Estrogen Signaling Induces Mitochondrial Dysfunction-Associated Autophagy and Senescence in Breast Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
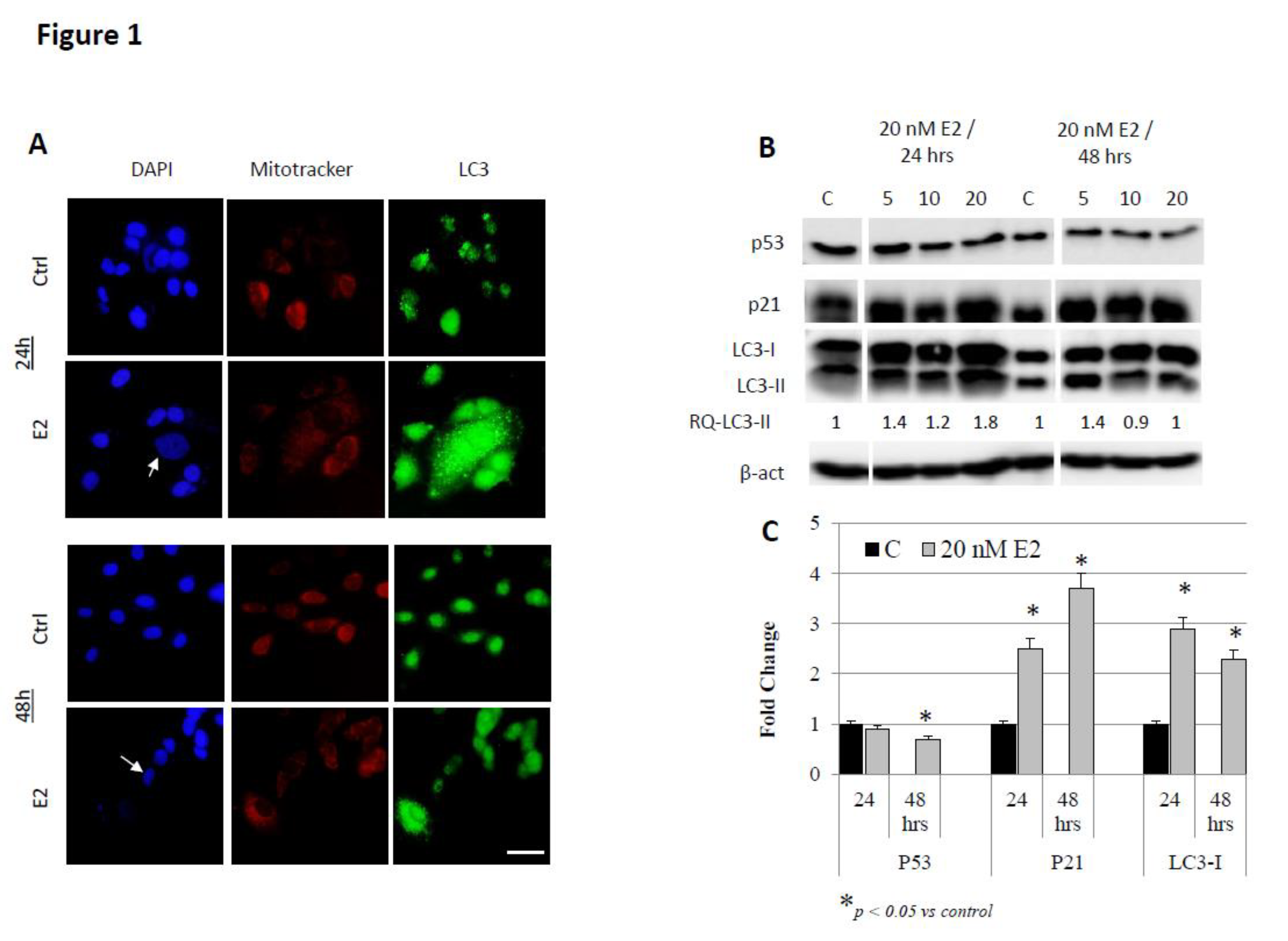
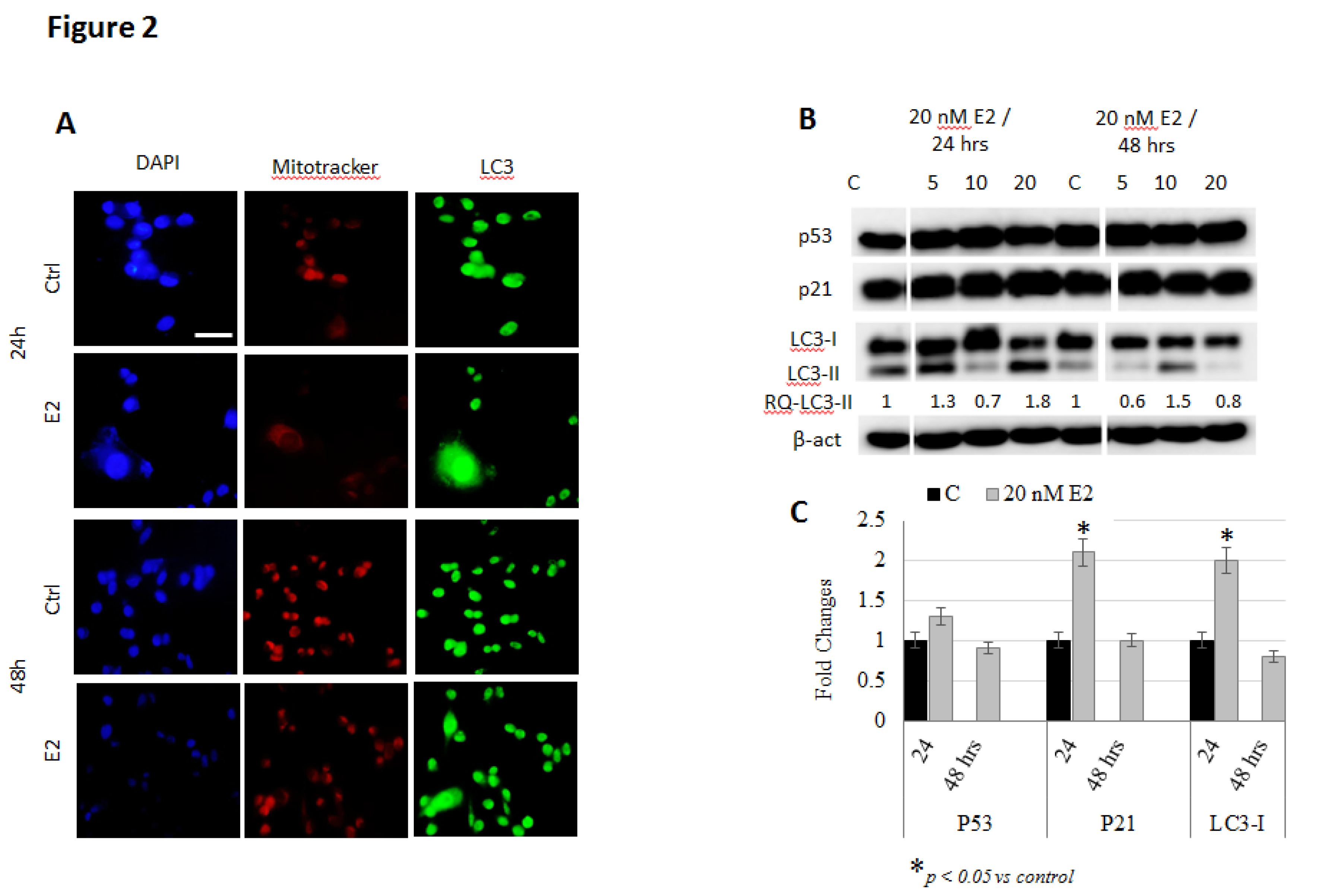
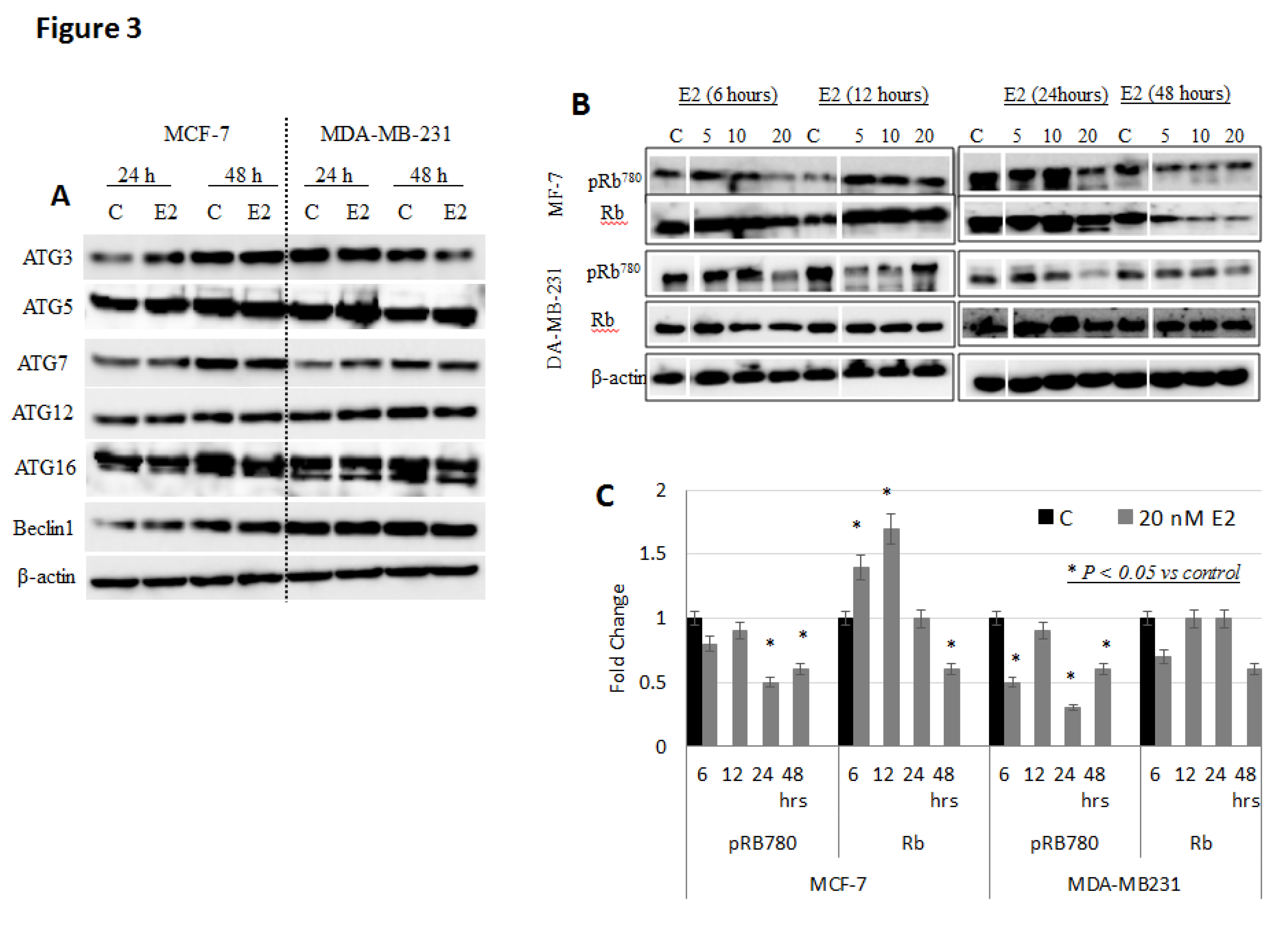
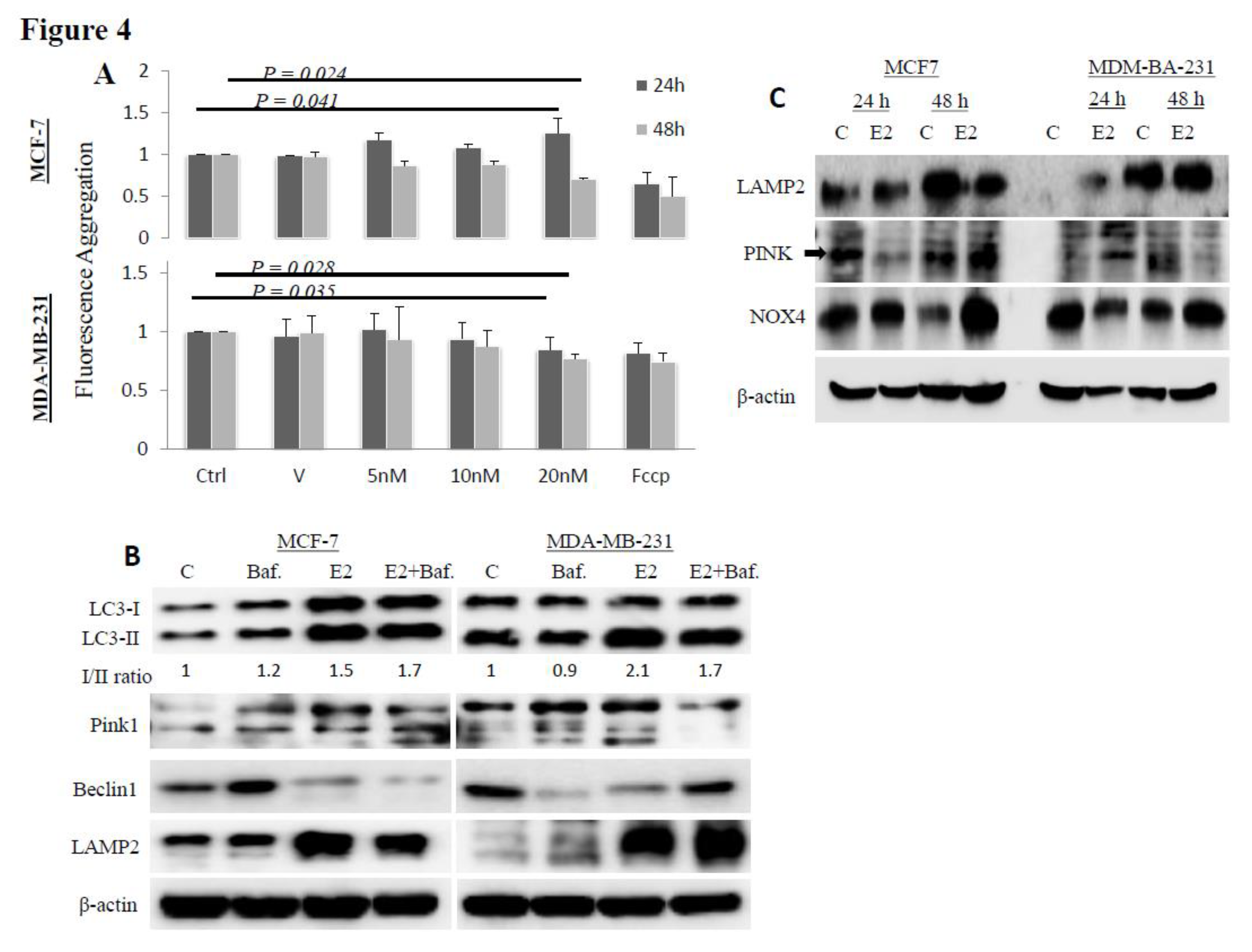
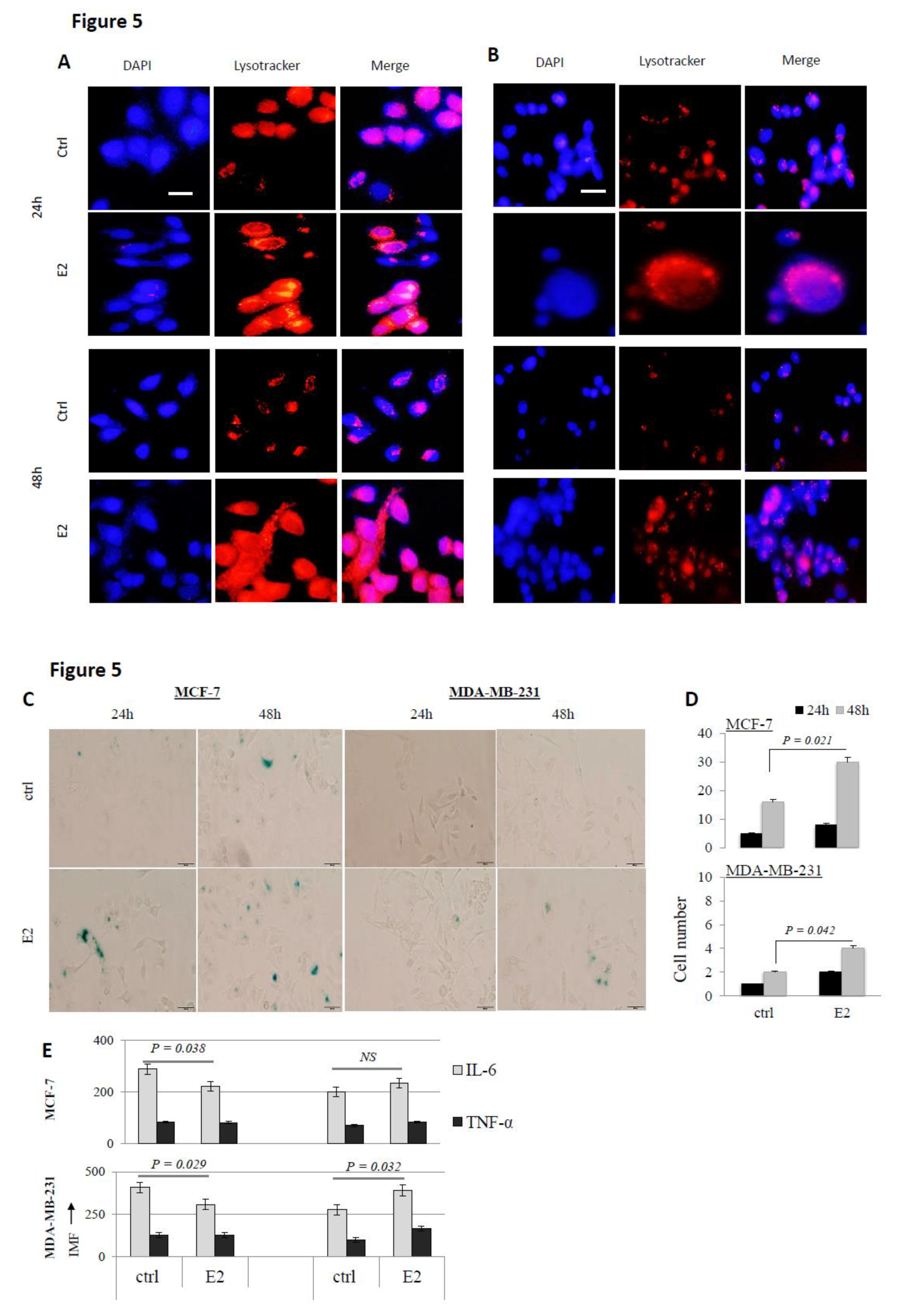
3. Results
4. Discussion
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Liehr, J.G. Is estradiol a genotoxic mutagenic carcinogen? Endocr. Rev. 2000, 21, 40–54. [Google Scholar] [PubMed]
- Yager, J.D.; Davidson, N.E. E2 carcinogenesis in breast cancer. N. Engl. J. Med. 2006, 354, 270–282. [Google Scholar] [CrossRef] [PubMed]
- Levenson, A.S.; Jordan, V.C. MCF-7: The first hormone-responsive breast cancer cell line. Cancer Res. 1997, 57, 3071–3078. [Google Scholar] [PubMed]
- Jordan, V.C. A century of deciphering the control mechanisms of sex steroid action in breast and prostate cancer: The origins of targeted therapy and chemoprevention. Cancer Res. 2009, 69, 124–154. [Google Scholar] [CrossRef]
- Haddow, A.; Watkinson, J.M.; Paterson, E.; Koller, P.C. Influence of synthetic oE2s on advanced malignant disease. Br. Med. J. 1944, 2, 393–398. [Google Scholar] [CrossRef]
- Ingle, J.N.; Ahmann, D.L.; Green, S.J.; Edmonson, J.H.; Bisel, H.F.; Kvols, L.K.; Nichols, W.C.; Creagan, E.T.; Hahn, R.G.; Rubin, J.; et al. Randomized clinical trial of diethylstilbestrol versus tamoxifen in postmenopausal women with advanced breast cancer. N. Engl. J. Med. 1981, 304, 16–21. [Google Scholar] [CrossRef]
- Peethambaram, P.P.; Ingle, J.N.; Suman, V.J.; Hartmann, L.C.; Loprinzi, C.L. Randomized trial of diethylstilbestrol vs. tamoxifen in postmenopausal women with metastatic breast cancer. An updated analysis. Breast Cancer Res. Treat. 1999, 54, 117–122. [Google Scholar] [CrossRef]
- La Croix, A.Z.; Chlebowski, R.T.; Manson, J.E.; Aragaki, A.K.; Johnson, K.C.; Martin, L.; Margolis, K.L.; Stefanick, M.L.; Brzyski, R.; Curb, J.D.; et al. WHI Investigators, Health outcomes after stopping conjugated equine E2s among postmenopausal women with prior hysterectomy: A randomized controlled trial. JAMA 2011, 305, 1305–1314. [Google Scholar] [CrossRef] [PubMed]
- Wands, J. Hepatocellular carcinoma and sex. N. Engl. J. Med. 2007, 357, 1974–1976. [Google Scholar] [CrossRef]
- Sophonnithiprasert, T.; Saelee, P.; Pongtheerat, T. GSTM1 and GSTT1 copy number variants and the risk to Thai females of hepatocellular carcinoma. J. Gastrointest. Oncol. 2019, 10, 324–329. [Google Scholar] [CrossRef]
- Yeh, S.H.; Chen, P.J. Gender disparity of hepatocellular carcinoma: The roles of sex hormones. Oncology 2010, 78, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Qiu, S.; Vazquez, J.T.; Boulger, E.; Liu, H.; Xue, P.; Hussain, M.A.; Wolfe, A. Hepatic estrogen receptor α is critical for regulation of gluconeogenesis and lipid metabolism in males. Sci. Rep. 2017, 7, 1661. [Google Scholar] [CrossRef] [PubMed]
- Jordan, V.C. The 38th David A. Karnofsky Lecture: The paradoxical actions of E2 in breast cancer—Survival or death? J. Clin. Oncol. 2008, 26, 307–382. [Google Scholar]
- Fan, P.; Griffith, O.L.; Agboke, F.A.; Anur, P.; Zou, X.; McDaniel, R.E.; Creswell, K.; Kim, S.H.; Katzenellenbogen, J.A.; Gray, J.W.; et al. c-Src modulates E2-induced stress and apoptosis in E2-deprived breast cancer cells. Cancer Res. 2013, 73, 4510–4520. [Google Scholar] [PubMed]
- Bajbouj, K.; Shafarin, J.; Abdalla, M.Y.; Ahmad, I.M.; Hamad, M. Estrogen-induced disruption of intracellular iron metabolism leads to oxidative stress, membrane damage, and cell cycle arrest in MCF-7 cells. Tumour Biol. 2017, 39. [Google Scholar] [CrossRef] [PubMed]
- Shafarin, J.; Bajbouj, K.; El-Serafy, A.; Sandeep, S.; Hamad, M. Estrogen-dependent downregulation of hepcidin synthesis induces intracellular iron efflux in cancer cells in vitro. Biol. Med. 2016, 8, 356–363. [Google Scholar] [CrossRef]
- Bajbouj, K.; Shafarin, J.; Hamad, M. Estrogen-dependent disruption of intracellular iron metabolism augments the cytotoxic effects of doxorubicin in select breast and ovarian cancer cells. Cancer Manag. Res. 2019, 11, 4655–4668. [Google Scholar]
- Xiang, J.; Liu, X.; Ren, J.; Chen, K.; Wang, H.; Miao, Y.; Qi, M. How does estrogen work on autophagy? Autophagy 2019, 15, 197–211. [Google Scholar] [CrossRef]
- Sabatino, M.E.; Petiti, J.P.; del Valle Sosa, L.; Pe’rez, P.A.; Gutie´rrez, S.; Leimgruber, C.; Latini, A.; Torres, A.I.; De Paul, A.L. Evidence of cellular senescence during the development of estrogen-induced pituitary tumors. Endocr. Relat. Cancer 2015, 22, 299–317. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, L.; Yang, J.; Bandyopadhyay, A.; Kaklamani, V.; Wang, S.; Sun, L. Estrogen receptor alpha inhibits senescence-like phenotype and facilitates transformation induced by oncogenic ras in human mammary epithelial cells. Oncotarget 2016, 28, 39097–39107. [Google Scholar]
- Qin, S.; Schulte, B.A.; Wang, G.Y. Role of senescence induction in cancer treatment. World J. Clin. Oncol. 2018, 9, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Sastre-Serra, J.; Nadal-Serrano, M.; Pons, D.G.; Roca, P.; Oliver, J. Mitochondrial dynamics is affected by 17β-estradiol in the MCF-7 breast cancer cell line. Effects on fusion and fission related genes. Int. J. Biochem. Cell Biol. 2012, 44, 1901–1905. [Google Scholar] [CrossRef] [PubMed]
- Radde, B.N.; Ivanova, M.M.; Mai, H.X.; Salabei, J.K.; Hill, B.G.; Klinge, C.M. Bioenergetic differences between MCF-7 and T47D breast cancer cells and their regulation by oestradiol and tamoxifen. Biochem. J. 2015, 465, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Oo, P.S.; Yamaguchi, Y.; Sawaguchi, A.; Kyaw, M.T.; Choijookhuu, N.; Ali, M.N.; Srisowanna, N.; Hino, S.; Hishikawa, Y. Estrogen regulates mitochondrial morphology through phosphorylation of dynamin-related protein 1 in MCF7 human breast cancer cells. Acta Histochem. Cytochem. 2018, 51, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Thiede, A.; Gellerich, F.N.; Schönfeld, P.; Siemen, D. Complex effects of 17β-estradiol on mitochondrial function. Biochim. Biophys. Acta 2012, 1817, 1747–1753. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kahlem, P.; Dörken, B.; Schmitt, C.A. Cellular senescence in cancer treatment: Friend or foe? J. Clin. Investig. 2004, 113, 169–174. [Google Scholar] [CrossRef]
- Yang, L.; Fang, J.; Chen, J. Tumor cell senescence response produces aggressive variants. Cell Death Discov. 2017, 3, 17049. [Google Scholar] [CrossRef]
- Kuilman, T.; Michaloglou, C.; Mooi, W.J.; Deeper, P.S. The essence of senescence. Genes Dev. 2010, 24, 2463–2479. [Google Scholar] [CrossRef]
- Xue, W.; Zender, L.; Miething, C.; Dickins, R.A.; Hernando, E.; Krizhanovsky, V.; Cordon-Cardo, C.; Lowe, S.W. Senescence and tumour clearance is triggered by P53 restoration in murine liver carcinomas. Nature 2007, 445, 656–660. [Google Scholar] [CrossRef]
- Gewirtzad, D.A.; Holtabcd, S.E.; Elmorecd, L.W. Accelerated senescence: An emerging role in tumor cell response to chemotherapy and radiation. Biochem. Pharmacol. 2008, 76, 947–957. [Google Scholar] [CrossRef]
- Schmitt, C.A. Cellular senescence and cancer treatment. Biochim. Biophys. Acta 2007, 1775, 5–20. [Google Scholar] [CrossRef]
- Moats, R.K.; Ramirez, V.D. Rapid uptake and binding of estradiol-17β-6-(O-carboxymethyl) oxime: 125I-labeled BSA by female rat liver. Biol. Reprod. 1998, 58, 531–538. [Google Scholar] [CrossRef][Green Version]
- Moats, R.K.; Ramirez, V.D. Electron microscopic visualization of membrane-mediated uptake and translocation of estrogen-BSA: Colloidal gold by hep G2 cells. J. Endocrinol. 2000, 166, 631–647. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, J.Q.; Delannoy, M.; Cooke, C.; Yager, J.D. Mitochondrial localization of ERalpha and ERbeta in human MCF7 cells. Am. J. Physiol. Endocrinol. Metab. 2004, 286, E1011–E1022. [Google Scholar] [CrossRef] [PubMed]
- Felty, Q.; Roy, D. Estrogen, mitochondria, and growth of cancer and non-cancer cells. J. Carcinog. 2005, 4. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Borrás, C.; Gambini, J.; López-Grueso, R.; Pallardó, F.V.; Viña, J. Direct antioxidant and protective effect of estradiol on isolated mitochondria. Biochim. Biophys. Acta 2010, 1802, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Dykens, J.A.; Simpkins, J.W.; Wang, J.; Gordon, K. Polycyclic phenols, estrogens and neuroprotection: A proposed mitochondrial mechanism. Exp. Gerontol. 2003, 38, 101–107. [Google Scholar] [CrossRef]
- O’Lone, R.; Knorr, K.; Jaffe, I.Z.; Schaffer, M.E.; Martini, G.V.P.; Karas, R.H.; Bienkowska, J.; Mendelsohn, M.E.; Hansen, U. Estrogen receptors α and β mediate distinct pathways of vascular gene expression, including genes involved in mitochondrial electron transport and generation of reactive oxygen species. Mol. Endocrinol. 2007, 21, 1281–1296. [Google Scholar]
- Chen, J.Q.; Cammarata, P.R.; Baines, C.P.; Yager, J.D. Regulation of mitochondrial respiratory chain biogenesis by estrogens/estrogen receptors and physiological, pathological and pharmacological implications. Biochim. Biophys. Acta 2009, 1793, 1540–1570. [Google Scholar] [CrossRef]
- Pedram, A.; Razandi, M.; Wallace, D.C.; Levin, E.R. Functional Estrogen Receptors in the Mitochondria of Breast Cancer Cells. Mol. Biol. Cell 2006, 17, 2125–2137. [Google Scholar] [CrossRef]
- Tuttle, R.; Miller, K.R.; Maiorano, J.N.; Termuhlen, P.M.; Gao, Y.; Berberich, S.J. Novel senescence associated gene, YPEL3, is repressed by estrogen in ER1 mammary tumor cells and required for tamoxifen-induced cellular senescence. Int. J. Cancer 2012, 130, 2291–2299. [Google Scholar] [CrossRef] [PubMed]
- Imanishi, T.; Hano, T.; Nishio, I. Estrogen reduces endothelial progenitor cell senescence through augmentation of telomerase activity. J. Hypertens. 2005, 23, 1699–1706. [Google Scholar] [CrossRef]
- Breu, A.; Sprinzing, B.; Merkl, K.; Bechmann, V.; Kujat, R.; Jenei-Lanzl, Z.; Prantl, L.; Angele, P. Estrogen reduces cellular aging in human mesenchymal stem cells and chondrocytes. J. Orthop. Res. 2011, 29, 1563–1571. [Google Scholar] [CrossRef]
- Lejri, I.; Grimm, A.; Eckert, A. Mitochondria, estrogen and female brain aging. Front. Aging Neurosci. 2018, 10, 124. [Google Scholar] [CrossRef] [PubMed]
- Malavolta, M.; Costarelli, L.; Giacconi, R.; Piacenza, F.; Basso, A.; Pierpaoli, E.; Marchegiani, F.; Cardelli, M.; Provinciali, M.; Mocchegiani, E. Modulators of cellular senescence: Mechanisms, promises, and challenges from in vitro studies with dietary bioactive compounds. Nutr. Res. 2014, 34, 1017–1035. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.C.; McDonald, P.R.; Liu, J.; Klaassen, C.D. Screening of natural compounds as activators of the keap1-nrf2 pathway. Planta Medica 2014, 80, 97–104. [Google Scholar] [CrossRef]
- Costa, G.; Francisco, V.; Lopes, M.C.; Cruz, M.T.; Batista, M.T. Intracellular signaling pathways modulated by phenolic compounds: Application for new anti-inflammatory drugs discovery. Curr. Med. Chem. 2012, 19, 2876–2900. [Google Scholar] [CrossRef]
- Su, P.; Zhang, J.; Wang, S.; Aschner, M.; Cao, Z.; Zhao, F.; Wang, D.; Chen, J.; Luo, W. Genistein alleviates lead induced neurotoxicity in vitro and in vivo: Involvement of multiple signaling pathways. Neurotoxicology 2016, 53, 153–164. [Google Scholar] [CrossRef]
- Giudice, A.; Arra, C.; Turco, M.C. Review of molecular mechanisms involved in the activation of the Nrf2-ARE signaling pathway by chemopreventive agents. Methods Mol. Biol. 2010, 647, 37–74. [Google Scholar]
- Wu, J.; Williams, D.; Walter, G.A.; Thompson, W.E.; Sidell, N. Estrogen increases Nrf2 activity through activation of the PI3K pathway in MCF-7 breast cancer cells. Exp. Cell Res. 2014, 328, 351–360. [Google Scholar] [CrossRef]
- Young, A.R.; Narita, M.; Ferreira, M.; Kirschner, K.; Sadaie, M.; Darot, J.F.; Tavaré, S.; Arakawa, S.; Shimizu, S.; Watt, F.M.; et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009, 23, 798–803. [Google Scholar] [CrossRef] [PubMed]
- Webb, A.E.; Brunet, A. FOXO transcription factors: Key regulators of cellular quality control. Trends Biochem. Sci. 2014, 39, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Mandell, M.A.; Jain, A.; Arko-Mensah, J.; Chauhan, S.; Kimura, T.; Dinkins, C.; Silvestri, G.; Münch, J.; Kirchhoff, F.; Simonsen, A.; et al. TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Dev. Cell 2014, 30, 394–409. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Katzenellenbogen, B.S. Estrogen Receptor-β modulation of the ERα-P53 loop regulating gene expression, proliferation, and apoptosis in breast cancer. Horm. Cancer 2017, 8, 230–242. [Google Scholar] [CrossRef]
- Campisi, J. Senescent cells, tumor suppression, and organismal aging: Good citizens, bad neighbors. Cell 2005, 120, 513–522. [Google Scholar] [CrossRef]
- Zheng, K.; He1, Z.; Kitazato, K.; Wang, Y. Selective Autophagy Regulates Cell Cycle in Cancer Therapy. Theranostics 2019, 9, 104–125. [Google Scholar] [CrossRef]
- Bernhart, E.; Damm, S.; Heffeter, P.; Wintersperger, A.; Asslaber, M.; Frank, S.; Hammer, A.; Strohmaier, H.; DeVaney, T.; Mrfka, M.; et al. Silencing of protein kinase D2 induces glioma cell senescence via P53-dependent and -independent pathways. Neuro Oncol. 2014, 16, 933–945. [Google Scholar] [CrossRef]
- Ciavarra, G.; Zacksenhaus, F. Multiple pathways counteract cell death induced by RB1 loss: Implications for cancer. Cell Cycle 2011, 10, 1533–1539. [Google Scholar] [CrossRef]
- White, E. Autophagy and P53, Cold Spring Harb. Perspect. Med. 2016, 6, a026120. [Google Scholar]
- Chakradeo, S.; Elmore, L.W.; Gewirtz, D.A. Is senescence reversible? Curr. Drug Targets 2016, 17, 460–466. [Google Scholar] [CrossRef]
- Alexander, K.; Yang, H.S.; Hinds, P.W. pRb inactivation in senescent cells leads to an E2F-dependent apoptosis requiring p73. Mol. Cancer Res. 2003, 1, 716–728. [Google Scholar] [PubMed]
- Takahashi, A.; Ohtani, N.; Hara, E. Irreversibility of cellular senescence: Dual roles of p16INK4a/Rb-pathway in cell cycle control. Cell Div. 2007, 2, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Beauséjour, C.M.; Krtolica, A.; Galimi, F.; Narita, M.; Lowe, S.W.; Yaswen, P.; Campisi, J. Reversal of human cellular senescence: Roles of the P53 and p16 pathways. EMBO J. 2003, 22, 4212–4222. [Google Scholar] [CrossRef]
- Shiloh, Y.; Ziv, Y. The ATM protein kinase: Regulating the cellular response to genotoxic stress and more. Nat. Rev. Mol. Cell Biol. 2013, 14, 197–210. [Google Scholar] [CrossRef]
- Parekh, A.; Das, D.; Das, S.; Dhara, S.; Biswas, K.; Mandal, M.; Das, S. Bioimpedimetric analysis in conjunction with growth dynamics to differentiate aggressiveness of cancer cells. Sci. Rep. 2018, 8, 783. [Google Scholar] [CrossRef]
- Jia, T.; Zhang, L.; Duan, Y.; Zhang, M.; Wang, G.; Zhang, J.; Zhao, Z. The differential susceptibilities of MCF-7 and MDA-MB-231 cells to the cytotoxic effects of curcumin are associated with the PI3K/Akt-SKP2- Cip/Kips pathway. Cancer Cell Int. 2014, 14, 126. [Google Scholar] [CrossRef] [PubMed]
- Garbar, C.; Mascaux, C.; Giustiniani, J.; Merrouche, Y.; Bensussan, A. Chemotherapy treatment induces an increase of autophagy in the luminal breast cancer cell MCF7, but not in the triple-negative MDA-MB231. Sci. Rep. 2017, 7, 7201. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Song, T.; Chen, L.; Soliman, H.; Chen, J. Nucleolar repression facilitates initiation and maintenance of senescence. Cell Cycle 2015, 14, 3613–3623. [Google Scholar] [CrossRef]
- Galbiati, A.; Penzo, M.; Bacalini, M.G.; Onofrillo, C.; Guerrieri, A.N.; Garagnani, P.; Franceschi, C.; Treré, D.; Montanar, L. Epigenetic up-regulation of ribosome biogenesis and more aggressive phenotype triggered by the lack of the histone demethylase JHDM1B in mammary epithelial cells. Oncotarget 2017, 8, 37091–37103. [Google Scholar] [CrossRef]
- Hsieh, D.H.; Kuo, W.W.; Lai, Y.P. 17β-estradiol and/or estrogen receptor β attenuate the autophagic and apoptotic effects induced by prolonged hypoxia through HIF-1α-Mediated BNIP3 and IGFBP-3 signaling blockage. Cell Physiol. Biochem. 2015, 36, 274–284. [Google Scholar] [CrossRef]
- Lee, M.; Lee, J.S. Exploiting tumor cell senescence in anticancer therapy. BMB Rep. 2014, 47, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Ruhland, M.; Coussens, L.M.; Stewart, S. Senescence and cancer: An evolving inflammatory paradox. Biochim. Biophys. Acta 2016, 1865, 14–22. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bajbouj, K.; Shafarin, J.; Taneera, J.; Hamad, M. Estrogen Signaling Induces Mitochondrial Dysfunction-Associated Autophagy and Senescence in Breast Cancer Cells. Biology 2020, 9, 68. https://doi.org/10.3390/biology9040068
Bajbouj K, Shafarin J, Taneera J, Hamad M. Estrogen Signaling Induces Mitochondrial Dysfunction-Associated Autophagy and Senescence in Breast Cancer Cells. Biology. 2020; 9(4):68. https://doi.org/10.3390/biology9040068
Chicago/Turabian StyleBajbouj, Khuloud, Jasmin Shafarin, Jalal Taneera, and Mawieh Hamad. 2020. "Estrogen Signaling Induces Mitochondrial Dysfunction-Associated Autophagy and Senescence in Breast Cancer Cells" Biology 9, no. 4: 68. https://doi.org/10.3390/biology9040068
APA StyleBajbouj, K., Shafarin, J., Taneera, J., & Hamad, M. (2020). Estrogen Signaling Induces Mitochondrial Dysfunction-Associated Autophagy and Senescence in Breast Cancer Cells. Biology, 9(4), 68. https://doi.org/10.3390/biology9040068




