Assessing Autophagy in Archived Tissue or How to Capture Autophagic Flux from a Tissue Snapshot
Abstract
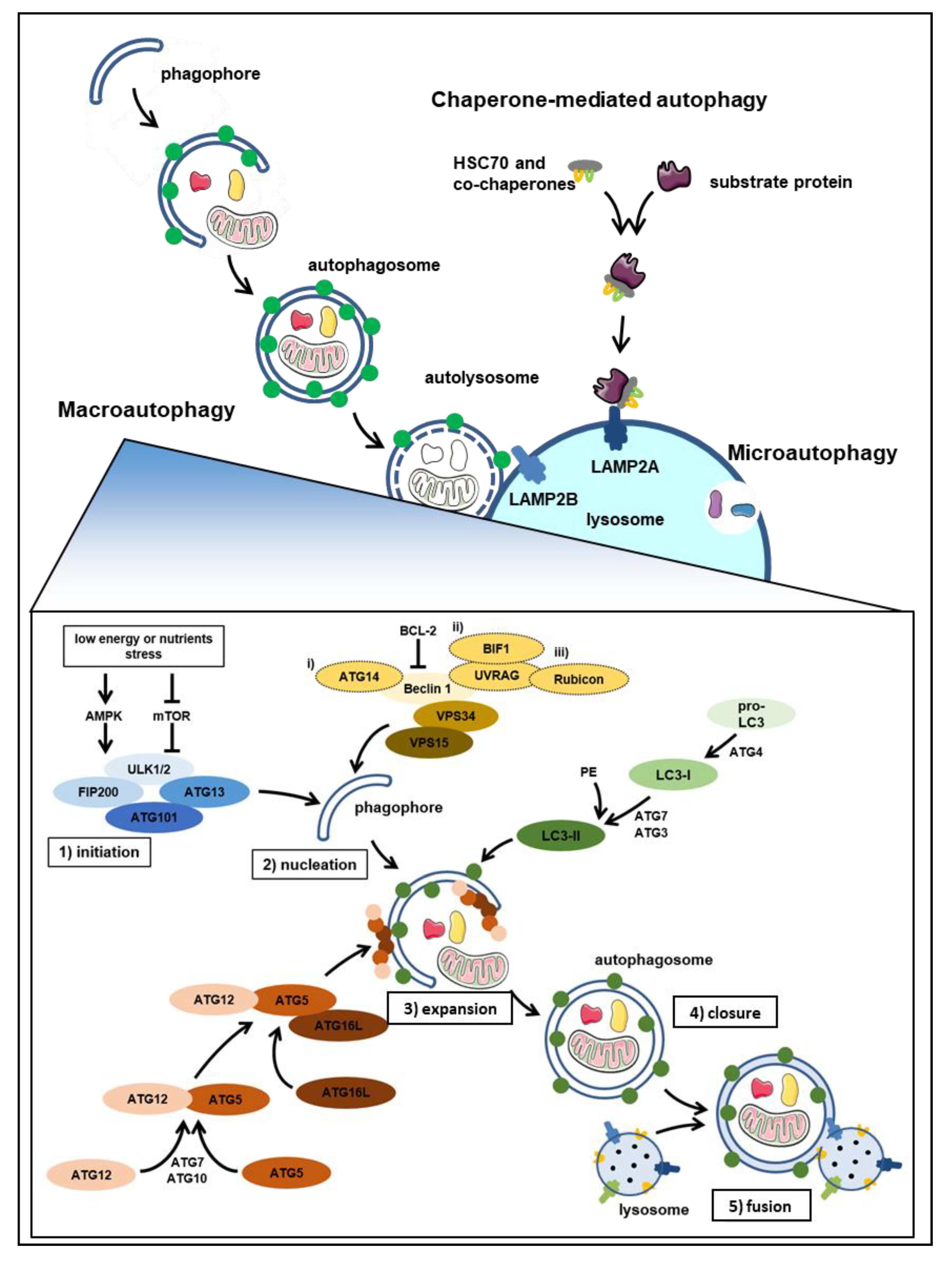
1. Autophagy at Glance
2. Autophagy and Human Disease
2.1. Autophagy in Cancer
2.1.1. Solid Tumors
2.1.2. Leukemia and Lymphomas
2.2. Autophagy in other Diseases (Neurodegenerative Conditions, Metabolic Disorders, Myopathies, and Heart Conditions)
2.2.1. Neurodegenerative Disorders
2.2.2. Metabolic Syndrome Diseases
2.2.3. Autoimmune Diseases
2.2.4. Mitochondrial Diseases
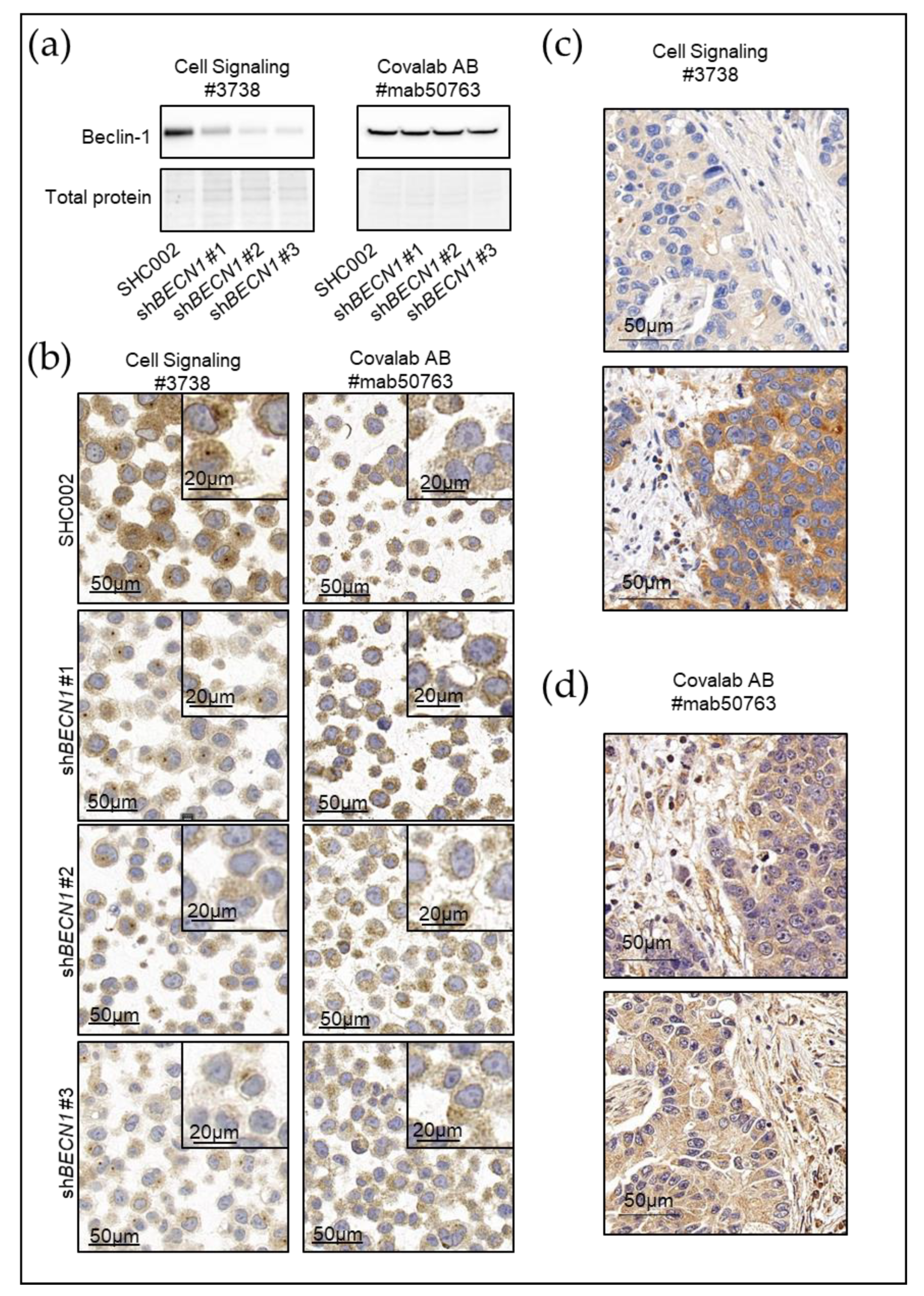
3. Current Autophagy Markers and Their Limitations
4. New Avenues for Better Autophagy Markers
5. The Pathologists’ Corner
6. Gathering New Knowledge and Tools for a Better Autophagy Assessment
7. Improving Autophagy Assessment by Sharing Knowledge
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Levine, B.; Klionsky, D.J. Autophagy wins the 2016 Nobel Prize in Physiology or Medicine: Breakthroughs in baker’s yeast fuel advances in biomedical research. Proc. Natl. Acad. Sci. USA 2017, 114, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cecconi, F.; Choi, A.M.; Chu, C.T.; Codogno, P.; Colombo, M.I.; et al. Molecular definitions of autophagy and related processes. EMBO J. 2017, 36, 1811–1836. [Google Scholar] [CrossRef] [PubMed]
- Tanida, I. Autophagy basics. Microbiol. Immunol. 2011, 55, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J. Look people, “Atg” is an abbreviation for “autophagy-related.” That’s it. Autophagy 2012, 8, 1281–1282. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Klionsky, D.J. The regulation of autophagy—Unanswered questions. J. Cell Sci. 2011, 124, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Parzych, K.R.; Klionsky, D.J. An Overview of Autophagy: Morphology, Mechanism, and Regulation. Antioxid. Redox Signal. 2013, 20, 460–473. [Google Scholar] [CrossRef]
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The Role of Atg Proteins in Autophagosome Formation. Annu. Rev. Cell Dev. Biol. 2011, 27, 107–132. [Google Scholar] [CrossRef]
- Yin, Z.; Pascual, C.; Klionsky, D.J. Autophagy: Machinery and regulation. Microb. Cell 2016, 3, 588–596. [Google Scholar] [CrossRef]
- Yu, L.; Chen, Y.; Tooze, S.A. Autophagy pathway: Cellular and molecular mechanisms. Autophagy 2018, 14, 207–215. [Google Scholar] [CrossRef]
- Suzuki, K.; Kubota, Y.; Sekito, T.; Ohsumi, Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 2007, 12, 209–218. [Google Scholar] [CrossRef]
- Tang, Z.; Takahashi, Y.; He, H.; Hattori, T.; Chen, C.; Liang, X.; Chen, H.; Young, M.M.; Wang, H.-G. TOM40 Targets Atg2 to Mitochondria-Associated ER Membranes for Phagophore Expansion. Cell Rep. 2019, 28, 1744–1757.e5. [Google Scholar] [CrossRef] [PubMed]
- Valverde, D.P.; Yu, S.; Boggavarapu, V.; Kumar, N.; Lees, J.A.; Walz, T.; Reinisch, K.M.; Melia, T.J. ATG2 transports lipids to promote autophagosome biogenesis. J. Cell Biol. 2019, 218, 1787–1798. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Sánchez, R.; Rose, J.; Guimarães, R.; Mari, M.; Papinski, D.; Rieter, E.; Geerts, W.J.; Hardenberg, R.; Kraft, C.; Ungermann, C.; et al. Atg9 establishes Atg2-dependent contact sites between the endoplasmic reticulum and phagophores. J. Cell Biol. 2018, 217, 2743–2763. [Google Scholar] [CrossRef] [PubMed]
- Bozic, M.; van den Bekerom, L.; Milne, B.A.; Goodman, N.; Roberston, L.; Prescott, A.R.; Macartney, T.J.; Dawe, N.; McEwan, D.G. A conserved ATG2-GABARAP family interaction is critical for phagophore formation. EMBO Rep. 2020, 21, e48412. [Google Scholar] [CrossRef] [PubMed]
- Amaya, C.; Fader, C.M.; Colombo, M.I. Autophagy and proteins involved in vesicular trafficking. FEBS Lett. 2015, 589, 3343–3353. [Google Scholar] [CrossRef] [PubMed]
- Fraser, J.; Cabodevilla, A.G.; Simpson, J.; Gammoh, N. Interplay of autophagy, receptor tyrosine kinase signalling and endocytic trafficking. Essays Biochem. 2017, 61, 597–607. [Google Scholar] [PubMed]
- Shibutani, S.; Okazaki, H.; Iwata, H. Dynamin-dependent amino acid endocytosis activates mechanistic target of rapamycin complex 1 (mTORC1). J. Biol. Chem. 2017, 292, 18052–18061. [Google Scholar] [CrossRef]
- Dice, J.F. Chaperone-mediated autophagy. Autophagy 2007, 3, 295–299. [Google Scholar] [CrossRef]
- Kaushik, S.; Cuervo, A.M. The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 365–381. [Google Scholar] [CrossRef]
- Yang, Q.; Wang, R.; Zhu, L. Chaperone-Mediated Autophagy. In Autophagy: Biology and Diseases: Basic Science; Qin, Z.-H., Ed.; Advances in Experimental Medicine and Biology; Springer: Singapore, 2019; pp. 435–452. ISBN 9789811506024. [Google Scholar]
- Kirchner, P.; Bourdenx, M.; Madrigal-Matute, J.; Tiano, S.; Diaz, A.; Bartholdy, B.A.; Will, B.; Cuervo, A.M. Proteome-wide analysis of chaperone-mediated autophagy targeting motifs. PLoS Biol. 2019, 17, e3000301. [Google Scholar] [CrossRef]
- Olsvik, H.L.; Svenning, S.; Abudu, Y.P.; Brech, A.; Stenmark, H.; Johansen, T.; Mejlvang, J. Endosomal microautophagy is an integrated part of the autophagic response to amino acid starvation. Autophagy 2019, 15, 182–183. [Google Scholar] [CrossRef] [PubMed]
- Mijaljica, D.; Prescott, M.; Devenish, R.J. Microautophagy in mammalian cells: Revisiting a 40-year-old conundrum. Autophagy 2011, 7, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Oku, M.; Sakai, Y. Three Distinct Types of Microautophagy Based on Membrane Dynamics and Molecular Machineries. BioEssays 2018, 40, 1800008. [Google Scholar] [CrossRef] [PubMed]
- Doherty, J.; Baehrecke, E.H. Life, death and autophagy. Nat. Cell Biol. 2018, 20, 1110–1117. [Google Scholar] [CrossRef] [PubMed]
- Denton, D.; Kumar, S. Autophagy-dependent cell death. Cell Death Differ. 2019, 26, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Betin, V.M.S.; Lane, J.D. Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. J. Cell Sci. 2009, 122, 2554–2566. [Google Scholar] [CrossRef]
- Yousefi, S.; Perozzo, R.; Schmid, I.; Ziemiecki, A.; Schaffner, T.; Scapozza, L.; Brunner, T.; Simon, H.-U. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat. Cell Biol. 2006, 8, 1124–1132. [Google Scholar] [CrossRef]
- Wirawan, E.; Walle, L.V.; Kersse, K.; Cornelis, S.; Claerhout, S.; Vanoverberghe, I.; Roelandt, R.; Rycke, R.D.; Verspurten, J.; Declercq, W.; et al. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 2010, 1. [Google Scholar] [CrossRef]
- Lindner, P.; Christensen, S.B.; Nissen, P.; Møller, J.V.; Engedal, N. Cell death induced by the ER stressor thapsigargin involves death receptor 5, a non-autophagic function of MAP1LC3B, and distinct contributions from unfolded protein response components. Cell Commun. Signal. 2020, 18, 1–23. [Google Scholar] [CrossRef]
- Young, M.M.; Takahashi, Y.; Khan, O.; Park, S.; Hori, T.; Yun, J.; Sharma, A.K.; Amin, S.; Hu, C.-D.; Zhang, J.; et al. Autophagosomal Membrane Serves as Platform for Intracellular Death-inducing Signaling Complex (iDISC)-mediated Caspase-8 Activation and Apoptosis. J. Biol. Chem. 2012, 287, 12455–12468. [Google Scholar] [CrossRef]
- Pan, J.-A.; Ullman, E.; Dou, Z.; Zong, W.-X. Inhibition of Protein Degradation Induces Apoptosis through a Microtubule-Associated Protein 1 Light Chain 3-Mediated Activation of Caspase-8 at Intracellular Membranes. Mol. Cell. Biol. 2011, 31, 3158–3170. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, S.; Kanaseki, T.; Mizushima, N.; Mizuta, T.; Arakawa-Kobayashi, S.; Thompson, C.B.; Tsujimoto, Y. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat. Cell Biol. 2004, 6, 1221–1228. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.; Xie, Y.; Song, X.; Sun, X.; Lotze, M.T.; Zeh, H.J., III; Kang, R.; Tang, D. Autophagy promotes ferroptosis by degradation of ferritin. Autophagy 2016, 12, 1425–1428. [Google Scholar] [CrossRef] [PubMed]
- Basit, F.; Cristofanon, S.; Fulda, S. Obatoclax (GX15-070) triggers necroptosis by promoting the assembly of the necrosome on autophagosomal membranes. Cell Death Differ. 2013, 20, 1161–1173. [Google Scholar] [CrossRef] [PubMed]
- Sosna, J.; Philipp, S.; Chico, J.F.; Saggau, C.; Fritsch, J.; Föll, A.; Plenge, J.; Arenz, C.; Pinkert, T.; Kalthoff, H.; et al. Differences and Similarities in TRAIL and Tumor Necrosis Factor-Mediated Necroptotic Signaling in Cancer Cells. Mol. Cell. Biol. 2016, 36, 2626–2644. [Google Scholar] [CrossRef]
- Vakifahmetoglu-Norberg, H.; Xia, H.; Yuan, J. Pharmacologic agents targeting autophagy. J. Clin. Investig. 2015, 125, 5–13. [Google Scholar] [CrossRef]
- Arias, E.; Cuervo, A.M. Pros and Cons of Chaperone-Mediated Autophagy in Cancer Biology. Trends Endocrinol. Metab. TEM 2020, 31, 53–66. [Google Scholar] [CrossRef]
- Bonam, S.R.; Wang, F.; Muller, S. Autophagy: A new concept in autoimmunity regulation and a novel therapeutic option. J. Autoimmun. 2018, 94, 16–32. [Google Scholar] [CrossRef]
- Condello, M.; Pellegrini, E.; Caraglia, M.; Meschini, S. Targeting Autophagy to Overcome Human Diseases. Int. J. Mol. Sci. 2019, 20, 725. [Google Scholar] [CrossRef]
- Djavaheri-Mergny, M.; Giuriato, S.; Tschan, M.P.; Humbert, M. Therapeutic Modulation of Autophagy in Leukaemia and Lymphoma. Cells 2019, 8, 103. [Google Scholar] [CrossRef]
- Janji, B.; Berchem, G.; Chouaib, S. Targeting Autophagy in the Tumor Microenvironment: New Challenges and Opportunities for Regulating Tumor Immunity. Front. Immunol. 2018, 9, 887. [Google Scholar] [CrossRef]
- Jiang, P.; Mizushima, N. Autophagy and human diseases. Cell Res. 2014, 24, 69–79. [Google Scholar] [CrossRef]
- Nixon, R.A. The role of autophagy in neurodegenerative disease. Nat. Med. 2013, 19, 983–997. [Google Scholar] [CrossRef] [PubMed]
- Robert, G.; Jacquel, A.; Auberger, P. Chaperone-Mediated Autophagy and Its Emerging Role in Hematological Malignancies. Cells 2019, 8, 1260. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.L.; Cuervo, A.M. Autophagy and human disease: Emerging themes. Curr. Opin. Genet. Dev. 2014, 26, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Ward, C.; Martinez-Lopez, N.; Otten, E.G.; Carroll, B.; Maetzel, D.; Singh, R.; Sarkar, S.; Korolchuk, V.I. Autophagy, lipophagy and lysosomal lipid storage disorders. Biochim. Biophys. Acta 2016, 1861, 269–284. [Google Scholar] [CrossRef]
- Qu, X.; Yu, J.; Bhagat, G.; Furuya, N.; Hibshoosh, H.; Troxel, A.; Rosen, J.; Eskelinen, E.-L.; Mizushima, N.; Ohsumi, Y.; et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Investig. 2003, 112, 1809–1820. [Google Scholar] [CrossRef]
- Galluzzi, L.; Pietrocola, F.; Bravo-San Pedro, J.M.; Amaravadi, R.K.; Baehrecke, E.H.; Cecconi, F.; Codogno, P.; Debnath, J.; Gewirtz, D.A.; Karantza, V.; et al. Autophagy in malignant transformation and cancer progression. EMBO J. 2015, 34, 856–880. [Google Scholar] [CrossRef]
- Galluzzi, L.; Bravo-San Pedro, J.M.; Levine, B.; Green, D.R.; Kroemer, G. Pharmacological modulation of autophagy: Therapeutic potential and persisting obstacles. Nat. Rev. Drug Discov. 2017, 16, 487–511. [Google Scholar] [CrossRef]
- Hoare, M.; Young, A.R.J.; Narita, M. Autophagy in cancer: Having your cake and eating it. Semin. Cancer Biol. 2011, 21, 397–404. [Google Scholar] [CrossRef]
- Kon, M.; Kiffin, R.; Koga, H.; Chapochnick, J.; Macian, F.; Varticovski, L.; Cuervo, A.M. Chaperone-Mediated Autophagy Is Required for Tumor Growth. Sci. Transl. Med. 2011, 3, 109ra117. [Google Scholar] [CrossRef] [PubMed]
- Tasset, I.; Cuervo, A.M. Role of chaperone-mediated autophagy in metabolism. FEBS J. 2016, 283, 2403–2413. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Li, D.; Zhao, D.; Lin, R.; Chu, Y.; Zhang, H.; Zha, Z.; Liu, Y.; Li, Z.; Xu, Y.; et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol. Cell 2011, 42, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Najafov, A.; Geng, J.; Galan-Acosta, L.; Han, X.; Guo, Y.; Shan, B.; Zhang, Y.; Norberg, E.; Zhang, T.; et al. Degradation of HK2 by chaperone-mediated autophagy promotes metabolic catastrophe and cell death. J. Cell Biol. 2015, 210, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Saha, T. LAMP2A overexpression in breast tumors promotes cancer cell survival via chaperone-mediated autophagy. Autophagy 2012, 8, 1643–1656. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.B.; Nin, D.S.; Tam, J.; Khan, M. Role of chaperone mediated autophagy (CMA) in the degradation of misfolded N-CoR protein in non-small cell lung cancer (NSCLC) cells. PLoS ONE 2011, 6, e25268. [Google Scholar] [CrossRef]
- Ferreira, J.V.; Soares, A.R.; Ramalho, J.S.; Pereira, P.; Girao, H. K63 linked ubiquitin chain formation is a signal for HIF1A degradation by Chaperone-Mediated Autophagy. Sci. Rep. 2015, 5, 10210. [Google Scholar] [CrossRef]
- Hubbi, M.E.; Hu, H.; Ahmed, I.; Levchenko, A.; Semenza, G.L. Chaperone-mediated autophagy targets hypoxia-inducible factor-1α (HIF-1α) for lysosomal degradation. J. Biol. Chem. 2013, 288, 10703–10714. [Google Scholar] [CrossRef]
- Han, Q.; Deng, Y.; Chen, S.; Chen, R.; Yang, M.; Zhang, Z.; Sun, X.; Wang, W.; He, Y.; Wang, F.; et al. Downregulation of ATG5-dependent macroautophagy by chaperone-mediated autophagy promotes breast cancer cell metastasis. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Y.-Y.; Yao, C.-B.; Li, J.-T.; Zhao, X.-N.; Yang, H.-B.; Zhang, M.; Yin, M.; Chen, J.; Lei, Q.-Y. Acetylation targets HSD17B4 for degradation via the CMA pathway in response to estrone. Autophagy 2017, 13, 538–553. [Google Scholar] [CrossRef]
- Zhou, J.; Yang, J.; Fan, X.; Hu, S.; Zhou, F.; Dong, J.; Zhang, S.; Shang, Y.; Jiang, X.; Guo, H.; et al. Chaperone-mediated autophagy regulates proliferation by targeting RND3 in gastric cancer. Autophagy 2016, 12, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.L.; Suh, Y.; Cuervo, A.M. Deficient chaperone-mediated autophagy in liver leads to metabolic dysregulation. Cell Metab. 2014, 20, 417–432. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-Y.; White, E. Role of Autophagy in Cancer Prevention. Cancer Prev. Res. 2011, 4, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Miracco, C.; Cosci, E.; Oliveri, G.; Luzi, P.; Pacenti, L.; Monciatti, I.; Mannucci, S.; De Nisi, M.C.; Toscano, M.; Malagnino, V.; et al. Protein and mRNA expression of autophagy gene Beclin 1 in human brain tumours. Int. J. Oncol. 2007, 30, 429–436. [Google Scholar] [PubMed]
- Tang, H.; Sebti, S.; Titone, R.; Zhou, Y.; Isidoro, C.; Ross, T.S.; Hibshoosh, H.; Xiao, G.; Packer, M.; Xie, Y.; et al. Decreased BECN1 mRNA Expression in Human Breast Cancer is Associated with Estrogen Receptor-Negative Subtypes and Poor Prognosis. EBioMedicine 2015, 2, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Coppola, D.; Matsushita, N.; Cualing, H.D.; Sun, M.; Sato, Y.; Liang, C.; Jung, J.U.; Cheng, J.Q.; Mulé, J.J.; et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat. Cell Biol. 2007, 9, 1142–1151. [Google Scholar] [CrossRef] [PubMed]
- Coppola, D.; Khalil, F.; Eschrich, S.A.; Boulware, D.; Yeatman, T.; Wang, H.-G. Down-regulation of Bax-interacting factor-1 in colorectal adenocarcinoma. Cancer 2008, 113, 2665–2670. [Google Scholar] [CrossRef]
- Levy, J.M.M.; Towers, C.G.; Thorburn, A. Targeting autophagy in cancer. Nat. Rev. Cancer 2017, 17, 528–542. [Google Scholar] [CrossRef]
- Guo, J.Y.; White, E. Autophagy is required for mitochondrial function, lipid metabolism, growth, and fate of KRAS(G12D)-driven lung tumors. Autophagy 2013, 9, 1636–1638. [Google Scholar] [CrossRef]
- Kim, M.-J.; Woo, S.-J.; Yoon, C.-H.; Lee, J.-S.; An, S.; Choi, Y.-H.; Hwang, S.-G.; Yoon, G.; Lee, S.-J. Involvement of autophagy in oncogenic K-Ras-induced malignant cell transformation. J. Biol. Chem. 2011, 286, 12924–12932. [Google Scholar] [CrossRef]
- Yang, S.; Wang, X.; Contino, G.; Liesa, M.; Sahin, E.; Ying, H.; Bause, A.; Li, Y.; Stommel, J.M.; Dell’antonio, G.; et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011, 25, 717–729. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Liu, H.; Ming, L. Multiple Roles of Autophagy in the Sorafenib Resistance of Hepatocellular Carcinoma. Cell. Physiol. Biochem. 2017, 44, 716–727. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Li, X.; Liu, H.; Jiang, R.; Yang, M.; Zhang, M.; Wang, Y.; Zhao, Y.; Li, H. GATA6-upregulating autophagy promotes TKI resistance in nonsmall cell lung cancer. Cancer Biol. Ther. 2019, 20, 1206–1212. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Ye, T.; Ge, M.; Yang, M.; Zhang, L.; Jin, S.; Ye, X.; Long, B.; Li, L. Inhibition of autophagy enhances the targeted therapeutic effect of sorafenib in thyroid cancer. Oncol. Rep. 2018, 39, 711–720. [Google Scholar] [CrossRef]
- Zai, W.; Chen, W.; Han, Y.; Wu, Z.; Fan, J.; Zhang, X.; Luan, J.; Tang, S.; Jin, X.; Fu, X.; et al. Targeting PARP and autophagy evoked synergistic lethality in hepatocellular carcinoma. Carcinogenesis 2019. [Google Scholar] [CrossRef]
- Cheng, G.; Zhang, Q.; Pan, J.; Lee, Y.; Ouari, O.; Hardy, M.; Zielonka, M.; Myers, C.R.; Zielonka, J.; Weh, K.; et al. Targeting lonidamine to mitochondria mitigates lung tumorigenesis and brain metastasis. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Mele, L.; la Noce, M.; Paino, F.; Regad, T.; Wagner, S.; Liccardo, D.; Papaccio, G.; Lombardi, A.; Caraglia, M.; Tirino, V.; et al. Glucose-6-phosphate dehydrogenase blockade potentiates tyrosine kinase inhibitor effect on breast cancer cells through autophagy perturbation. J. Exp. Clin. Cancer Res. 2019, 38, 160. [Google Scholar] [CrossRef]
- Martins, I.; Michaud, M.; Sukkurwala, A.Q.; Adjemian, S.; Ma, Y.; Shen, S.; Kepp, O.; Menger, L.; Vacchelli, E.; Galluzzi, L.; et al. Premortem autophagy determines the immunogenicity of chemotherapy-induced cancer cell death. Autophagy 2012, 8, 413–415. [Google Scholar] [CrossRef]
- Martins, I.; Wang, Y.; Michaud, M.; Ma, Y.; Sukkurwala, A.Q.; Shen, S.; Kepp, O.; Métivier, D.; Galluzzi, L.; Perfettini, J.-L.; et al. Molecular mechanisms of ATP secretion during immunogenic cell death. Cell Death Differ. 2014, 21, 79–91. [Google Scholar] [CrossRef]
- Kroemer, G.; Galluzzi, L.; Kepp, O.; Zitvogel, L. Immunogenic cell death in cancer therapy. Annu. Rev. Immunol. 2013, 31, 51–72. [Google Scholar] [CrossRef]
- Ko, A.; Kanehisa, A.; Martins, I.; Senovilla, L.; Chargari, C.; Dugue, D.; Mariño, G.; Kepp, O.; Michaud, M.; Perfettini, J.-L.; et al. Autophagy inhibition radiosensitizes in vitro, yet reduces radioresponses in vivo due to deficient immunogenic signalling. Cell Death Differ. 2014, 21, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Michaud, M.; Martins, I.; Sukkurwala, A.Q.; Adjemian, S.; Ma, Y.; Pellegatti, P.; Shen, S.; Kepp, O.; Scoazec, M.; Mignot, G.; et al. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science 2011, 334, 1573–1577. [Google Scholar] [CrossRef] [PubMed]
- Pietrocola, F.; Pol, J.; Vacchelli, E.; Rao, S.; Enot, D.P.; Baracco, E.E.; Levesque, S.; Castoldi, F.; Jacquelot, N.; Yamazaki, T.; et al. Caloric Restriction Mimetics Enhance Anticancer Immunosurveillance. Cancer Cell 2016, 30, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Page, D.B.; Hulett, T.W.; Hilton, T.L.; Hu, H.-M.; Urba, W.J.; Fox, B.A. Glimpse into the future: Harnessing autophagy to promote anti-tumor immunity with the DRibbles vaccine. J. Immunother. Cancer 2016, 4, 25. [Google Scholar] [CrossRef]
- Cao, Y.; Cai, J.; Zhang, S.; Yuan, N.; Li, X.; Fang, Y.; Song, L.; Shang, M.; Liu, S.; Zhao, W.; et al. Loss of autophagy leads to failure in megakaryopoiesis, megakaryocyte differentiation, and thrombopoiesis in mice. Exp. Hematol. 2015, 43, 488–494. [Google Scholar] [CrossRef]
- Cao, Y.; Cai, J.; Li, X.; Yuan, N.; Zhang, S. Autophagy governs erythroid differentiation both in vitro and in vivo. Hematology 2016, 21, 225–233. [Google Scholar] [CrossRef]
- Ho, T.T.; Warr, M.R.; Adelman, E.R.; Lansinger, O.M.; Flach, J.; Verovskaya, E.V.; Figueroa, M.E.; Passegué, E. Autophagy maintains the metabolism and function of young and old stem cells. Nature 2017, 543, 205–210. [Google Scholar] [CrossRef]
- Liu, F.; Lee, J.Y.; Wei, H.; Tanabe, O.; Engel, J.D.; Morrison, S.J.; Guan, J.-L. FIP200 is required for the cell-autonomous maintenance of fetal hematopoietic stem cells. Blood 2010, 116, 4806–4814. [Google Scholar] [CrossRef]
- Mortensen, M.; Soilleux, E.J.; Djordjevic, G.; Tripp, R.; Lutteropp, M.; Sadighi-Akha, E.; Stranks, A.J.; Glanville, J.; Knight, S.; Jacobsen, S.-E.W.; et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J. Exp. Med. 2011, 208, 455–467. [Google Scholar] [CrossRef]
- Warr, M.R.; Binnewies, M.; Flach, J.; Reynaud, D.; Garg, T.; Malhotra, R.; Debnath, J.; Passegué, E. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature 2013, 494, 323–327. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef] [PubMed]
- Auberger, P.; Puissant, A. Autophagy, a key mechanism of oncogenesis and resistance in leukemia. Blood 2017, 129, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, Â.; Azevedo, M.M.; Pereira, O.; Sampaio-Marques, B.; Paiva, A.; Correia-Neves, M.; Castro, I.; Ludovico, P. Proteolytic systems and AMP-activated protein kinase are critical targets of acute myeloid leukemia therapeutic approaches. Oncotarget 2015, 6, 31428–31440. [Google Scholar] [CrossRef][Green Version]
- Jin, J.; Britschgi, A.; Schläfli, A.M.; Humbert, M.; Shan-Krauer, D.; Batliner, J.; Federzoni, E.A.; Ernst, M.; Torbett, B.E.; Yousefi, S.; et al. Low Autophagy (ATG) Gene Expression Is Associated with an Immature AML Blast Cell Phenotype and Can Be Restored during AML Differentiation Therapy. Oxidative Med. Cell. Longev. 2018, 2018. [Google Scholar] [CrossRef]
- Watson, A.S.; Riffelmacher, T.; Stranks, A.; Williams, O.; De Boer, J.; Cain, K.; MacFarlane, M.; McGouran, J.; Kessler, B.; Khandwala, S.; et al. Autophagy limits proliferation and glycolytic metabolism in acute myeloid leukemia. Cell Death Discov. 2015, 1, 15008. [Google Scholar] [CrossRef]
- Piya, S.; Kornblau, S.M.; Ruvolo, V.R.; Mu, H.; Ruvolo, P.P.; Mcqueen, T.; Davis, R.E.; Hail, N., Jr.; Kantarjian, H.; Andreeff, M.; et al. Atg7 suppression enhances chemotherapeutic agent sensitivity and overcomes stroma-mediated chemoresistance in acute myeloid leukemia. Blood 2016, 128, 1260–1270. [Google Scholar] [CrossRef]
- Rudat, S.; Pfaus, A.; Cheng, Y.Y.; Holtmann, J.; Ellegast, J.M.; Bühler, C.; Marcantonio, D.D.; Martinez, E.; Göllner, S.; Wickenhauser, C.; et al. RET-mediated autophagy suppression as targetable co-dependence in acute myeloid leukemia. Leukemia 2018, 32, 2189–2202. [Google Scholar] [CrossRef]
- Visconte, V.; Przychodzen, B.; Han, Y.; Nawrocki, S.T.; Thota, S.; Kelly, K.R.; Patel, B.J.; Hirsch, C.; Advani, A.S.; Carraway, H.E.; et al. Complete mutational spectrum of the autophagy interactome: A novel class of tumor suppressor genes in myeloid neoplasms. Leukemia 2017, 31, 505–510. [Google Scholar] [CrossRef]
- Walter, M.J.; Payton, J.E.; Ries, R.E.; Shannon, W.D.; Deshmukh, H.; Zhao, Y.; Baty, J.; Heath, S.; Westervelt, P.; Watson, M.A.; et al. Acquired copy number alterations in adult acute myeloid leukemia genomes. Proc. Natl. Acad. Sci. USA 2009, 106, 12950–12955. [Google Scholar] [CrossRef]
- Heydt, Q.; Larrue, C.; Saland, E.; Bertoli, S.; Sarry, J.-E.; Besson, A.; Manenti, S.; Joffre, C.; Mansat-De Mas, V. Oncogenic FLT3-ITD supports autophagy via ATF4 in acute myeloid leukemia. Oncogene 2018, 37, 787–797. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Chen, L.; Atkinson, J.M.; Claxton, D.F.; Wang, H.-G. Atg5-dependent autophagy contributes to the development of acute myeloid leukemia in an MLL-AF9-driven mouse model. Cell Death Dis. 2016, 7, e2361. [Google Scholar] [CrossRef] [PubMed]
- Rothe, K.; Porter, V.; Jiang, X. Current Outlook on Autophagy in Human Leukemia: Foe in Cancer Stem Cells and Drug Resistance, Friend in New Therapeutic Interventions. Int. J. Mol. Sci. 2019, 20, 461. [Google Scholar] [CrossRef]
- Bosnjak, M.; Ristic, B.; Arsikin, K.; Mircic, A.; Suzin-Zivkovic, V.; Perovic, V.; Bogdanovic, A.; Paunovic, V.; Markovic, I.; Bumbasirevic, V.; et al. Inhibition of mTOR-dependent autophagy sensitizes leukemic cells to cytarabine-induced apoptotic death. PLoS ONE 2014, 9, e94374. [Google Scholar] [CrossRef] [PubMed]
- Isakson, P.; Bjørås, M.; Bøe, S.O.; Simonsen, A. Autophagy contributes to therapy-induced degradation of the PML/RARA oncoprotein. Blood 2010, 116, 2324–2331. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Cao, L.; Kang, R.; Yang, M.; Liu, L.; Zhao, Y.; Yu, Y.; Xie, M.; Yin, X.; Livesey, K.M.; et al. Autophagy regulates myeloid cell differentiation by p62/SQSTM1-mediated degradation of PML-RARα oncoprotein. Autophagy 2011, 7, 401–411. [Google Scholar] [CrossRef]
- Elzinga, B.M.; Nyhan, M.J.; Crowley, L.C.; O’Donovan, T.R.; Cahill, M.R.; McKenna, S.L. Induction of autophagy by Imatinib sequesters Bcr-Abl in autophagosomes and down-regulates Bcr-Abl protein. Am. J. Hematol. 2013, 88, 455–462. [Google Scholar] [CrossRef]
- Goussetis, D.J.; Gounaris, E.; Wu, E.J.; Vakana, E.; Sharma, B.; Bogyo, M.; Altman, J.K.; Platanias, L.C. Autophagic degradation of the BCR-ABL oncoprotein and generation of antileukemic responses by arsenic trioxide. Blood 2012, 120, 3555–3562. [Google Scholar] [CrossRef]
- Torgersen, M.L.; Engedal, N.; Bøe, S.-O.; Hokland, P.; Simonsen, A. Targeting autophagy potentiates the apoptotic effect of histone deacetylase inhibitors in t (8; 21) AML cells. Blood 2013, 122, 2467–2476. [Google Scholar] [CrossRef]
- Botbol, Y.; Guerrero-Ros, I.; Macian, F. Key roles of autophagy in regulating T-cell function. Eur. J. Immunol. 2016, 46, 1326–1334. [Google Scholar] [CrossRef]
- Cenci, S. Autophagy, a new determinant of plasma cell differentiation and antibody responses. Mol. Immunol. 2014, 62, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Gros, F.; Muller, S. Pharmacological regulators of autophagy and their link with modulators of lupus disease. Br. J. Pharmacol. 2014, 171, 4337–4359. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Mizushima, N.; Virgin, H.W. Autophagy in immunity and inflammation. Nature 2011, 469, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of cells and tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef]
- Münz, C. Autophagy beyond Intracellular MHC Class II Antigen Presentation. Trends Immunol. 2016, 37, 755–763. [Google Scholar] [CrossRef]
- Valdor, R.; Mocholi, E.; Botbol, Y.; Guerrero-Ros, I.; Chandra, D.; Koga, H.; Gravekamp, C.; Cuervo, A.M.; Macian, F. Chaperone-mediated autophagy regulates T cell responses through targeted degradation of negative regulators of T cell activation. Nat. Immunol. 2014, 15, 1046–1054. [Google Scholar] [CrossRef]
- Zhong, Z.; Sanchez-Lopez, E.; Karin, M. Autophagy, Inflammation, and Immunity: A Troika Governing Cancer and Its Treatment. Cell 2016, 166, 288–298. [Google Scholar] [CrossRef]
- Dugger, B.N.; Dickson, D.W. Pathology of Neurodegenerative Diseases. Cold Spring Harb. Perspect. Biol. 2017, 9, a028035. [Google Scholar] [CrossRef]
- Frake, R.A.; Ricketts, T.; Menzies, F.M.; Rubinsztein, D.C. Autophagy and neurodegeneration. J. Clin. Investig. 2015, 125, 65–74. [Google Scholar] [CrossRef]
- Hara, T.; Nakamura, K.; Matsui, M.; Yamamoto, A.; Nakahara, Y.; Suzuki-Migishima, R.; Yokoyama, M.; Mishima, K.; Saito, I.; Okano, H.; et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006, 441, 885–889. [Google Scholar] [CrossRef]
- Komatsu, M.; Waguri, S.; Chiba, T.; Murata, S.; Iwata, J.; Tanida, I.; Ueno, T.; Koike, M.; Uchiyama, Y.; Kominami, E.; et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006, 441, 880–884. [Google Scholar] [CrossRef] [PubMed]
- Menzies, F.M.; Fleming, A.; Rubinsztein, D.C. Compromised autophagy and neurodegenerative diseases. Nat. Rev. Neurosci. 2015, 16, 345–357. [Google Scholar] [CrossRef] [PubMed]
- Hunn, B.H.M.; Vingill, S.; Threlfell, S.; Alegre-Abarrategui, J.; Magdelyns, M.; Deltheil, T.; Bengoa-Vergniory, N.; Oliver, P.L.; Cioroch, M.; Doig, N.M.; et al. Impairment of Macroautophagy in Dopamine Neurons has Opposing Effects on Parkinsonian Pathology and Behavior. Cell Rep. 2019, 29, 920–931.e7. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Green, D.R. Autophagy-Independent Functions of the Autophagy Machinery. Cell 2019, 177, 1682–1699. [Google Scholar] [CrossRef] [PubMed]
- McCracken, E.; Monaghan, M.; Sreenivasan, S. Pathophysiology of the metabolic syndrome. Clin. Dermatol. 2018, 36, 14–20. [Google Scholar] [CrossRef]
- Eisenberg, T.; Abdellatif, M.; Schroeder, S.; Primessnig, U.; Stekovic, S.; Pendl, T.; Harger, A.; Schipke, J.; Zimmermann, A.; Schmidt, A.; et al. Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat. Med. 2016, 22, 1428–1438. [Google Scholar] [CrossRef]
- Pyo, J.-O.; Yoo, S.-M.; Ahn, H.-H.; Nah, J.; Hong, S.-H.; Kam, T.-I.; Jung, S.; Jung, Y.-K. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat. Commun. 2013, 4, 1–9. [Google Scholar] [CrossRef]
- Lim, H.; Lim, Y.-M.; Kim, K.H.; Jeon, Y.E.; Park, K.; Kim, J.; Hwang, H.-Y.; Lee, D.J.; Pagire, H.; Kwon, H.J.; et al. A novel autophagy enhancer as a therapeutic agent against metabolic syndrome and diabetes. Nat. Commun. 2018, 9, 1–14. [Google Scholar] [CrossRef]
- Avrameas, S.; Alexopoulos, H.; Moutsopoulos, H.M. Natural Autoantibodies: An Undersugn Hero of the Immune System and Autoimmune Disorders—A Point of View. Front. Immunol. 2018, 9, 1320. [Google Scholar] [CrossRef]
- Macri, C.; Wang, F.; Tasset, I.; Schall, N.; Page, N.; Briand, J.-P.; Cuervo, A.M.; Muller, S. Modulation of deregulated chaperone-mediated autophagy by a phosphopeptide. Autophagy 2015, 11, 472–486. [Google Scholar] [CrossRef]
- Wang, F.; Muller, S. Manipulating Autophagic Processes in Autoimmune Diseases: A Special Focus on Modulating Chaperone-Mediated Autophagy, an Emerging Therapeutic Target. Front. Immunol. 2015, 6, 252. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, R.; Scherbarth, H.R.; Rillo, O.L.; Gomez-Reino, J.J.; Muller, S. Lupuzor/P140 peptide in patients with systemic lupus erythematosus: A randomised, double-blind, placebo-controlled phase IIb clinical trial. Ann. Rheum. Dis. 2013, 72, 1830–1835. [Google Scholar] [CrossRef] [PubMed]
- Cotán, D.; Cordero, M.D.; Garrido-Maraver, J.; Oropesa-Ávila, M.; Rodríguez-Hernández, Á.; Gómez Izquierdo, L.; De la Mata, M.; De Miguel, M.; Bautista Lorite, J.; Rivas Infante, E.; et al. Secondary coenzyme Q10 deficiency triggers mitochondria degradation by mitophagy in MELAS fibroblasts. FASEB J. 2011, 25, 2669–2687. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Hernández, A.; Cordero, M.D.; Salviati, L.; Artuch, R.; Pineda, M.; Briones, P.; Gómez Izquierdo, L.; Cotán, D.; Navas, P.; Sánchez-Alcázar, J.A. Coenzyme Q deficiency triggers mitochondria degradation by mitophagy. Autophagy 2009, 5, 19–32. [Google Scholar] [CrossRef] [PubMed]
- de la Mata, M.; Garrido-Maraver, J.; Cotán, D.; Cordero, M.D.; Oropesa-Ávila, M.; Izquierdo, L.G.; de Miguel, M.S.; Lorite, J.B.; Infante, E.R.; Ybot, P.; et al. Recovery of MERRF Fibroblasts and Cybrids Pathophysiology by Coenzyme Q10. Neurotherapeutics 2012, 9, 446–463. [Google Scholar] [CrossRef] [PubMed]
- Morán, M.; Delmiro, A.; Blázquez, A.; Ugalde, C.; Arenas, J.; Martín, M.A. Bulk autophagy, but not mitophagy, is increased in cellular model of mitochondrial disease. Biochim. Biophys. Acta 2014, 1842, 1059–1070. [Google Scholar] [CrossRef]
- Villanueva-Paz, M.; Povea-Cabello, S.; Villalón-García, I.; Álvarez-Córdoba, M.; Suárez-Rivero, J.M.; Talaverón-Rey, M.; Jackson, S.; Falcón-Moya, R.; Rodríguez-Moreno, A.; Sánchez-Alcázar, J.A. Parkin-mediated mitophagy and autophagy flux disruption in cellular models of MERRF syndrome. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165726. [Google Scholar] [CrossRef]
- Emperador, S.; Garrido-Pérez, N.; Amezcua-Gil, J.; Gaudó, P.; Andrés-Sanz, J.A.; Yubero, D.; Fernández-Marmiesse, A.; O’Callaghan, M.M.; Ortigoza-Escobar, J.D.; Iriondo, M.; et al. Molecular Characterization of New FBXL4 Mutations in Patients with mtDNA Depletion Syndrome. Front. Genet. 2019, 10, 1300. [Google Scholar] [CrossRef]
- Tyynismaa, H.; Mjosund, K.P.; Wanrooij, S.; Lappalainen, I.; Ylikallio, E.; Jalanko, A.; Spelbrink, J.N.; Paetau, A.; Suomalainen, A. Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice. Proc. Natl. Acad. Sci. USA 2005, 102, 17687–17692. [Google Scholar] [CrossRef]
- Tyynismaa, H.; Carroll, C.J.; Raimundo, N.; Ahola-Erkkilä, S.; Wenz, T.; Ruhanen, H.; Guse, K.; Hemminki, A.; Peltola-Mjøsund, K.E.; Tulkki, V.; et al. Mitochondrial myopathy induces a starvation-like response. Hum. Mol. Genet. 2010, 19, 3948–3958. [Google Scholar] [CrossRef]
- White, K.E.; Davies, V.J.; Hogan, V.E.; Piechota, M.J.; Nichols, P.P.; Turnbull, D.M.; Votruba, M. OPA1 deficiency associated with increased autophagy in retinal ganglion cells in a murine model of dominant optic atrophy. Investig. Ophthalmol. Vis. Sci. 2009, 50, 2567–2571. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo Arozena, A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef] [PubMed]
- Patel, B.; Cuervo, A.M. Methods to study chaperone-mediated autophagy. Methods 2015, 75, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Anka, P. Uses of Transmission Electron Microscope in Microscopy and its Advantages and Disadvantages. Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 743–747. [Google Scholar] [CrossRef]
- Fortunato, F.; Hackert, T.; Büchler, M.W.; Kroemer, G. Retrospective electron microscopy: Preservation of fine structure by freezing and aldehyde fixation. Mol. Cell. Oncol. 2016, 3, e1251382. [Google Scholar] [CrossRef]
- Adams, O.; Dislich, B.; Berezowska, S.; Schläfli, A.M.; Seiler, C.A.; Kröll, D.; Tschan, M.P.; Langer, R. Prognostic relevance of autophagy markers LC3B and p62 in esophageal adenocarcinomas. Oncotarget 2016, 7, 39241–39255. [Google Scholar] [CrossRef]
- Adams, O.; Janser, F.A.; Dislich, B.; Berezowska, S.; Humbert, M.; Seiler, C.A.; Kroell, D.; Slotta-Huspenina, J.; Feith, M.; Ott, K.; et al. A specific expression profile of LC3B and p62 is associated with nonresponse to neoadjuvant chemotherapy in esophageal adenocarcinomas. PLoS ONE 2018, 13, e0197610. [Google Scholar] [CrossRef]
- Holt, S.V.; Wyspianska, B.; Randall, K.J.; James, D.; Foster, J.R.; Wilkinson, R.W. The development of an immunohistochemical method to detect the autophagy-associated protein LC3-II in human tumor xenografts. Toxicol. Pathol. 2011, 39, 516–523. [Google Scholar] [CrossRef]
- Janser, F.A.; Adams, O.; Bütler, V.; Schläfli, A.M.; Dislich, B.; Seiler, C.A.; Kröll, D.; Langer, R.; Tschan, M.P. Her2-Targeted Therapy Induces Autophagy in Esophageal Adenocarcinoma Cells. Int. J. Mol. Sci. 2018, 19, 3069. [Google Scholar] [CrossRef]
- Ladoire, S.; Chaba, K.; Martins, I.; Sukkurwala, A.Q.; Adjemian, S.; Michaud, M.; Poirier-Colame, V.; Andreiuolo, F.; Galluzzi, L.; White, E.; et al. Immunohistochemical detection of cytoplasmic LC3 puncta in human cancer specimens. Autophagy 2012, 8, 1175–1184. [Google Scholar] [CrossRef]
- Rosenfeldt, M.T.; Nixon, C.; Liu, E.; Mah, L.Y.; Ryan, K.M. Analysis of macroautophagy by immunohistochemistry. Autophagy 2012, 8, 963–969. [Google Scholar] [CrossRef] [PubMed]
- Schläfli, A.M.; Berezowska, S.; Adams, O.; Langer, R.; Tschan, M.P. Reliable LC3 and p62 autophagy marker detection in formalin fixed paraffin embedded human tissue by immunohistochemistry. Eur. J. Histochem. EJH 2015, 59. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Yang, M.; Zhao, B. Beclin 1 and LC3 as predictive biomarkers for metastatic colorectal carcinoma. Oncotarget 2017, 8, 59058–59067. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Yue, C.; Deng, J.; Hu, R.; Xu, J.; Feng, L.; Lan, Q.; Zhang, W.; Ji, D.; Wu, J.; et al. Autophagic Protein Beclin 1 Serves as an Independent Positive Prognostic Biomarker for Non-Small Cell Lung Cancer. PLoS ONE 2013, 8, e80338. [Google Scholar] [CrossRef] [PubMed]
- Ueno, T.; Saji, S.; Sugimoto, M.; Masuda, N.; Kuroi, K.; Sato, N.; Takei, H.; Yamamoto, Y.; Ohno, S.; Yamashita, H.; et al. Clinical significance of the expression of autophagy-associated marker, beclin 1, in breast cancer patients who received neoadjuvant endocrine therapy. BMC Cancer 2016, 16, 230. [Google Scholar] [CrossRef]
- Willenbacher, W.; Thangavadivel, S.; Greil, R.; Willenbacher, E.; Weger, R.; Manzl, C.; Jöhrer, K.; Brunner, A. Low Beclin-1 expression predicts improved overall survival in patients treated with immunomodulatory drugs for multiple myeloma and identifies autophagy inhibition as a promising potentially druggable new therapeutic target: An analysis from The Austrian Myeloma Registry (AMR). Leuk. Lymphoma 2016, 57, 2330–2341. [Google Scholar]
- Yu, S.; Li, G.; Wang, Z.; Wang, Z.; Chen, C.; Cai, S.; He, Y. Low expression of MAP1LC3B, associated with low Beclin-1, predicts lymph node metastasis and poor prognosis of gastric cancer. Tumor Biol. 2016, 37, 15007–15017. [Google Scholar] [CrossRef]
- Zheng, T.; Li, D.; He, Z.; Feng, S.; Zhao, S. Prognostic and Clinicopathological Significance of Beclin-1 in Non-Small-Cell Lung Cancer: A Meta-Analysis. Available online: https://www.dovepress.com/prognostic-and-clinicopathological-significance-of-beclin-1-in-non-sma-peer-reviewed-article-OTT (accessed on 13 March 2020).
- Minamoto, T.; Nakayama, K.; Nakamura, K.; Katagiri, H.; Sultana, R.; Ishibashi, T.; Ishikawa, M.; Yamashita, H.; Sanuki, K.; Iida, K.; et al. Loss of beclin 1 expression in ovarian cancer: A potential biomarker for predicting unfavorable outcomes. Oncol. Lett. 2018, 15, 1170–1176. [Google Scholar] [CrossRef]
- Malik, S.A.; Orhon, I.; Morselli, E.; Criollo, A.; Shen, S.; Mariño, G.; BenYounes, A.; Bénit, P.; Rustin, P.; Maiuri, M.C.; et al. BH3 mimetics activate multiple pro-autophagic pathways. Oncogene 2011, 30, 3918–3929. [Google Scholar] [CrossRef]
- Liang, X.H.; Kleeman, L.K.; Jiang, H.H.; Gordon, G.; Goldman, J.E.; Berry, G.; Herman, B.; Levine, B. Protection against Fatal Sindbis Virus Encephalitis by Beclin, a Novel Bcl-2-Interacting Protein. J. Virol. 1998, 72, 8586–8596. [Google Scholar] [CrossRef]
- Li, H.; Wang, P.; Sun, Q.; Ding, W.-X.; Yin, X.-M.; Sobol, R.W.; Stolz, D.B.; Yu, J.; Zhang, L. Following Cytochrome c Release, Autophagy Is Inhibited during Chemotherapy-Induced Apoptosis by Caspase 8–Mediated Cleavage of Beclin 1. Cancer Res. 2011, 71, 3625–3634. [Google Scholar] [CrossRef]
- Subramani, S.; Malhotra, V. Non-autophagic roles of autophagy-related proteins. EMBO Rep. 2013, 14, 143–151. [Google Scholar] [CrossRef]
- Loos, B.; du Toit, A.; Hofmeyr, J.-H.S. Defining and measuring autophagosome flux—Concept and reality. Autophagy 2014, 10, 2087–2096. [Google Scholar] [CrossRef] [PubMed]
- Yoshii, S.R.; Mizushima, N. Monitoring and Measuring Autophagy. Int. J. Mol. Sci. 2017, 18, 1865. [Google Scholar] [CrossRef] [PubMed]
- Szalai, P.; Hagen, L.K.; Sætre, F.; Luhr, M.; Sponheim, M.; Øverbye, A.; Mills, I.G.; Seglen, P.O.; Engedal, N. Autophagic bulk sequestration of cytosolic cargo is independent of LC3, but requires GABARAPs. Exp. Cell Res. 2015, 333, 21–38. [Google Scholar] [CrossRef] [PubMed]
- Scicchitano, M.S.; Dalmas, D.A.; Boyce, R.W.; Thomas, H.C.; Frazier, K.S. Protein Extraction of Formalin-fixed, Paraffin-embedded Tissue Enables Robust Proteomic Profiles by Mass Spectrometry. J. Histochem. Cytochem. 2009, 57, 849–860. [Google Scholar] [CrossRef]
- Eng, K.E.; Panas, M.D.; Hedestam, G.B.K.; McInerney, G.M. A novel quantitative flow cytometry-based assay for autophagy. Autophagy 2010, 6, 634–641. [Google Scholar] [CrossRef]
- Corver, W.E.; Haar, N.T. High-Resolution Multiparameter DNA Flow Cytometry for the Detection and Sorting of Tumor and Stromal Subpopulations from Paraffin-Embedded Tissues. Curr. Protoc. Cytom. 2011, 55, 6–27. [Google Scholar] [CrossRef]
- Cadwell, K.; Debnath, J. Beyond self-eating: The control of nonautophagic functions and signaling pathways by autophagy-related proteins. J. Cell Biol. 2018, 217, 813–822. [Google Scholar] [CrossRef]
- Levine, B.; Kroemer, G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 2019, 176, 11–42. [Google Scholar] [CrossRef]
- Aits, S. Methods to Detect Loss of Lysosomal Membrane Integrity. In Autophagy: Methods and Protocols; Ktistakis, N., Florey, O., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2019; pp. 315–329. ISBN 978-1-4939-8873-0. [Google Scholar]
- Aits, S.; Jäättelä, M.; Nylandsted, J. Chapter 13—Methods for the quantification of lysosomal membrane permeabilization: A hallmark of lysosomal cell death. In Methods in Cell Biology; Platt, F., Platt, N., Eds.; Lysosomes and Lysosomal Diseases; Academic Press: Cambridge, MA, USA, 2015; Volume 126, pp. 261–285. [Google Scholar]
- Aits, S.; Kricker, J.; Liu, B.; Ellegaard, A.-M.; Hämälistö, S.; Tvingsholm, S.; Corcelle-Termeau, E.; Høgh, S.; Farkas, T.; Holm Jonassen, A.; et al. Sensitive detection of lysosomal membrane permeabilization by lysosomal galectin puncta assay. Autophagy 2015, 11, 1408–1424. [Google Scholar] [CrossRef] [PubMed]
- Ellegaard, A.-M.; Jäättelä, M.; Nylandsted, J. Visualizing Lysosomal Membrane Permeabilization by Fluorescent Dextran Release. Cold Spring Harb. Protoc. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Maulucci, G.; Chiarpotto, M.; Papi, M.; Samengo, D.; Pani, G.; De Spirito, M. Quantitative analysis of autophagic flux by confocal pH-imaging of autophagic intermediates. Autophagy 2015, 11, 1905–1916. [Google Scholar] [CrossRef] [PubMed]
- Leyva-Paredes, K.; Castrejón-Jiménez, N.S.; IvánArrieta-Oliva, H.; Baltierra-Uribe, S.L.; EstelaGarcía-Pérez, B. Choosing Lunch: The Role of Selective Autophagy Adaptor Proteins. Autophagy Curr. Trends Cell. Physiol. Pathol. 2016. [Google Scholar] [CrossRef]
- Mancias, J.D.; Pontano Vaites, L.; Nissim, S.; Biancur, D.E.; Kim, A.J.; Wang, X.; Liu, Y.; Goessling, W.; Kimmelman, A.C.; Harper, J.W. Ferritinophagy via NCOA4 is required for erythropoiesis and is regulated by iron dependent HERC2-mediated proteolysis. eLife 2015, 4, e10308. [Google Scholar] [CrossRef]
- Jaeger, K.; Sukseree, S.; Zhong, S.; Phinney, B.S.; Mlitz, V.; Buchberger, M.; Narzt, M.S.; Gruber, F.; Tschachler, E.; Rice, R.H.; et al. Cornification of nail keratinocytes requires autophagy for bulk degradation of intracellular proteins while sparing components of the cytoskeleton. Apoptosis 2019, 24, 62–73. [Google Scholar] [CrossRef]
- Zhang, T.; Shen, S.; Qu, J.; Ghaemmaghami, S. Global Analysis of Cellular Protein Flux Quantifies the Selectivity of Basal Autophagy. Cell Rep. 2016, 14, 2426–2439. [Google Scholar] [CrossRef]
- Beine, B.; Diehl, H.C.; Meyer, H.E.; Henkel, C. Tissue MALDI Mass Spectrometry Imaging (MALDI MSI) of Peptides. In Proteomics in Systems Biology: Methods and Protocols; Reinders, J., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2016; pp. 129–150. ISBN 978-1-4939-3341-9. [Google Scholar]
- Coscia, F.; Doll, S.; Bech, J.M.; Schweizer, L.; Mund, A.; Lengyel, E.; Lindebjerg, J.; Madsen, G.I.; Moreira, J.M.A.; Mann, M. A streamlined mass spectrometry-based proteomics workflow for large scale FFPE tissue analysis. bioRxiv 2019. [Google Scholar] [CrossRef]
- Kang, C.; Xu, Q.; Martin, T.D.; Li, M.Z.; Demaria, M.; Aron, L.; Lu, T.; Yankner, B.A.; Campisi, J.; Elledge, S.J. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science 2015, 349, aaa5612. [Google Scholar] [CrossRef]
- Botti-Millet, J.; Nascimbeni, A.C.; Dupont, N.; Morel, E.; Codogno, P. Fine-tuning autophagy: From transcriptional to posttranslational regulation. Am. J. Physiol. Cell Physiol. 2016, 311, C351–C362. [Google Scholar] [CrossRef]
- Xie, Y.; Kang, R.; Sun, X.; Zhong, M.; Huang, J.; Klionsky, D.J.; Tang, D. Posttranslational modification of autophagy-related proteins in macroautophagy. Autophagy 2015, 11, 28–45. [Google Scholar] [CrossRef] [PubMed]
- Dragich, J.M.; Kuwajima, T.; Hirose-Ikeda, M.; Yoon, M.S.; Eenjes, E.; Bosco, J.R.; Fox, L.M.; Lystad, A.H.; Oo, T.F.; Yarygina, O.; et al. Autophagy Linked FYVE (Alfy/WDFY3) Is Required for Establishing Neuronal Connectivity in the Mammalian Brain. Available online: https://elifesciences.org/articles/14810/figures (accessed on 16 July 2019).
- Langer, R.; Streutker, C.J.; Swanson, P.E. Autophagy and its current relevance to the diagnosis and clinical management of esophageal diseases. Ann. N. Y. Acad. Sci. 2016, 1381, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Vieira, A.F.; Schmitt, F. An Update on Breast Cancer Multigene Prognostic Tests—Emergent Clinical Biomarkers. Front. Med. 2018, 5, 248. [Google Scholar] [CrossRef] [PubMed]
- Castellazzi, M.; Patergnani, S.; Donadio, M.; Giorgi, C.; Bonora, M.; Fainardi, E.; Casetta, I.; Granieri, E.; Pugliatti, M.; Pinton, P. Correlation between auto/mitophagic processes and magnetic resonance imaging activity in multiple sclerosis patients. J. Neuroinflamm. 2019, 16, 131. [Google Scholar] [CrossRef]
- Patergnani, S.; Castellazzi, M.; Bonora, M.; Marchi, S.; Casetta, I.; Pugliatti, M.; Giorgi, C.; Granieri, E.; Pinton, P. Autophagy and mitophagy elements are increased in body fluids of multiple sclerosis-affected individuals. J. Neurol. Neurosurg. Psychiatry 2018, 89, 439–441. [Google Scholar] [CrossRef]
- Naguib, M.; Rashed, L.A. Serum level of the autophagy biomarker Beclin-1 in patients with diabetic kidney disease. Diabetes Res. Clin. Pract. 2018, 143, 56–61. [Google Scholar] [CrossRef]
| Type of Autophagy | Level of Activity | Type of Tumor | Role of Autophagy |
|---|---|---|---|
| Macroautophagy | Low | Solid Tumors | Beclin-1 deficiency in mice increase tumor incidence [48] |
| Low Beclin-1 expression levels in human brain tumors [65] | |||
| Low Beclin1 mRNA levels iscorrelated with a poor prognosis in HER2-enriched breast tumors [66] | |||
| Low macroautophagy activity in HSCs increases risk of developing hematopoietic malignancies [86,87,88,89,90,91] | |||
| Leukemia and Lymphomas | Low ATG gene expression in AML [90,94,95,96,97] | ||
| Accelerated leukemia development in mouse model with impaired macroautophagy [96,97,98,99] | |||
| ATG gene mutations found in AML [97,100,101] | |||
| High | Solid Tumors | Therapy-induced macroautophagy enhances tumor cell survival [49,50,73,74,94,95] | |
| RAS-driven tumors are autophagy-dependent [66,70,71,72] | |||
| Leukemia and Lymphomas | High macroautophagy activity in FLT3-ITD mutant AML patients [102] | ||
| Important for the differentiation and activation of lymphocytes [39,111,112,113,114,115,116,117,118] | |||
| CMA | Low | Solid Tumors | Lamp2a deficiency in a mouse model increase liver tumor incidence [63] |
| High | Solid Tumors | High LAMP2A expression commonly found in cancers [52] | |
| Therapy-induced CMA promote tumor cell survival [56] | |||
| CMA activity contributes to tumor cell proliferation and metastatic potential [52,60,61,62] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Humbert, M.; Morán, M.; de la Cruz-Ojeda, P.; Muntané, J.; Wiedmer, T.; Apostolova, N.; McKenna, S.L.; Velasco, G.; Balduini, W.; Eckhart, L.; et al. Assessing Autophagy in Archived Tissue or How to Capture Autophagic Flux from a Tissue Snapshot. Biology 2020, 9, 59. https://doi.org/10.3390/biology9030059
Humbert M, Morán M, de la Cruz-Ojeda P, Muntané J, Wiedmer T, Apostolova N, McKenna SL, Velasco G, Balduini W, Eckhart L, et al. Assessing Autophagy in Archived Tissue or How to Capture Autophagic Flux from a Tissue Snapshot. Biology. 2020; 9(3):59. https://doi.org/10.3390/biology9030059
Chicago/Turabian StyleHumbert, Magali, María Morán, Patricia de la Cruz-Ojeda, Jordi Muntané, Tabea Wiedmer, Nadezda Apostolova, Sharon L. McKenna, Guillermo Velasco, Walter Balduini, Leopold Eckhart, and et al. 2020. "Assessing Autophagy in Archived Tissue or How to Capture Autophagic Flux from a Tissue Snapshot" Biology 9, no. 3: 59. https://doi.org/10.3390/biology9030059
APA StyleHumbert, M., Morán, M., de la Cruz-Ojeda, P., Muntané, J., Wiedmer, T., Apostolova, N., McKenna, S. L., Velasco, G., Balduini, W., Eckhart, L., Janji, B., Sampaio-Marques, B., Ludovico, P., Žerovnik, E., Langer, R., Perren, A., Engedal, N., & Tschan, M. P. (2020). Assessing Autophagy in Archived Tissue or How to Capture Autophagic Flux from a Tissue Snapshot. Biology, 9(3), 59. https://doi.org/10.3390/biology9030059
















