Data Analysis of Electrical Impedance Spectroscopy-Based Biosensors Using Artificial Neural Networks for Resource Constrained Devices
Abstract
1. Introduction
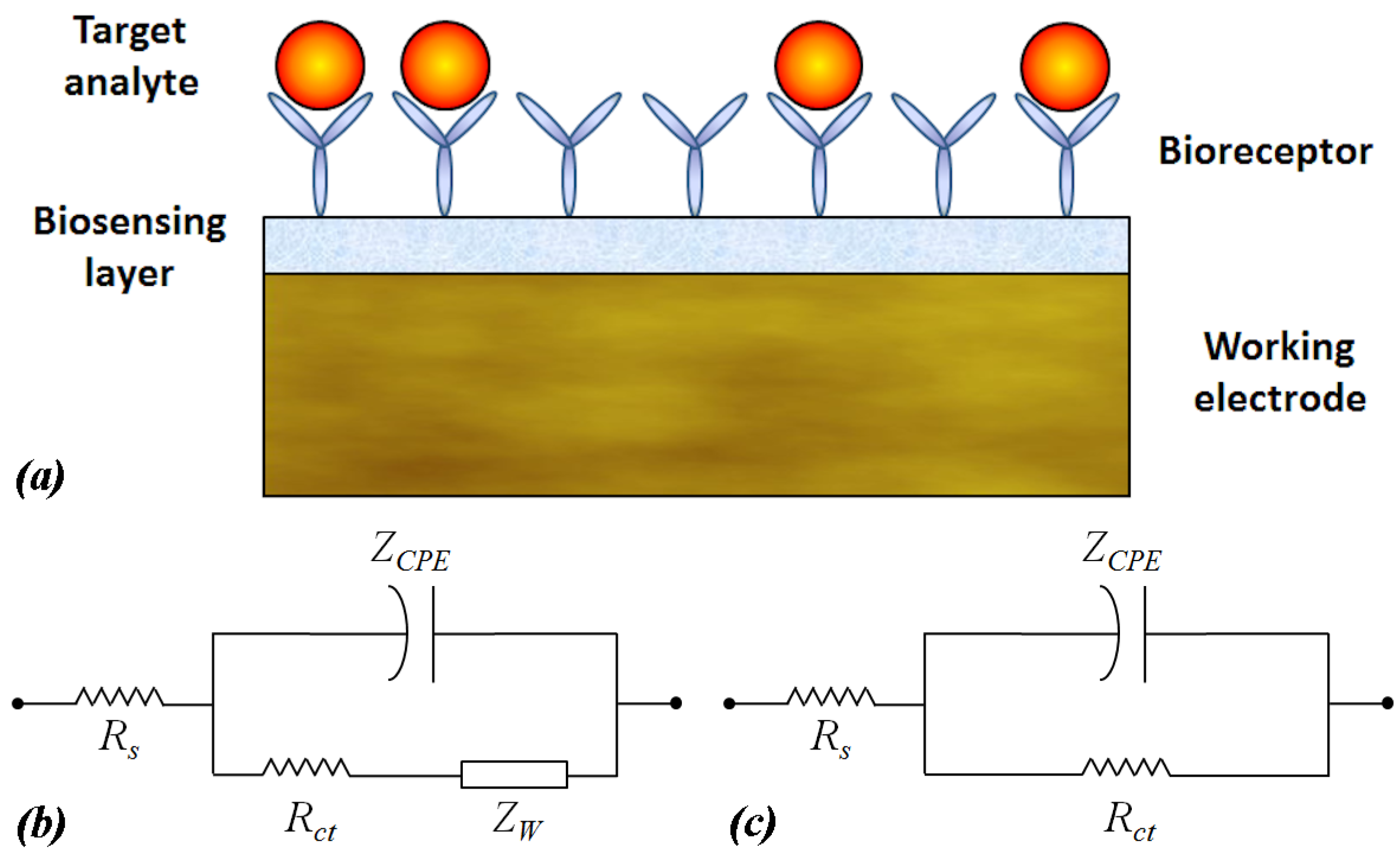
2. Equivalent Circuit for Electrochemical Biosensors
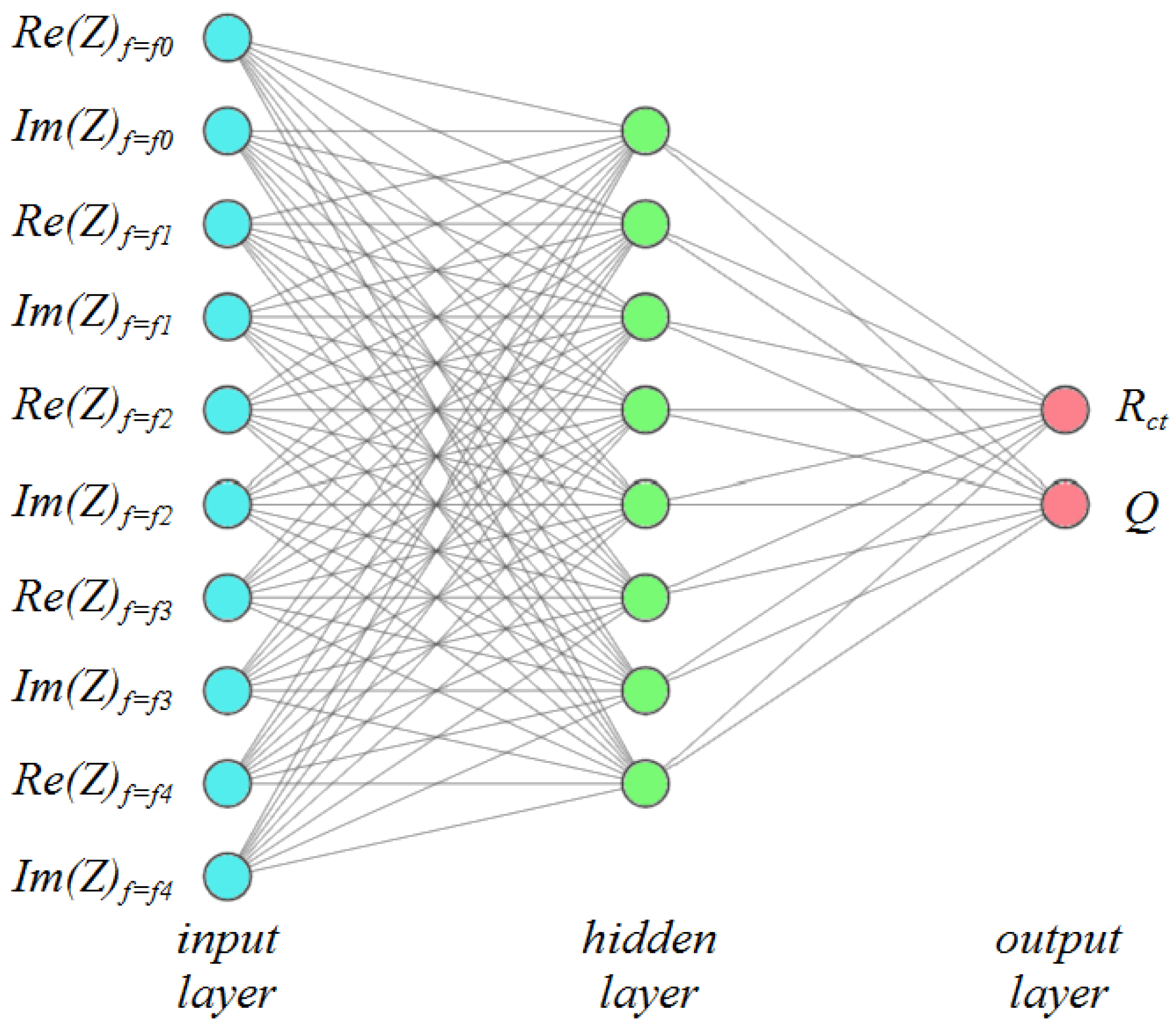
3. Artificial Neural Networks for EIS Data Fitting
3.1. Considered EIS Datasets
3.2. Neural Network Structures
3.3. Performance Metrics
3.4. Circuit Fitting Accuracy with a PC Software
4. Results and Discussion
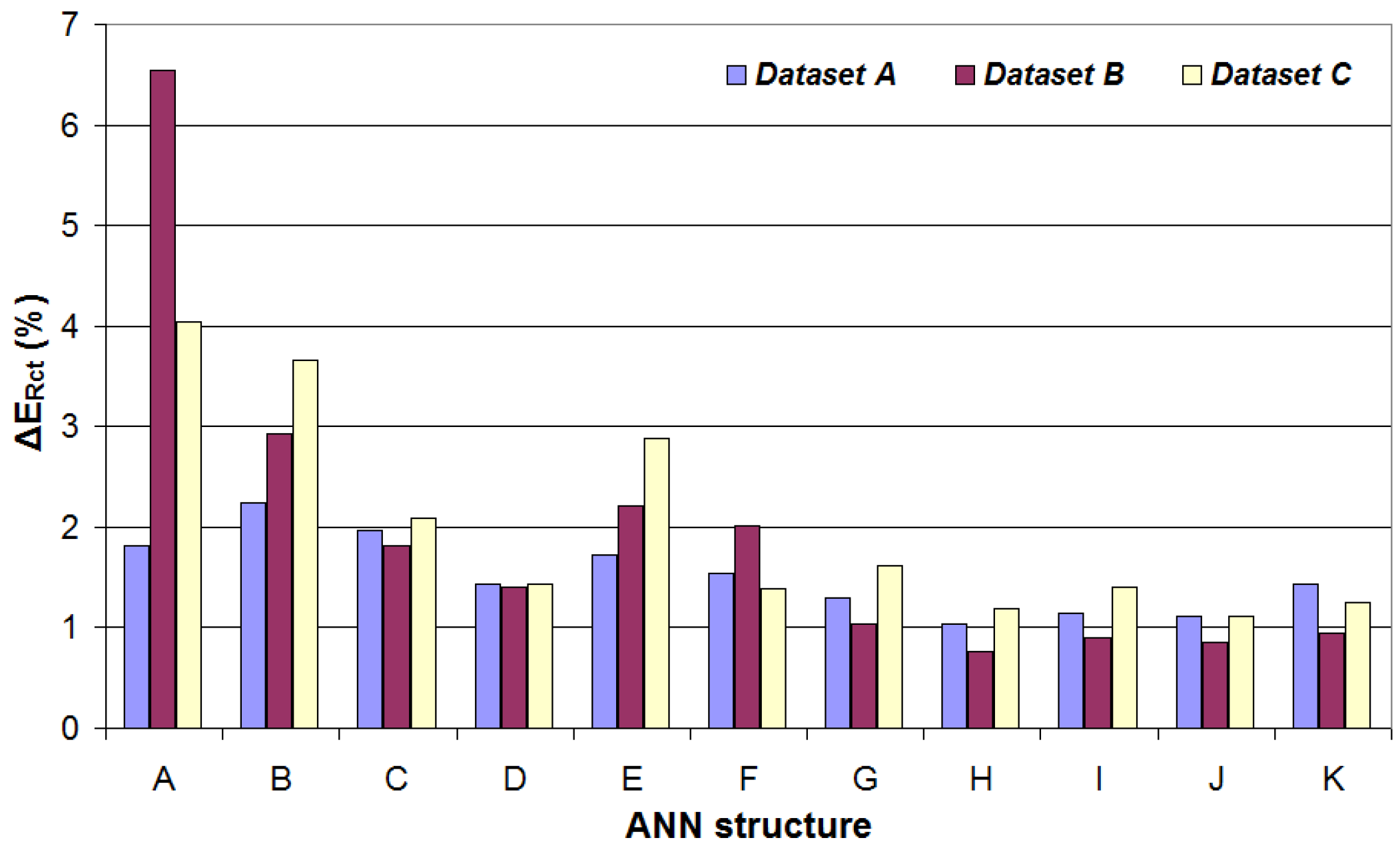
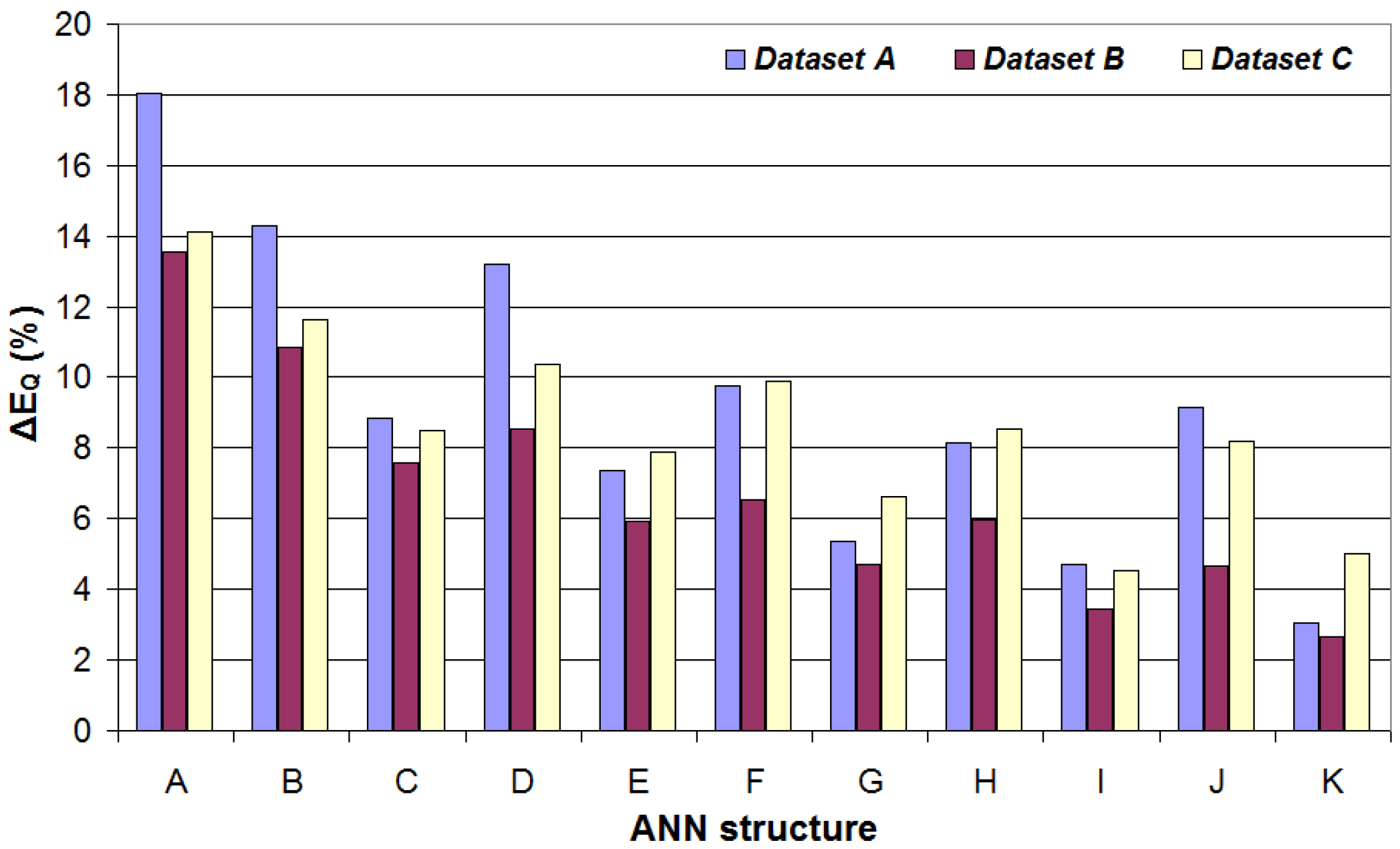
4.1. Performance Metrics for the Software-Generated Datasets
4.2. Discussion on the Required SRAM Size
- The memory needed to store the voltage sine-wave signals Vin (input test signal) and Vout (proportional to the current through the sensor), acquired with an ADC (either integrated in the microcontroller or external) and used to calculate the sensor impedance components Re(Z) and Im(Z). The signals Vin and Vout must be acquired for every test frequency. However, since the impedance components are calculated immediately after the signals’ acquisition, the same memory region can be reused for the signals’ acquisition for different test frequencies. Assuming that each sample is stored as a floating point number (4 bytes) and 100 samples are acquired for each of the two signals (Vin and Vout), 800 bytes are needed.
- The memory needed to store the sensor impedance components Re(Z) and Im(Z) for each frequency of the test signal. Assuming that each impedance component is represented by a floating-point number (4 bytes), this memory component requires 40 bytes for Dataset A (5 test frequencies), 120 bytes for Dataset B (15 test frequencies), and 200 bytes for Dataset C (25 test frequencies).
- The memory needed to execute the microcontroller code: acquisition of the sine-wave signals, calculation of the impedance components, implementation of the ANN sum of products and application of the activation function, information data transfer with the UART interface. This memory component was estimated by the implementation on a Nucleo-L152RE development board of the code written in C and compiled with the MBED Keil Studio Cloud online compiler. The SRAM size was estimated to be about 2 kB, but it can eventually be lowered by a more efficient assembly code.
4.3. Validation on a Real EIS Dataset
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Al Mamun, M.A.; Yuce, M.R. Sensors and systems for wearable environmental monitoring toward IoT-enabled applications: A review. IEEE Sens. J. 2019, 19, 7771–7788. [Google Scholar] [CrossRef]
- Cureau, R.J.; Pigliautile, I.; Pisello, A.L. A new wearable system for sensing outdoor environmental conditions for monitoring hyper-microclimate. Sensors 2022, 22, 502. [Google Scholar] [CrossRef]
- Anik, S.M.H.; Gao, X.; Meng, N.; Agee, P.R.; McCoy, A.P. A cost-effective, scalable, and portable IoT data infrastructure for indoor environment sensing. J. Build. Eng. 2022, 49, 104027. [Google Scholar] [CrossRef]
- Tiele, A.; Esfahani, S.; Covington, J. Design and Development of a Low-Cost, Portable Monitoring Device for Indoor Environment Quality. J. Sens. 2018, 2018, 5353816. [Google Scholar] [CrossRef]
- Bülbül, G.; Hayat, A.; Andreescu, S. Portable nanoparticle-based sensors for food safety assessment. Sensors 2015, 15, 30736–30758. [Google Scholar] [CrossRef] [PubMed]
- Grossi, M.; Bendini, A.; Valli, E.; Gallina Toschi, T. Field-deployable determinations of peroxide index and total phenolic content in olive oil using a promising portable sensor system. Sensors 2023, 23, 5002. [Google Scholar] [CrossRef] [PubMed]
- Wojnowski, W.; Majchrzak, T.; Dymerski, T.; Gębicki, J.; Namieśnik, J. Portable electronic nose based on electrochemical sensors for food quality assessment. Sensors 2017, 17, 2715. [Google Scholar] [CrossRef]
- Grossi, M.; Valli, E.; Glicerina, V.T.; Rocculi, P.; Gallina Toschi, T.; Riccò, B. Optical determination of solid fat content in fats and oils: Effects of wavelength on estimated accuracy. Eur. J. Lipid Sci. Technol. 2022, 124, 2100071. [Google Scholar] [CrossRef]
- Sony, S.; Laventure, S.; Sadhu, A. A literature review of next-generation smart sensing technology in structural health monitoring. Struct. Control Health Monit. 2019, 26, e2321. [Google Scholar] [CrossRef]
- Hassani, S.; Dackermann, U. A systematic review of advanced sensor technologies for non-destructive testing and structural health monitoring. Sensors 2023, 23, 2204. [Google Scholar] [CrossRef]
- Soman, R.; Wee, J.; Peters, K. Optical fiber sensors for ultrasonic structural health monitoring: A review. Sensors 2021, 21, 7345. [Google Scholar] [CrossRef]
- Liu, Z.; Cui, B.; Yang, Q.; Gu, X. Sensor-based structural health monitoring of asphalt pavements with semi-rigid bases combining accelerated pavement testing and a falling weight deflectometer test. Sensors 2024, 24, 994. [Google Scholar] [CrossRef]
- Kalasin, S.; Surareungchai, W. Challenges of emerging wearable sensors for remote monitoring toward telemedicine healthcare. Anal. Chem. 2023, 95, 1773–1784. [Google Scholar] [CrossRef]
- Rajendran, S.; Porwal, A.; Anjali, K.; Anvaya; Anuradha, R. Portable IoT Devices in Healthcare for Health Monitoring and Diagnostics. In Internet of Things in Bioelectronics: Emerging Technologies and Applications; Scrivener Publishing: Beverly, MA, USA, 2024; pp. 263–296. [Google Scholar]
- Vaghasiya, J.V.; Mayorga-Martinez, C.C.; Pumera, M. Telemedicine platform for health assessment remotely by an integrated nanoarchitectonics FePS3/rGO and Ti3C2-based wearable device. NPJ Flex. Electron. 2022, 6, 73. [Google Scholar] [CrossRef] [PubMed]
- Faham, S.; Salimi, A.; Ghavami, R. Electrochemical-based remote biomarker monitoring: Toward Internet of Wearable Things in telemedicine. Talanta 2023, 253, 123892. [Google Scholar] [CrossRef] [PubMed]
- Grossi, M.; Parolin, C.; Vitali, B.; Riccò, B. A portable sensor system for bacterial concentration monitoring in metalworking fluids. J. Sens. Sens. Syst. 2018, 7, 349–357. [Google Scholar] [CrossRef]
- Ezenarro, J.J.; Mas, J.; Muñoz-Berbel, X.; Uria, N. Advances in bacterial concentration methods and their integration in portable detection platforms: A review. Anal. Chim. Acta 2022, 1209, 339079. [Google Scholar] [CrossRef]
- Psotta, C.; Chaturvedi, V.; Gonzalez-Martinez, J.F.; Sotres, J.; Falk, M. Portable Prussian Blue-Based Sensor for Bacterial Detection in Urine. Sensors 2022, 23, 388. [Google Scholar] [CrossRef]
- Grossi, M.; Parolin, C.; Vitali, B.; Riccò, B. Computer vision approach for the determination of microbial concentration and growth kinetics using a low cost sensor system. Sensors 2019, 19, 5367. [Google Scholar] [CrossRef]
- Buscaglia, L.A.; Oliveira, O.N.; Carmo, J.P. Roadmap for electrical impedance spectroscopy for sensing: A tutorial. IEEE Sens. J. 2021, 21, 22246–22257. [Google Scholar] [CrossRef]
- Grossi, M.; Valli, E.; Bendini, A.; Gallina Toschi, T.; Riccò, B. A Portable Battery-Operated Sensor System for Simple and Rapid Assessment of Virgin Olive Oil Quality Grade. Chemosensors 2022, 10, 102. [Google Scholar] [CrossRef]
- Akhter, F.; Siddiquei, H.R.; Alahi, M.E.E.; Mukhopadhyay, S.C. An IoT-enabled portable sensing system with MWCNTs/PDMS sensor for nitrate detection in water. Measurement 2021, 178, 109424. [Google Scholar] [CrossRef]
- Buscaglia, L.A.; Carmo, J.P.; Oliveira, O.N. Simple-Z: A low-cost portable impedance analyzer. IEEE Sens. J. 2023, 23, 26067–26074. [Google Scholar] [CrossRef]
- Bigdeli, I.K.; Yeganeh, M.; Shoushtari, M.T.; Zadeh, M.K. Electrochemical impedance spectroscopy (EIS) for biosensing. Nanosensors Smart Manuf. 2021, 533–554. [Google Scholar]
- Van Haeverbeke, M.; Stock, M.; De Baets, B. Equivalent electrical circuits and their use across electrochemical impedance spectroscopy application domains. IEEE Access 2022, 10, 51363–51379. [Google Scholar] [CrossRef]
- Santoni, F.; De Angelis, A.; Moschitta, A.; Carbone, P.; Galeotti, M.; Cinà, L.; Giammanco, C.; Di Carlo, A. A guide to equivalent circuit fitting for impedance analysis and battery state estimation. J. Energy Storage 2024, 82, 110389. [Google Scholar] [CrossRef]
- Bandarenka, A.S. Development of hybrid algorithms for EIS data fitting. Lect. Notes Impedance Spectrosc. Meas. Model. Appl. 2013, 4, 29–36. [Google Scholar]
- Kappel, M.A.; Fabbri, R.; Domingos, R.P.; Bastos, I.N. Novel electrochemical impedance simulation design via stochastic algorithms for fitting equivalent circuits. Measurement 2016, 94, 344–354. [Google Scholar] [CrossRef]
- ZView Data Fit Software. Available online: https://www.scribner.com/software/68-general-electrochemistr376-zview-for-windows/ (accessed on 10 August 2025).
- Zahner Analysis Data Fit Software. Available online: https://zahner.de/products-details/software/zahner-analysis (accessed on 10 August 2025).
- ZSimpWin Data Fit Software. Available online: https://www.ameteksi.com/products/software/zsimpwin (accessed on 10 August 2025).
- EIS Studio on-Line Data Fit Software. Available online: https://app.circuitfitting.net/ (accessed on 10 August 2025).
- ESP32 Data Sheet. Available online: https://cdn.sparkfun.com/datasheets/IoT/esp32_datasheet_en.pdf (accessed on 11 August 2025).
- STM32WB5MMG Data Sheet. Available online: https://www.st.com/resource/en/datasheet/stm32wb5mmg.pdf (accessed on 11 August 2025).
- CY8C5888LTI-LP097 Data Sheet. Available online: https://files.ic-hongda.com/h-pdf/ds/4071/4071073-datasheet.pdf (accessed on 11 August 2025).
- MSP430FG6425 Data Sheet. Available online: https://www.ti.com/lit/ds/symlink/msp430fg6425.pdf (accessed on 11 August 2025).
- PIC18F2455 Data Sheet. Available online: https://ww1.microchip.com/downloads/en/devicedoc/39632b.pdf (accessed on 11 August 2025).
- STM32L073RZT6 Data Sheet. Available online: https://www.st.com/resource/en/datasheet/stm32l073v8.pdf (accessed on 11 August 2025).
- ATmega328P Data Sheet. Available online: https://ww1.microchip.com/downloads/en/DeviceDoc/Atmel-7810-Automotive-Microcontrollers-ATmega328P_Datasheet.pdf (accessed on 11 August 2025).
- Abdou, M.A. Literature review: Efficient deep neural networks techniques for medical image analysis. Neural Comput. Appl. 2022, 34, 5791–5812. [Google Scholar] [CrossRef]
- Mienye, I.D.; Swart, T.G.; Obaido, G.; Jordan, M.; Ilono, P. Deep convolutional neural networks in medical image analysis: A review. Information 2025, 16, 195. [Google Scholar] [CrossRef]
- Shaukat, N.; Ali, A.; Javed Iqbal, M.; Moinuddin, M.; Otero, P. Multi-sensor fusion for underwater vehicle localization by augmentation of rbf neural network and error-state kalman filter. Sensors 2021, 21, 1149. [Google Scholar] [CrossRef]
- Wang, J.; Wang, D.; Wang, S.; Li, W.; Song, K. Fault diagnosis of bearings based on multi-sensor information fusion and 2D convolutional neural network. IEEE Access 2021, 9, 23717–23725. [Google Scholar] [CrossRef]
- Kui, H.; Pan, J.; Zong, R.; Yang, H.; Wang, W. Heart sound classification based on log Mel-frequency spectral coefficients features and convolutional neural networks. Biomed. Signal Process. Control 2021, 69, 102893. [Google Scholar] [CrossRef]
- Nanni, L.; Maguolo, G.; Brahnam, S.; Paci, M. An ensemble of convolutional neural networks for audio classification. Appl. Sci. 2021, 11, 5796. [Google Scholar] [CrossRef]
- Buchicchio, E.; De Angelis, A.; Santoni, F.; Carbone, P.; Bianconi, F.; Smeraldi, F. Battery SOC estimation from EIS data based on machine learning and equivalent circuit model. Energy 2023, 283, 128461. [Google Scholar] [CrossRef]
- Luo, Y.F. A multi-frequency electrical impedance spectroscopy technique of artificial neural network-based for the static state of charge. Energies 2021, 14, 2526. [Google Scholar] [CrossRef]
- Saran, D.; Mishra, N.; Bathula, S.; Sahu, K.K. Machine learning assisted classification and interpretation of EIS data with experimental demonstration for chemical conversion coatings on Mg alloys. Electrochim. Acta 2025, 527, 146231. [Google Scholar] [CrossRef]
- Wang, C.; Li, W.; Wang, Y.; Yang, X.; Xu, S. Study of electrochemical corrosion on Q235A steel under stray current excitation using combined analysis by electrochemical impedance spectroscopy and artificial neural network. Constr. Build. Mater. 2020, 247, 118562. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, L.; Wu, J.; Bai, W.; Dai, H.; Lin, H.; Zhang, F.; Yang, Y. SOC estimation of lithium-ion batteries using equivalent circuit model and Nyquist plots from EIS data: A machine learning approach. J. Electroanal. Chem. 2025, 987, 119093. [Google Scholar] [CrossRef]
- Doonyapisut, D.; Kannan, P.K.; Kim, B.; Kim, J.K.; Lee, E.; Chung, C.H. Analysis of electrochemical impedance data: Use of deep neural networks. Adv. Intell. Syst. 2023, 5, 2300085. [Google Scholar] [CrossRef]
- Zulueta, A.; Zulueta, E.; Olarte, J.; Fernandez-Gamiz, U.; Lopez-Guede, J.M.; Etxeberria, S. Electrochemical Impedance Spectrum Equivalent Circuit Parameter Identification Using a Deep Learning Technique. Electronics 2023, 12, 5038. [Google Scholar] [CrossRef]
- Thévenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical biosensors: Recommended definitions and classification. Anal. Lett. 2001, 34, 635–659. [Google Scholar] [CrossRef]
- Lazanas, A.C.; Prodromidis, M.I. Electrochemical impedance spectroscopy—A tutorial. ACS Meas. Sci. Au 2023, 3, 162–193. [Google Scholar] [CrossRef]
- Ferro, L.M.M.; de Barros, A.; Falsetti, L.O.Z.; Corrêa, C.C.; Merces, L.; Bufon, C.C.B. Highly efficient electrochemical energy conversion in a 3D hollow microenvironment: Towards on-a-chip sensor applications. J. Mater. Chem. A 2020, 8, 19855–19865. [Google Scholar] [CrossRef]
- Forster, R.J. Microelectrodes: New dimensions in electrochemistry. Chem. Soc. Rev. 1994, 23, 289–297. [Google Scholar] [CrossRef]
- Fuku, X.; Iftikar, F.; Hess, E.; Iwuoha, E.; Baker, P. Cytochrome c biosensor for determination of trace levels of cyanide and arsenic compounds. Anal. Chim. Acta 2021, 730, 49–59. [Google Scholar] [CrossRef]
- Li, Y.; Afrasiabi, R.; Fathi, F.; Wang, N.; Xiang, C.; Love, R.; She, Z.; Kraatz, H.B. Impedance based detection of pathogenic E. coli O157: H7 using a ferrocene-antimicrobial peptide modified biosensor. Biosens. Bioelectron. 2014, 58, 193–199. [Google Scholar] [CrossRef]
- Gupta, V.K.; Yola, M.L.; Qureshi, M.S.; Solak, A.O.; Atar, N.; Üstündağ, Z. A novel impedimetric biosensor based on graphene oxide/gold nanoplatform for detection of DNA arrays. Sens. Actuators B Chem. 2013, 188, 1201–1211. [Google Scholar] [CrossRef]
- Rengaraj, S.; Cruz-Izquierdo, Á.; Scott, J.L.; Di Lorenzo, M. Impedimetric paper-based biosensor for the detection of bacterial contamination in water. Sens. Actuators B Chem. 2018, 265, 50–58. [Google Scholar] [CrossRef]
- Oliveira, M.D.; Correia, M.T.; Diniz, F.B. Concanavalin A and polyvinyl butyral use as a potential dengue electrochemical biosensor. Biosens. Bioelectron. 2009, 25, 728–732. [Google Scholar] [CrossRef] [PubMed]
- Qi, P.; Wan, Y.; Zhang, D. Impedimetric biosensor based on cell-mediated bioimprinted films for bacterial detection. Biosens. Bioelectron. 2013, 39, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Shervedani, R.K.; Mehrjardi, A.H.; Zamiri, N. A novel method for glucose determination based on electrochemical impedance spectroscopy using glucose oxidase self-assembled biosensor. Bioelectrochemistry 2006, 69, 201–208. [Google Scholar] [CrossRef]
- Wang, L.; Xu, M.; Han, L.; Zhou, M.; Zhu, C.; Dong, S. Graphene enhanced electron transfer at aptamer modified electrode and its application in biosensing. Anal. Chem. 2012, 84, 7301–7307. [Google Scholar] [CrossRef]
- Rivai, M.; Attamimi, M.; Firdaus, M.H. Fish quality recognition using electrochemical gas sensor array and neural network. In Proceedings of the 2019 International Conference on Computer Engineering, Network, and Intelligent Multimedia (CENIM), Surabaya, Indonesia, 19–20 November 2019; pp. 1–5. [Google Scholar]
- Spiro, J.C.K.; Mishra, K.K.; Dhamu, V.N.; Bhatia, A.; Muthukumar, S.; Prasad, S. Development of a portable electrochemical sensing platform for impedance spectroscopy-based biosensing using an ARM-based microcontroller. Sens. Diagn. 2024, 3, 1835–1842. [Google Scholar] [CrossRef]
- Sonoda, K.; Kishida, Y.; Tanaka, T.; Kanda, K.; Fujita, T.; Higuchi, K.; Maenaka, K. Wearable photoplethysmographic sensor system with PSoC microcontroller. Int. J. Intell. Comput. Med. Sci. Image Process. 2013, 5, 45–55. [Google Scholar] [CrossRef]
- Al Bassam, N.; Hussain, S.A.; Al Qaraghuli, A.; Khan, J.; Sumesh, E.P.; Lavanya, V. IoT based wearable device to monitor the signs of quarantined remote patients of COVID-19. Inform. Med. Unlocked 2021, 24, 100588. [Google Scholar] [CrossRef]
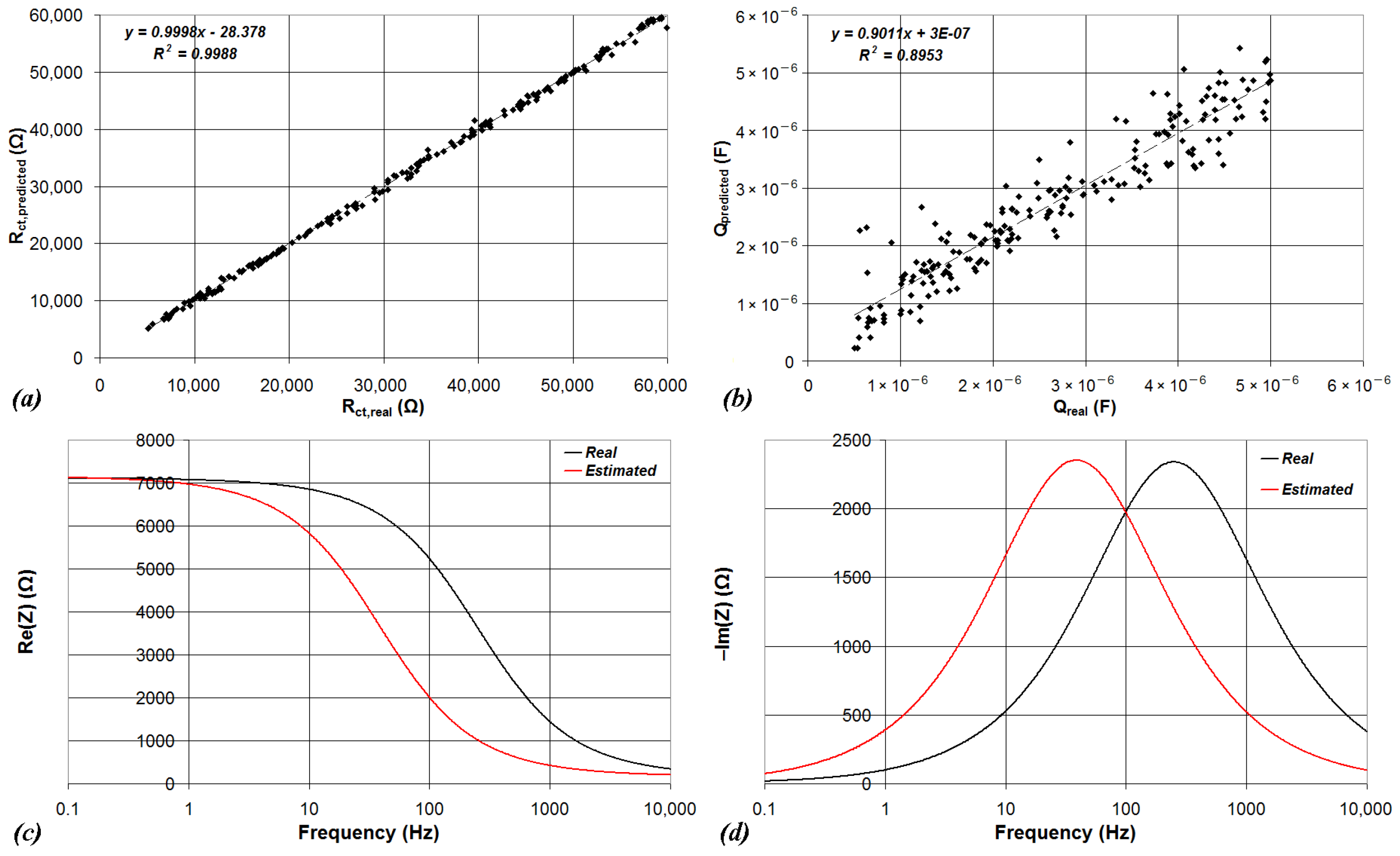
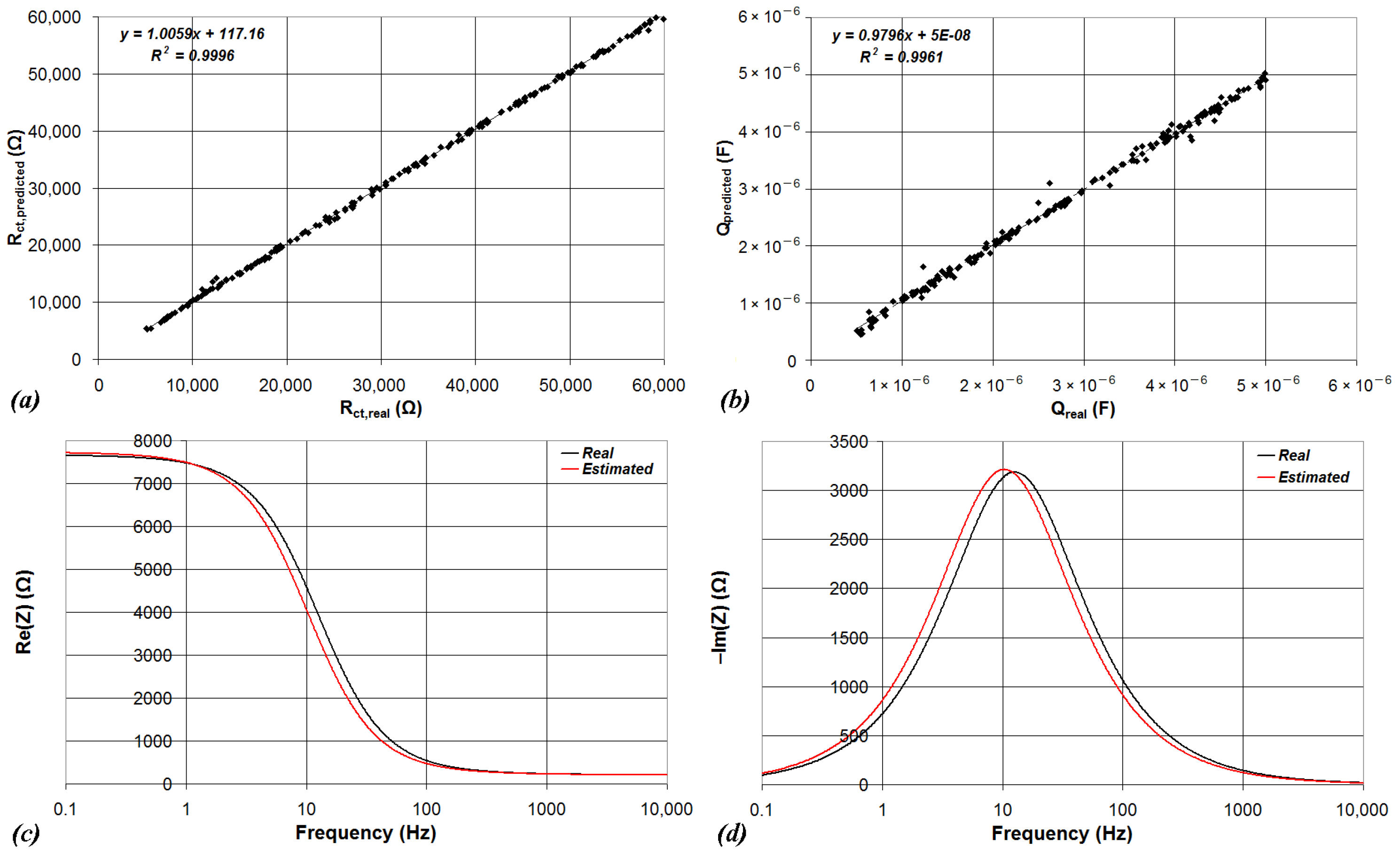
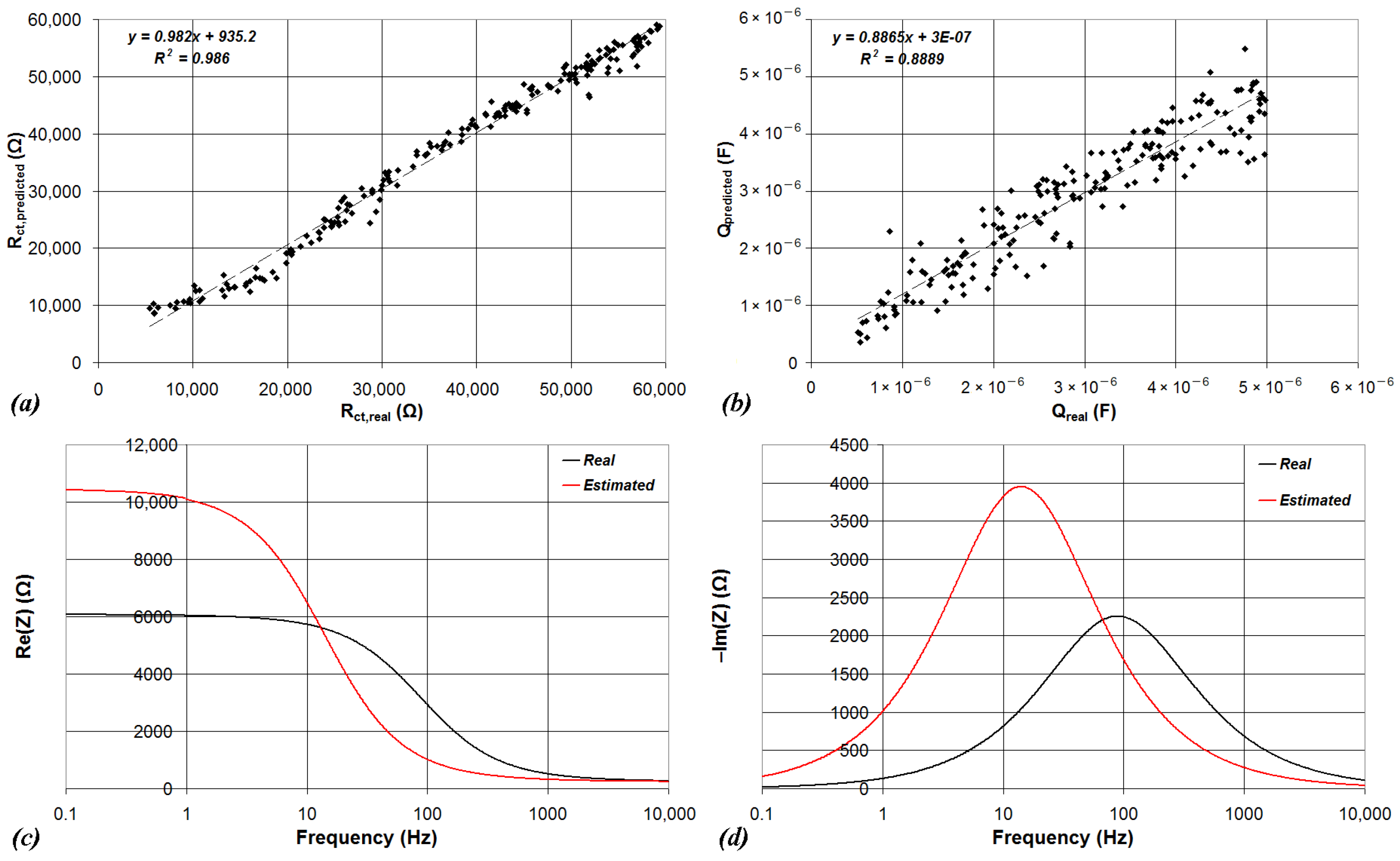
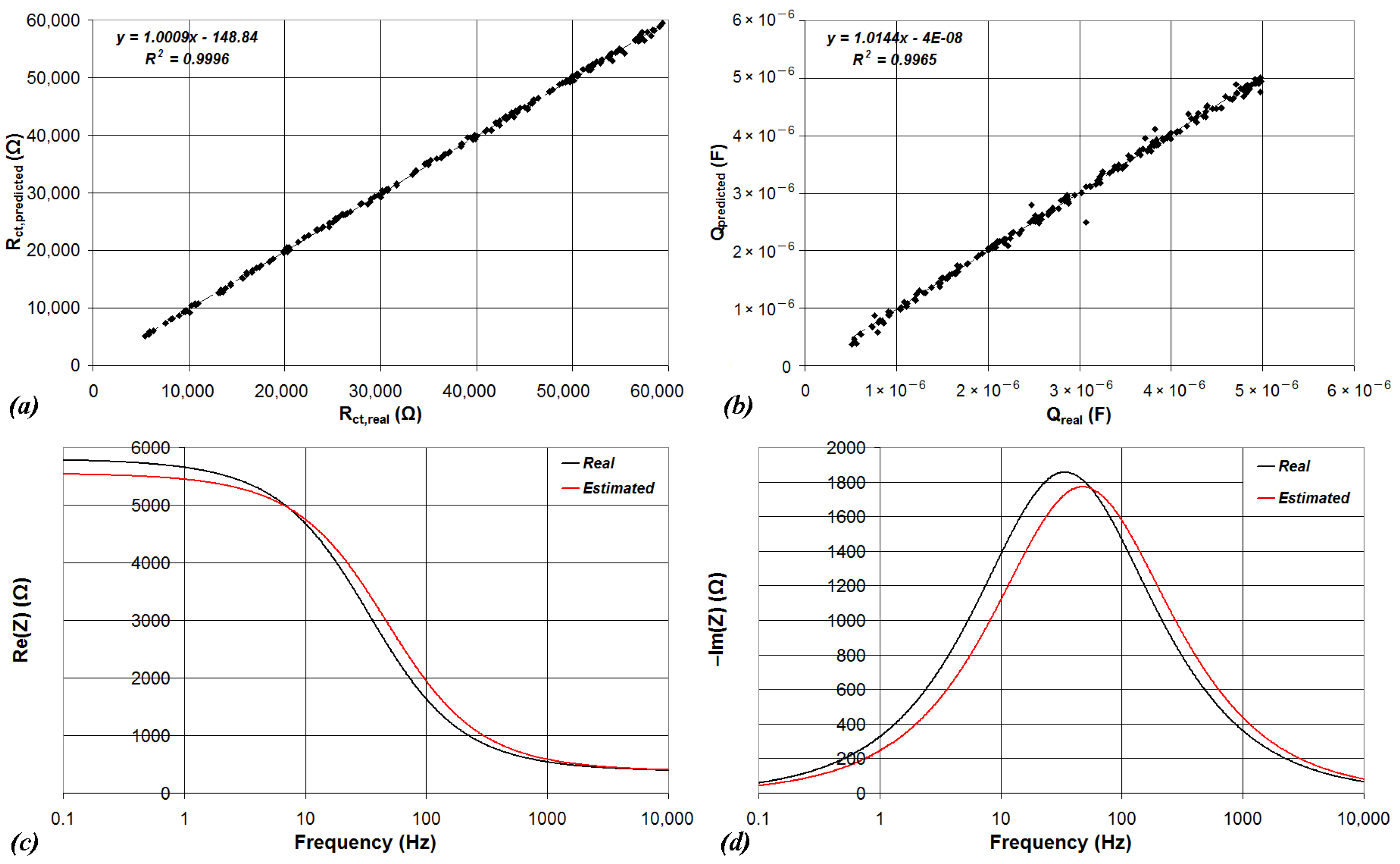
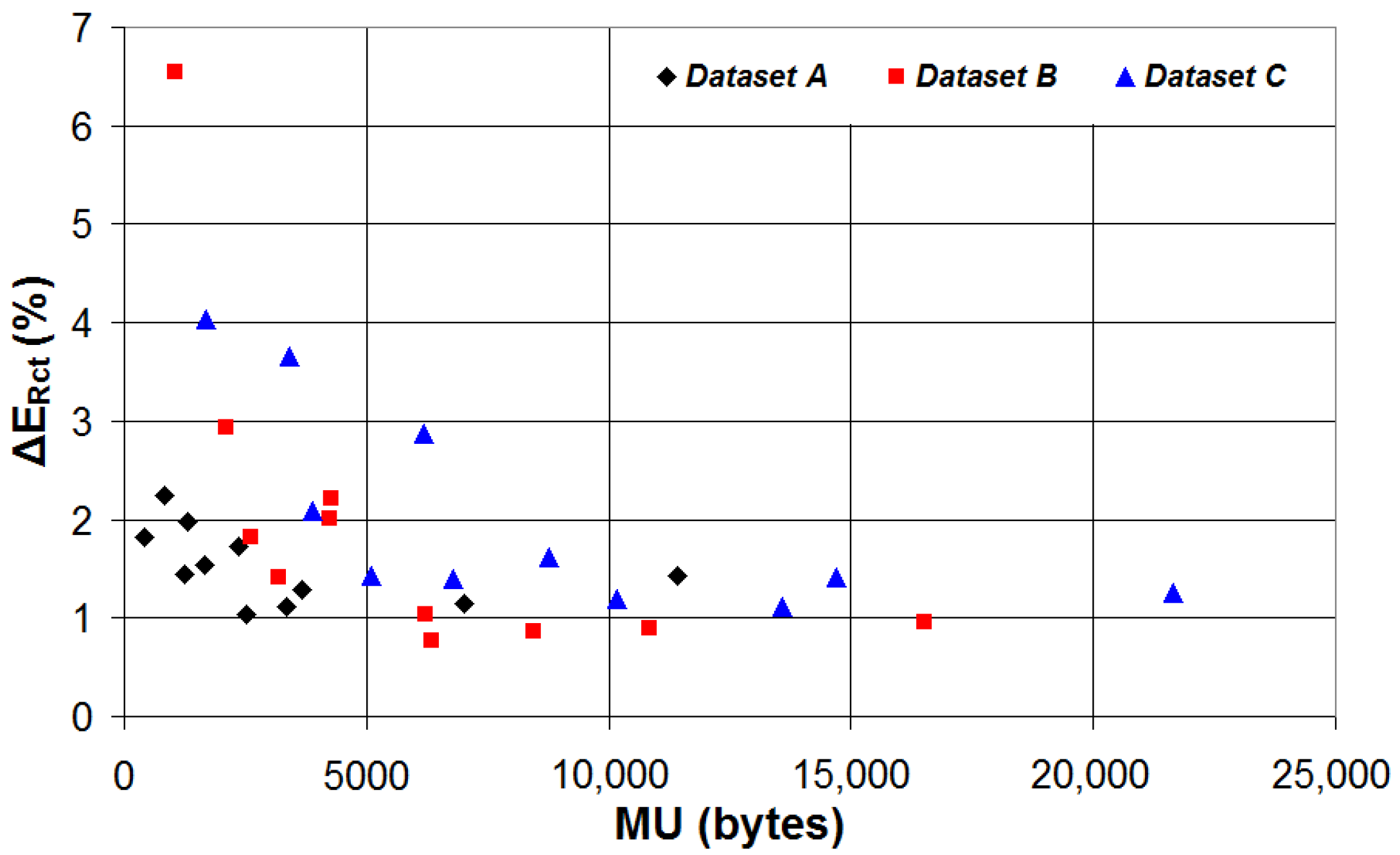
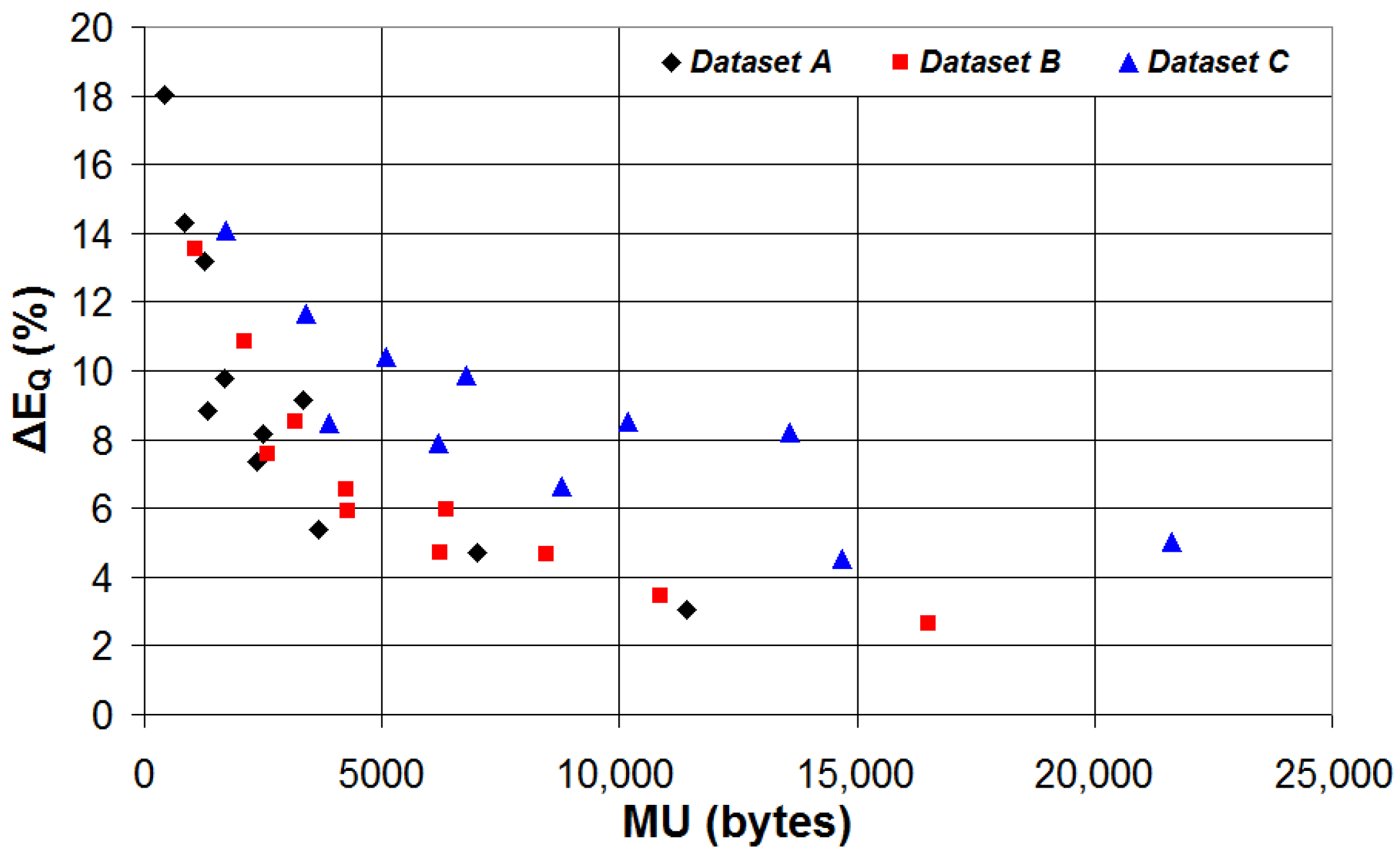
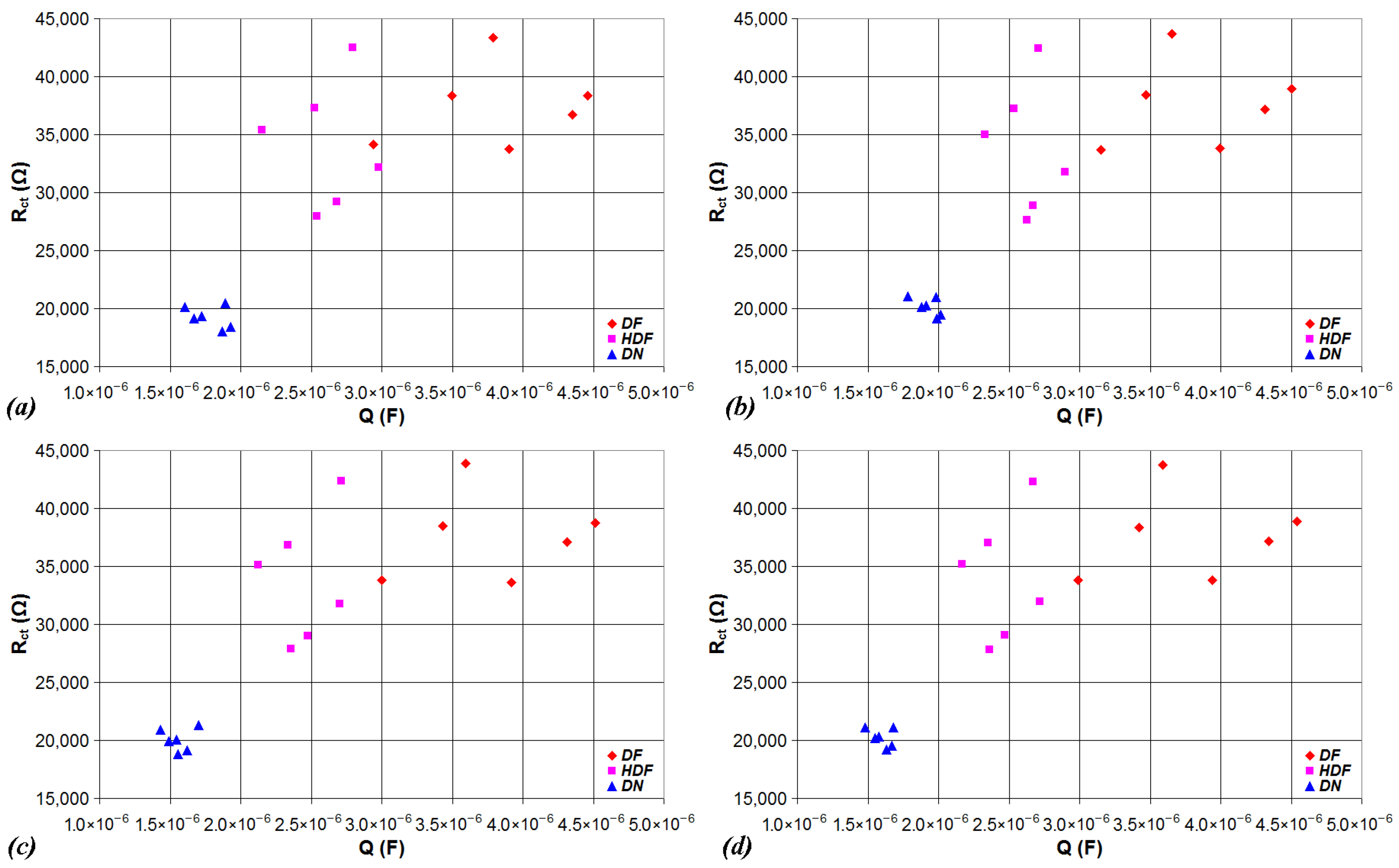
| Device | CPU | Flash | SRAM | ADC | DAC | Connectivity | Ref. |
|---|---|---|---|---|---|---|---|
| ESP32 | 32-bit LX6 CPU | 448 kB | 520 kB | 2 × 12-bit ADC | 2 × 8-bit DAC | Wi-fi, Bluetooth | [34] |
| STM32WB5MMG | 32-bit Cortex M4 | 1 MB | 256 kB | 12-bit ADC | NA | BLE, Zigbee, OT | [35] |
| CY8C5888LTI-LP097 | 32-bit Cortex M3 | 256 kB | 64 kB | 2 × 12-bit ADC | 4 × 8-bit DAC | NA | [36] |
| MSP430FG6425 | 16-bit RISC CPU | 64 kB | 10 kB | 16-bit ADC | 2 × 12-bit DAC | NA | [37] |
| PIC18F2455 | 8-bit RISC CPU | 24 kB | 2 kB | 10-bit ADC | NA | NA | [38] |
| STM32L073RZT6 | 32-bit Cortex M0+ | 192 kB | 20 kB | 12-bit ADC | 12-bit DAC | NA | [39] |
| ATmega328P | 8-bit RISC CPU | 32 kB | 2 kB | 10-bit ADC | NA | NA | [40] |
| Target Analyte | Detection Range | ΔRct | ΔQ | Δα | Ref. |
|---|---|---|---|---|---|
| Prussian blue | 0–8 μM | 4.15–14.9 MΩ | 0.82–1.8 μF | NA | [58] |
| KCN | 0–8 μM | 9–13 MΩ | 0.8–4.8 μF | NA | [58] |
| As2O3 | 0–8 μM | 1.96–4.95 MΩ | 0.8–0.89 μF | NA | [58] |
| E. coli O157:H7 | 103–107 cfu/mL | 1–15 kΩ | NA | NA | [59] |
| DNA | 10−13–10−7 M | 20–130 kΩ | NA | NA | [60] |
| Bacteria | 103–106 cfu/mL | 100 Ω–2.5 kΩ | NA | NA | [61] |
| Dengue virus | NA | 10–50 kΩ | 1–4 μF | 0.8–0.9 | [62] |
| Bacteria | 104–108 cfu/mL | 70–500 Ω | NA | NA | [63] |
| Glucose | NA | 100–600 kΩ | NA | NA | [64] |
| ATP | 15·10−9–4·10−3 M | 3–30 kΩ | NA | NA | [65] |
| Sample Type | Rct (kΩ) | Q (μ) | α |
|---|---|---|---|
| DF | 33.80 | 3.94 | 0.79 |
| DF | 37.20 | 4.34 | 0.79 |
| DF | 38.90 | 4.54 | 0.78 |
| DF | 33.79 | 2.99 | 0.80 |
| DF | 38.34 | 3.42 | 0.80 |
| DF | 43.73 | 3.59 | 0.79 |
| HDF | 32.00 | 2.72 | 0.85 |
| HDF | 29.10 | 2.47 | 0.86 |
| HDF | 27.86 | 2.36 | 0.86 |
| HDF | 42.31 | 2.67 | 0.86 |
| HDF | 37.01 | 2.35 | 0.86 |
| HDF | 35.20 | 2.17 | 0.86 |
| DN | 19.22 | 1.63 | 0.88 |
| DN | 21.11 | 1.48 | 0.88 |
| DN | 20.19 | 1.55 | 0.88 |
| DN | 19.51 | 1.67 | 0.88 |
| DN | 21.15 | 1.68 | 0.88 |
| DN | 20.34 | 1.58 | 0.88 |
| ANN Structure | ΔERct | ΔEQ | NMSERct | NMSEQ | MU (Bytes) |
|---|---|---|---|---|---|
| A | 1.82% | 18.04% | 3.07 × 10−4 | 2.66 × 10−2 | 424 |
| B | 2.24% | 14.31% | 3.97 × 10−4 | 1.87 × 10−2 | 840 |
| C | 1.97% | 8.83% | 4.24 × 10−4 | 8.44 × 10−3 | 1320 |
| D | 1.44% | 13.2% | 1.97 × 10−4 | 1.58 × 10−2 | 1256 |
| E | 1.73% | 7.37% | 3.26 × 10−4 | 6.11 × 10−3 | 2360 |
| F | 1.54% | 9.76% | 1.90 × 10−4 | 1.03 × 10−2 | 1672 |
| G | 1.29% | 5.38% | 1.50 × 10−4 | 2.74 × 10−3 | 3656 |
| H | 1.03% | 8.15% | 7.37 × 10−5 | 7.98 × 10−3 | 2504 |
| I | 1.15% | 4.71% | 1.04 × 10−4 | 2.24 × 10−3 | 7016 |
| J | 1.11% | 9.15% | 1.01 × 10−4 | 8.69 × 10−3 | 3336 |
| K | 1.43% | 3.04% | 1.99 × 10−4 | 1.05 × 10−3 | 11,400 |
| ANN Structure | ΔERct | ΔEQ | NMSERct | NMSEQ | MU (Bytes) |
|---|---|---|---|---|---|
| A | 6.55% | 13.56% | 2.58 × 10−3 | 2.19 × 10−2 | 1064 |
| B | 2.93% | 10.85% | 7.49 × 10−4 | 1.22 × 10−2 | 2120 |
| C | 1.82% | 7.57% | 3.40 × 10−4 | 5.13 × 10−3 | 2600 |
| D | 1.41% | 8.53% | 1.62 × 10−4 | 6.72 × 10−3 | 3176 |
| E | 2.21% | 5.94% | 4.71 × 10−4 | 3.52 × 10−3 | 4280 |
| F | 2.01% | 6.54% | 2.97 × 10−4 | 4.72 × 10−3 | 4232 |
| G | 1.04% | 4.72% | 7.96 × 10−5 | 3.02 × 10−3 | 6216 |
| H | 0.77% | 5.98% | 6.28 × 10−5 | 4.19 × 10−3 | 6344 |
| I | 0.90% | 3.44% | 7.28 × 10−5 | 1.08 × 10−3 | 10,856 |
| J | 0.86% | 4.68% | 6.61 × 10−5 | 2.48 × 10−3 | 8456 |
| K | 0.95% | 2.64% | 8.55 × 10−5 | 7.45 × 10−4 | 16,520 |
| ANN Structure | ΔERct | ΔEQ | NMSERct | NMSEQ | MU (Bytes) |
|---|---|---|---|---|---|
| A | 4.04% | 14.10% | 1.47 × 10−3 | 1.85 × 10−2 | 1704 |
| B | 3.66% | 11.64% | 5.99 × 10−4 | 1.34 × 10−2 | 3400 |
| C | 2.09% | 8.49% | 2.88 × 10−4 | 6.91 × 10−3 | 3880 |
| D | 1.43% | 10.39% | 1.75 × 10−4 | 9.70 × 10−3 | 5096 |
| E | 2.88% | 7.87% | 5.56 × 10−4 | 5.15 × 10−3 | 6200 |
| F | 1.39% | 9.87% | 1.33 × 10−4 | 1.03 × 10−2 | 6792 |
| G | 1.62% | 6.64% | 2.44 × 10−4 | 3.30 × 10−3 | 8776 |
| H | 1.19% | 8.53% | 1.24 × 10−4 | 7.97 × 10−3 | 10,184 |
| I | 1.41% | 4.55% | 1.69 × 10−4 | 2.40 × 10−3 | 14,696 |
| J | 1.11% | 8.19% | 8.95 × 10−5 | 6.23 × 10−3 | 13,576 |
| K | 1.25% | 5.01% | 1.19 × 10−4 | 1.98 × 10−3 | 21,640 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grossi, M.; Omaña, M. Data Analysis of Electrical Impedance Spectroscopy-Based Biosensors Using Artificial Neural Networks for Resource Constrained Devices. J. Low Power Electron. Appl. 2025, 15, 56. https://doi.org/10.3390/jlpea15040056
Grossi M, Omaña M. Data Analysis of Electrical Impedance Spectroscopy-Based Biosensors Using Artificial Neural Networks for Resource Constrained Devices. Journal of Low Power Electronics and Applications. 2025; 15(4):56. https://doi.org/10.3390/jlpea15040056
Chicago/Turabian StyleGrossi, Marco, and Martin Omaña. 2025. "Data Analysis of Electrical Impedance Spectroscopy-Based Biosensors Using Artificial Neural Networks for Resource Constrained Devices" Journal of Low Power Electronics and Applications 15, no. 4: 56. https://doi.org/10.3390/jlpea15040056
APA StyleGrossi, M., & Omaña, M. (2025). Data Analysis of Electrical Impedance Spectroscopy-Based Biosensors Using Artificial Neural Networks for Resource Constrained Devices. Journal of Low Power Electronics and Applications, 15(4), 56. https://doi.org/10.3390/jlpea15040056







