as an antifibrotic molecule [56], and its expression may be induced by estrogen [57]. Considering that the DSCs of this study were obtained from female donors, it can be speculated that the age-related decline of estrogen contributes to the observed decrease in CCN5. Moreover, the characteristic expression of CCN2 and CCN5 observed in the aged DSCs could be associated with age-related fibrotic processes. Interestingly, plant-derived molecules such as fisetin and curcumin have been found to benefit the skin via the regulation of the CCN pathways [58]. As such, the flavonoids present in the natural extract of this study could potentially modulate these pathways, leading to positive effects on aging skin.
GO functional enrichment analysis further identified changes in prostanoid and prostaglandin metabolic processes in the aged DSCs. Prostaglandins, particularly prostaglandin E2 (PGE2), play a significant role in skin aging [59]. In human dermal fibroblasts (HDFs), PGE2 inhibits collagen production and induces matrix metallopeptidase 1 (MMP1) expression, contributing to dermal matrix degradation and skin aging [60]. Additionally, PGE2 induces cellular senescence in HDFs, leading to decreased cell proliferation and increased senescence-associated β-galactosidase staining [61]. Furthermore, senescent cells synthesize prostaglandins, which further promote the senescence status and reinforce the proliferative arrest [62]. As such, blocking PGE2 is a favorable approach to combat skin aging. In line with this, several natural compounds have been reported to prevent PGE2 synthesis [63,64,65].
When evaluating the extract treatment in this study, a substantial effect of the SCT treatment was observed on functional outcome, despite only a small number of DEGs associated with RNA splicing having been identified. Alternative RNA splicing is a post-transcriptional regulatory mechanism by which a single pre-mRNA transcript can be differentially processed into diverse mRNA isoforms, thereby expanding proteomic complexity and functional diversity from a limited set of genes [66]. The dysregulation of this mechanism is described as an emerging hallmark of aging [67] and may serve as a predictor of biological age and life expectancy, offering potential for developing therapeutics to extend healthy lifespan in humans [66]. Indeed, a study evaluating gene expression changes in aging murine skin observed an increase in alternatively spliced genes [68]. These alternatively spliced genes were consistently allocated to functions, including RNA processing and splicing, and found to be enriched within cellular components such as the nucleus and the spliceosome. Targeting RNA splicing has, therefore, emerged as a promising approach for anti-aging interventions. One of the first studies addressing such a modulation found that treating human dermal fibroblasts (NHDFs) with novel small resveratrol analogs was associated with altered splicing factor levels and a reversal of cellular senescence [69]. Further, in models of accelerated aging including Cockayne syndrome, Hutchinson–Gilford Progeroid syndrome (HGPS), and Werner syndrome, all three progeroid dermal fibroblast types showed dysregulated splicing factor gene expression [70]. Intriguingly, treatment with the kinase inhibitor trametinib led to altered gene expression of a panel of splicing factors in the HGPS and Cockayne syndrome cells, which was accompanied by a reduced senescence burden. Our study found that the SCT treatment of aged DSCs affected genes associated with the spliceosomal complex, the prespliceosome, and the nuclear speckles. Nuclear speckles, also known as splicing factor compartments, are located in the interchromatin regions of the cell nucleoplasm and serve as reservoirs for pre-mRNA splicing factors [71]. Based on the modulations observed, it can be speculated that the SCT extract exerts its restorative effects by modulating splicing factors, which in turn leads to improved cell function in the aged DSCs. When evaluating the extended duration of treatment, from 2 days in 2D models to 10 days for the 3D dermis functional assay, it can be speculated that further downstream mechanisms are initiated after prolonged treatment with the SCT extract. Further investigations, for example, with the targeted transcriptomics and evaluation of senescence burden in appropriate cell models, will certainly provide more insight into this proposed mechanism.
Dermal stem cells play a crucial role in skin maintenance, for example, growth factors derived and secreted from human dermal stem/progenitor cells can ameliorate UV-A-induced damage in NHDF [2]. Preserving dermal stem cell function is critical for skin homeostasis, which could be achieved by regenerative treatment with natural sources such as callus extracts. Indeed, it was previously observed that the plant-callus-derived shikimic acid effectively regenerates human skin by converting human dermal fibroblasts into multipotent skin-derived precursor cells, facilitating wound healing and dermal reconstruction [72]. Furthermore, the Rhododendrum ferrugineum extract was shown to have a senolytic effect in senescent fibroblasts [13]. Building on this understanding, the SCT extract examined in this study presents a promising strategy for addressing skin aging, as it potentially modulates RNA splicing, thereby influencing the expression and function of proteins critical to the maintenance of skin health and vitality. The successful cultivation of human DSCs under xeno-free conditions and the identification of DEGs between young and aged DSCs further offer a foundation for the exploration into the mechanisms of skin aging and potential therapeutic interventions.
5. Conclusions
Our findings highlight the potential of natural extracts in modulating biological characteristics of aged dermal stem cells. Importantly, our data suggest that these extracts may exert their effects through possible senolytic activity and the modulation of RNA splicing, a process crucial to gene expression and cellular function and described as an emerging hallmark of aging. In conclusion, this study underscores the potential of integrating high-throughput transcriptomics and natural extracts in understanding and combating skin aging. The findings present new avenues for the development of innovative, sustainable, and potentially more effective anti-aging strategies.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/cosmetics11050167/s1. Figure S1: Validation of xeno-free media; Figure S2: Fluorescence-activated cell sorting (FACS) characterization of human DSC.
Author Contributions
Conceptualization, C.C., V.V., F.Z., J.B. and F.W.; methodology, C.C. and V.V.; software, C.C., V.V. and J.B.; validation, C.C., V.V. and J.B.; formal analysis, C.C., V.V. and J.B.; investigation, C.C. and V.V.; resources, C.C. and V.V.; data curation, C.C., V.V. and J.B.; writing—original draft preparation, J.B.; writing—review and editing, J.B., K.N., F.W. and F.Z.; visualization, J.B., K.N. and V.V.; supervision, C.C.; project administration, C.C. and V.V.; funding acquisition, C.C. and F.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was sponsored by Mibelle Group, Buchs AG, Switzerland, and performed independently and blinded by Curio Biotech, Visp, Switzerland. The funder had no role in the design, data collection, data analysis, and reporting of this study.
Institutional Review Board Statement
Human skin samples from the abdomen of Caucasian females were acquired from a tissue collection site in France with license number AC-2017 3030 from French Ministry of Higher Education and Research according to French laws including ethics regulations. The tissues had been collected during plastic surgery after informed consent had been obtained. The study was conducted in accordance with the Declaration of Helsinki.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All data generated or analyzed during this study are included in this article and its Supplementary Material Files. Further enquiries can be directed to the corresponding author.
Acknowledgments
The authors thank the R&D Team of Mibelle Biochemistry for the preparation of the extracts. The authors further thank Roberto Salvi for providing support in experimental design and for performing experiments and analysis.
Conflicts of Interest
J.B., K.N., F.Z. and F.W. are employed by the Mibelle Biochemistry/Mibelle Group. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The funders had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Tobin, D.J. Introduction to skin aging. J. Tissue Viability 2017, 26, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.H.; Park, J.Y.; Lee, M.G.; Kang, H.H.; Lee, T.R.; Shin, D.W. Human dermal stem/progenitor cell-derived conditioned medium ameliorates ultraviolet a-induced damage of normal human dermal fibroblasts. PLoS ONE 2013, 8, e67604. [Google Scholar] [CrossRef] [PubMed]
- Zouboulis, C.C.; Adjaye, J.; Akamatsu, H.; Moe-Behrens, G.; Niemann, C. Human skin stem cells and the ageing process. Exp. Gerontol. 2008, 43, 986–997. [Google Scholar] [CrossRef] [PubMed]
- Flores, I.; Cayuela, M.L.; Blasco, M.A. Effects of Telomerase and Telomere Length on Epidermal Stem Cell Behavior. Science 2005, 309, 1253–1256. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Xuan, M.; Leung, V.Y.; Cheng, B. Stem cells and aberrant signaling of molecular systems in skin aging. Ageing Res. Rev. 2015, 19, 8–21. [Google Scholar] [CrossRef]
- Lyu, Y.; Ge, Y. Toward Elucidating Epigenetic and Metabolic Regulation of Stem Cell Lineage Plasticity in Skin Aging. Front. Cell Dev. Biol. 2022, 10, 903904. [Google Scholar] [CrossRef]
- Kyo, S.; Takakura, M.; Kanaya, T.; Zhuo, W.; Fujimoto, K.; Nishio, Y.; Orimo, A.; Inoue, M. Estrogen activates telomerase. Res. C 1999, 59, 5917–5921. [Google Scholar]
- Barnes, R.P.; Fouquerel, E.; Opresko, P.L. The impact of oxidative DNA damage and stress on telomere homeostasis. Mech. Ageing Dev. 2019, 177, 37–45. [Google Scholar] [CrossRef]
- Su, X.; Paris, M.; Gi, Y.J.; Tsai, K.Y.; Cho, M.S.; Lin, Y.L.; Biernaskie, J.A.; Sinha, S.; Prives, C.; Pevny, L.H.; et al. TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell Stem Cell 2009, 5, 64–75. [Google Scholar] [CrossRef]
- Naska, S.; Yuzwa, S.A.; Johnston, A.P.; Paul, S.; Smith, K.M.; Paris, M.; Sefton, M.V.; Datti, A.; Miller, F.D.; Kaplan, D.R. Identification of Drugs that Regulate Dermal Stem Cells and Enhance Skin Repair. Stem Cell Rep. 2016, 6, 74–84. [Google Scholar] [CrossRef][Green Version]
- Maity, P.; Singh, K.; Krug, L.; Koroma, A.; Hainzl, A.; Bloch, W.; Kochanek, S.; Wlaschek, M.; Schorpp-Kistner, M.; Angel, P.; et al. Persistent JunB activation in fibroblasts disrupts stem cell niche interactions enforcing skin aging. Cell Rep. 2021, 36, 109634. [Google Scholar] [CrossRef] [PubMed]
- Saud, B.; Malla, R.; Shrestha, K. A Review on the Effect of Plant Extract on Mesenchymal Stem Cell Proliferation and Differentiation. Stem Cells Int. 2019, 2019, 7513404. [Google Scholar] [CrossRef] [PubMed]
- Hubert, J.; Kotland, A.; Henes, B.; Poigny, S.; Wandrey, F. Deciphering the Phytochemical Profile of an Alpine Rose (Rhododendron ferrugineum L.) Leaf Extract for a Better Understanding of Its Senolytic and Skin-Rejuvenation Effects. Cosmetics 2022, 9, 37. [Google Scholar] [CrossRef]
- Chemat, F.; Abert-Vian, M.; Fabiano-Tixier, A.S.; Strube, J.; Uhlenbrock, L.; Gunjevic, V.; Cravotto, G. Green extraction of natural products. Origins, current status, and future challenges. TrAC Trends Anal. Chem. 2019, 118, 248–263. [Google Scholar] [CrossRef]
- Efferth, T. Biotechnology Applications of Plant Callus Cultures. Engineering 2019, 5, 50–59. [Google Scholar] [CrossRef]
- Buranasudja, V.; Rani, D.; Malla, A.; Kobtrakul, K.; Vimolmangkang, S. Insights into antioxidant activities and anti-skin-aging potential of callus extract from Centella asiatica (L.). Sci. Rep. 2021, 11, 13459. [Google Scholar] [CrossRef]
- Park, J.S.; Lim, Y.M.; Baik, J.; Jeong, J.O.; An, S.J.; Jeong, S.I.; Gwon, H.J.; Khil, M.S. Preparation and evaluation of beta-glucan hydrogel prepared by the radiation technique for drug carrier applications. Int. J. Biol. Macromol. 2018, 118, 333–339. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Collen, L.; Field, M.; Eran, A.; Mitsialis, V.; Barends, J.; Saccocia, G.; Bresnahan, M.; Yang, J.; Buijsen, F.; Combs, A.; et al. Bulk RNA-Sequencing of blood informs molecular diagnoses in very early onset inflammatory bowel disease. Inflamm. Bowel Dis. 2023, 29, S56. [Google Scholar] [CrossRef]
- Zhao, S.; Ji, W.; Shen, Y.; Fan, Y.; Huang, H.; Huang, J.; Lai, G.; Yuan, K.; Cheng, C. Expression of hub genes of endothelial cells in glioblastoma-A prognostic model for GBM patients integrating single-cell RNA sequencing and bulk RNA sequencing. BMC Cancer 2022, 22, 1274. [Google Scholar] [CrossRef]
- Kobayashi, S.; Nagafuchi, Y.; Okubo, M.; Sugimori, Y.; Shirai, H.; Hatano, H.; Junko, M.; Yanaoka, H.; Takeshima, Y.; Ota, M.; et al. Integrated bulk and single-cell RNA-sequencing identified disease-relevant monocytes and a gene network module underlying systemic sclerosis. J. Autoimmun. 2021, 116, 102547. [Google Scholar] [CrossRef] [PubMed]
- Alpern, D.; Gardeux, V.; Russeil, J.; Mangeat, B.; Meireles-Filho, A.C.A.; Breysse, R.; Hacker, D.; Deplancke, B. BRB-seq: Ultra-affordable high-throughput transcriptomics enabled by bulk RNA barcoding and sequencing. Genome Biol. 2019, 20, 71. [Google Scholar] [CrossRef] [PubMed]
- McCoy, R.C.; Taylor, R.W.; Blauwkamp, T.A.; Kelley, J.L.; Kertesz, M.; Pushkarev, D.; Petrov, D.A.; Fiston-Lavier, A.S. Illumina TruSeq synthetic long-reads empower de novo assembly and resolve complex, highly-repetitive transposable elements. PLoS ONE 2014, 9, e106689. [Google Scholar] [CrossRef] [PubMed]
- Blum, C.S.P.; Schmid, D.; Zulli, F. Cosmetic Preparation and Method for Preparing the Same. United States Patent Application US 12/148,241, 4 December 2008. [Google Scholar]
- Kaminow, B.; Yunusov, D.; Dobin, A. STARsolo: Accurate, fast and versatile mapping/quantification of single-cell and single-nucleus RNA-seq data. bioRxiv 2021. [Google Scholar] [CrossRef]
- Jabbarpour, Z.; Aghayan, S.; Arjmand, B.; Fallahzadeh, K.; Alavi-Moghadam, S.; Larijani, B.; Aghayan, H.R. Xeno-free protocol for GMP-compliant manufacturing of human fetal pancreas-derived mesenchymal stem cells. Stem Cell Res. Ther. 2022, 13, 268. [Google Scholar] [CrossRef] [PubMed]
- Newman, P.J.; Berndt, M.C.; Gorski, J.; White, G.C.; Lyman, S.; Paddock, C.; Muller, W.A. PECAM-1 (CD31) Cloning and Relation to Adhesion Molecules of the Immunoglobulin Gene Superfamily. Science 1990, 247, 1219–1222. [Google Scholar] [CrossRef]
- Sidney, L.E.; Branch, M.J.; Dunphy, S.E.; Dua, H.S.; Hopkinson, A. Concise review: Evidence for CD34 as a common marker for diverse progenitors. Stem Cells 2014, 32, 1380–1389. [Google Scholar] [CrossRef]
- Niu, X.; Li, J.; Zhao, X.; Wang, Q.; Wang, G.; Hou, R.; Li, X.; An, P.; Yin, G.; Zhang, K. Dermal mesenchymal stem cells: A resource of migration-associated function in psoriasis? Stem Cell Res. Ther. 2019, 10, 54. [Google Scholar] [CrossRef]
- Zhang, L.; Cen, Y.; Huang, Q.; Li, H.; Mo, X.; Meng, W.; Chen, J. Computational flow cytometric analysis to detect epidermal subpopulations in human skin. Biomed. Eng. Online 2021, 20, 22. [Google Scholar] [CrossRef]
- Denu, R.A.; Nemcek, S.; Bloom, D.D.; Goodrich, A.D.; Kim, J.; Mosher, D.F.; Hematti, P. Fibroblasts and Mesenchymal Stromal/Stem Cells Are Phenotypically Indistinguishable. Acta Haematol. 2016, 136, 85–97. [Google Scholar] [CrossRef]
- Rong, X.; Li, J.; Yang, Y.; Shi, L.; Jiang, T. Human fetal skin-derived stem cell secretome enhances radiation-induced skin injury therapeutic effects by promoting angiogenesis. Stem Cell Res. Ther. 2019, 10, 383. [Google Scholar] [CrossRef] [PubMed]
- Pfisterer, K.; Lipnik, K.M.; Hofer, E.; Elbe-Burger, A. CD90(+) human dermal stromal cells are potent inducers of FoxP3(+) regulatory T cells. J. Investig. Dermatol. 2015, 135, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, G.; Koul, S.; Ohki, R.; Maurer, M.; Borczuk, A.; Halmos, B. PHLDA2 is a key oncogene-induced negative feedback inhibitor of EGFR/ErbB2 signaling via interference with AKT signaling. Oncotarget 2018, 9, 24914–24926. [Google Scholar] [CrossRef] [PubMed]
- Elzi, D.J.; Song, M.; Hakala, K.; Weintraub, S.T.; Shiio, Y. Wnt antagonist SFRP1 functions as a secreted mediator of senescence. Mol. Cell Biol. 2012, 32, 4388–4399. [Google Scholar] [CrossRef] [PubMed]
- Janig, E.; Haslbeck, M.; Aigelsreiter, A.; Braun, N.; Unterthor, D.; Wolf, P.; Khaskhely, N.M.; Buchner, J.; Denk, H.; Zatloukal, K. Clusterin associates with altered elastic fibers in human photoaged skin and prevents elastin from ultraviolet-induced aggregation in vitro. Am. J. Pathol. 2007, 171, 1474–1482. [Google Scholar] [CrossRef] [PubMed]
- Pastori, D.; Pignatelli, P.; Farcomeni, A.; Menichelli, D.; Nocella, C.; Carnevale, R.; Violi, F. Aging-Related Decline of Glutathione Peroxidase 3 and Risk of Cardiovascular Events in Patients With Atrial Fibrillation. J. Am. Heart Assoc. 2016, 5, e003682. [Google Scholar] [CrossRef]
- Zhu, L.; Wang, L.; Liu, D.; Chen, C.; Mo, K.; Lan, X.; Liu, J.; Huang, Y.; Guo, D.; Huang, H.; et al. Single-cell transcriptomics implicates the FEZ1-DKK1 axis in the regulation of corneal epithelial cell proliferation and senescence. Cell Prolif. 2023, 56, e13433. [Google Scholar] [CrossRef]
- Dai, D.; Chen, B.; Feng, Y.; Wang, W.; Jiang, Y.; Huang, H.; Liu, J. Prognostic value of prostaglandin I2 synthase and its correlation with tumor-infiltrating immune cells in lung cancer, ovarian cancer, and gastric cancer. Aging 2020, 12, 9658–9685. [Google Scholar] [CrossRef] [PubMed]
- Swystun, L.L.; Ogiwara, K.; Lai, J.D.; Ojala, J.R.M.; Rawley, O.; Lassalle, F.; Notley, C.; Rengby, O.; Michels, A.; Nesbitt, K.; et al. The scavenger receptor SCARA5 is an endocytic receptor for von Willebrand factor expressed by littoral cells in the human spleen. J. Thromb. Haemost. 2019, 17, 1384–1396. [Google Scholar] [CrossRef]
- Roy, N.S.; Kim, J.A.; Choi, A.Y.; Ban, Y.W.; Park, N.I.; Park, K.C.; Yang, H.S.; Choi, I.Y.; Kim, S. RNA-Seq De Novo Assembly and Differential Transcriptome Analysis of Korean Medicinal Herb Cirsium japonicum var. spinossimum. Genomics Inform. 2018, 16, e34. [Google Scholar] [CrossRef]
- Klein, A.; Wrulich, O.A.; Jenny, M.; Gruber, P.; Becker, K.; Fuchs, D.; Gostner, J.M.; Uberall, F. Pathway-focused bioassays and transcriptome analysis contribute to a better activity monitoring of complex herbal remedies. BMC Genom. 2013, 14, 133. [Google Scholar] [CrossRef] [PubMed]
- Holly, A.C.; Melzer, D.; Pilling, L.C.; Fellows, A.C.; Tanaka, T.; Ferrucci, L.; Harries, L.W. Changes in splicing factor expression are associated with advancing age in man. Mech. Ageing Dev. 2013, 134, 356–366. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Sugrue, S.P. Dissection of protein linkage between keratins and pinin, a protein with dual location at desmosome-intermediate filament complex and in the nucleus. J. Biol. Chem. 2000, 275, 14910–14915. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Fukunaga-Kalabis, M.; Herlyn, M. Isolation and Cultivation of Dermal Stem Cells that Differentiate into Functional Epidermal Melanocytes. In Human Cell Culture Protocols; Mitry, R.R., Hughes, R.D., Eds.; Humana Press: Totowa, NJ, USA, 2012; pp. 15–29. [Google Scholar]
- McCabe, M.C.; Hill, R.C.; Calderone, K.; Cui, Y.; Yan, Y.; Quan, T.; Fisher, G.J.; Hansen, K.C. Alterations in extracellular matrix composition during aging and photoaging of the skin. Matrix Biol. Plus 2020, 8, 100041. [Google Scholar] [CrossRef] [PubMed]
- Leask, A.; Abraham, D.J. All in the CCN family: Essential matricellular signaling modulators emerge from the bunker. J. Cell Sci. 2006, 119, 4803–4810. [Google Scholar] [CrossRef]
- Xu, H.; Li, P.; Liu, M.; Liu, C.; Sun, Z.; Guo, X.; Zhang, Y. CCN2 and CCN5 exerts opposing effect on fibroblast proliferation and transdifferentiation induced by TGF-beta. Clin. Exp. Pharmacol. Physiol. 2015, 42, 1207–1219. [Google Scholar] [CrossRef]
- Rittie, L.; Perbal, B.; Castellot, J.J., Jr.; Orringer, J.S.; Voorhees, J.J.; Fisher, G.J. Spatial-temporal modulation of CCN proteins during wound healing in human skin in vivo. J. Cell Commun. Signal 2011, 5, 69–80. [Google Scholar] [CrossRef]
- Quan, T.; Shin, S.; Qin, Z.; Fisher, G.J. Expression of CCN family of genes in human skin in vivo and alterations by solar-simulated ultraviolet irradiation. J. Cell Commun. Signal 2009, 3, 19–23. [Google Scholar] [CrossRef]
- Jun, J.I.; Lau, L.F. CCN2 induces cellular senescence in fibroblasts. J. Cell Commun. Signal 2017, 11, 15–23. [Google Scholar] [CrossRef]
- Qin, Z.; He, T.; Guo, C.; Quan, T. Age-Related Downregulation of CCN2 Is Regulated by Cell Size in a YAP/TAZ-Dependent Manner in Human Dermal Fibroblasts: Impact on Dermal Aging. JID Innov. 2022, 2, 100111. [Google Scholar] [CrossRef]
- Quan, T.; Shao, Y.; He, T.; Voorhees, J.J.; Fisher, G.J. Reduced expression of connective tissue growth factor (CTGF/CCN2) mediates collagen loss in chronologically aged human skin. J. Investig. Dermatol. 2010, 130, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Leask, A. CCN2 in Skin Fibrosis. Methods Mol. Biol. 2017, 1489, 417–421. [Google Scholar] [PubMed]
- Makino, K.; Makino, T.; Stawski, L.; Lipson, K.E.; Leask, A.; Trojanowska, M. Anti-connective tissue growth factor (CTGF/CCN2) monoclonal antibody attenuates skin fibrosis in mice models of systemic sclerosis. Arthritis Res. Ther. 2017, 19, 134. [Google Scholar] [CrossRef] [PubMed]
- Jeong, D.; Lee, M.A.; Li, Y.; Yang, D.K.; Kho, C.; Oh, J.G.; Hong, G.; Lee, A.; Song, M.H.; LaRocca, T.J.; et al. Matricellular Protein CCN5 Reverses Established Cardiac Fibrosis. J. Am. Coll. Cardiol. 2016, 67, 1556–1568. [Google Scholar] [CrossRef]
- Jones, J.A.; Gray, M.R.; Oliveira, B.E.; Koch, M.; Castellot, J.J., Jr. CCN5 expression in mammals: I. Embryonic and fetal tissues of mouse and human. J. Cell Commun. Signal 2007, 1, 127–143. [Google Scholar] [CrossRef]
- Shon, M.S.; Kim, R.H.; Kwon, O.J.; Roh, S.S.; Kim, G.N. Beneficial role and function of fisetin in skin health via regulation of the CCN2/TGF-beta signaling pathway. Food Sci. Biotechnol. 2016, 25, 133–141. [Google Scholar] [CrossRef]
- Fuller, B. Role of PGE-2 and Other Inflammatory Mediators in Skin Aging and Their Inhibition by Topical Natural Anti-Inflammatories. Cosmetics 2019, 6, 6. [Google Scholar] [CrossRef]
- Shim, J.H. Prostaglandin E2 Induces Skin Aging via E-Prostanoid 1 in Normal Human Dermal Fibroblasts. Int. J. Mol. Sci. 2019, 20, 5555. [Google Scholar] [CrossRef]
- Yang, H.H.; Kim, C.; Jung, B.; Kim, K.S.; Kim, J.R. Involvement of IGF binding protein 5 in prostaglandin E(2)-induced cellular senescence in human fibroblasts. Biogerontology 2011, 12, 239–252. [Google Scholar] [CrossRef]
- Wiley, C.D.; Sharma, R.; Davis, S.S.; Lopez-Dominguez, J.A.; Mitchell, K.P.; Wiley, S.; Alimirah, F.; Kim, D.E.; Payne, T.; Rosko, A.; et al. Oxylipin biosynthesis reinforces cellular senescence and allows detection of senolysis. Cell Metab. 2021, 33, 1124–1136.e5. [Google Scholar] [CrossRef]
- Yu, T.; Lee, Y.J.; Yang, H.M.; Han, S.; Kim, J.H.; Lee, Y.; Kim, C.; Han, M.H.; Kim, M.Y.; Lee, J.; et al. Inhibitory effect of Sanguisorba officinalis ethanol extract on NO and PGE(2) production is mediated by suppression of NF-kappaB and AP-1 activation signaling cascade. J. Ethnopharmacol. 2011, 134, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Hammer, K.D.; Hillwig, M.L.; Solco, A.K.; Dixon, P.M.; Delate, K.; Murphy, P.A.; Wurtele, E.S.; Birt, D.F. Inhibition of prostaglandin E(2) production by anti-inflammatory hypericum perforatum extracts and constituents in RAW264.7 Mouse Macrophage Cells. J. Agric. Food Chem. 2007, 55, 7323–7331. [Google Scholar] [CrossRef] [PubMed]
- Koeberle, A.; Werz, O. Natural products as inhibitors of prostaglandin E2 and pro-inflammatory 5-lipoxygenase-derived lipid mediator biosynthesis. Biotechnol. Adv. 2018, 36, 1709–1723. [Google Scholar] [CrossRef] [PubMed]
- Bhadra, M.; Howell, P.; Dutta, S.; Heintz, C.; Mair, W.B. Alternative splicing in aging and longevity. Hum. Genet. 2020, 139, 357–369. [Google Scholar] [CrossRef]
- Lopez-Otin, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. Hallmarks of aging: An expanding universe. Cell 2023, 186, 243–278. [Google Scholar] [CrossRef]
- Rodriguez, S.A.; Grochova, D.; McKenna, T.; Borate, B.; Trivedi, N.S.; Erdos, M.R.; Eriksson, M. Global genome splicing analysis reveals an increased number of alternatively spliced genes with aging. Aging Cell 2016, 15, 267–278. [Google Scholar] [CrossRef]
- Latorre, E.; Birar, V.C.; Sheerin, A.N.; Jeynes, J.C.C.; Hooper, A.; Dawe, H.R.; Melzer, D.; Cox, L.S.; Faragher, R.G.A.; Ostler, E.L.; et al. Small molecule modulation of splicing factor expression is associated with rescue from cellular senescence. BMC Cell Biol. 2017, 18, 31. [Google Scholar] [CrossRef]
- Bramwell, L.R.; Harries, L.W. Senescence, regulators of alternative splicing and effects of trametinib treatment in progeroid syndromes. Geroscience 2024, 46, 1861–1879. [Google Scholar] [CrossRef]
- Spector, D.L.; Lamond, A.I. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 2011, 3, a000646. [Google Scholar] [CrossRef]
- Kwon, Y.W.; Lee, S.H.; Kim, A.R.; Kim, B.J.; Park, W.S.; Hur, J.; Jang, H.; Yang, H.M.; Cho, H.J.; Kim, H.S. Plant callus-derived shikimic acid regenerates human skin through converting human dermal fibroblasts into multipotent skin-derived precursor cells. Stem Cell Res. Ther. 2021, 12, 346. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).