Identification of a Novel Thermostable Alkaline Protease from Bacillus megaterium-TK1 for the Detergent and Leather Industry
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Microbial Strains and Growth Conditions
2.3. Protein Content Quantification and Protease Activity
2.4. Purification Procedure
2.5. Sodium Dodecyl Sulphate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) and Zymogram Analyses of Purified Protease
2.6. Effect of pH and Temperature on Protease Activity
2.7. Protease Substrate Specificity
2.8. Effect of Various Inhibitors, Metal Ions, Solvents, Surfactants, and Commercial Detergents on Purified Protease Activity
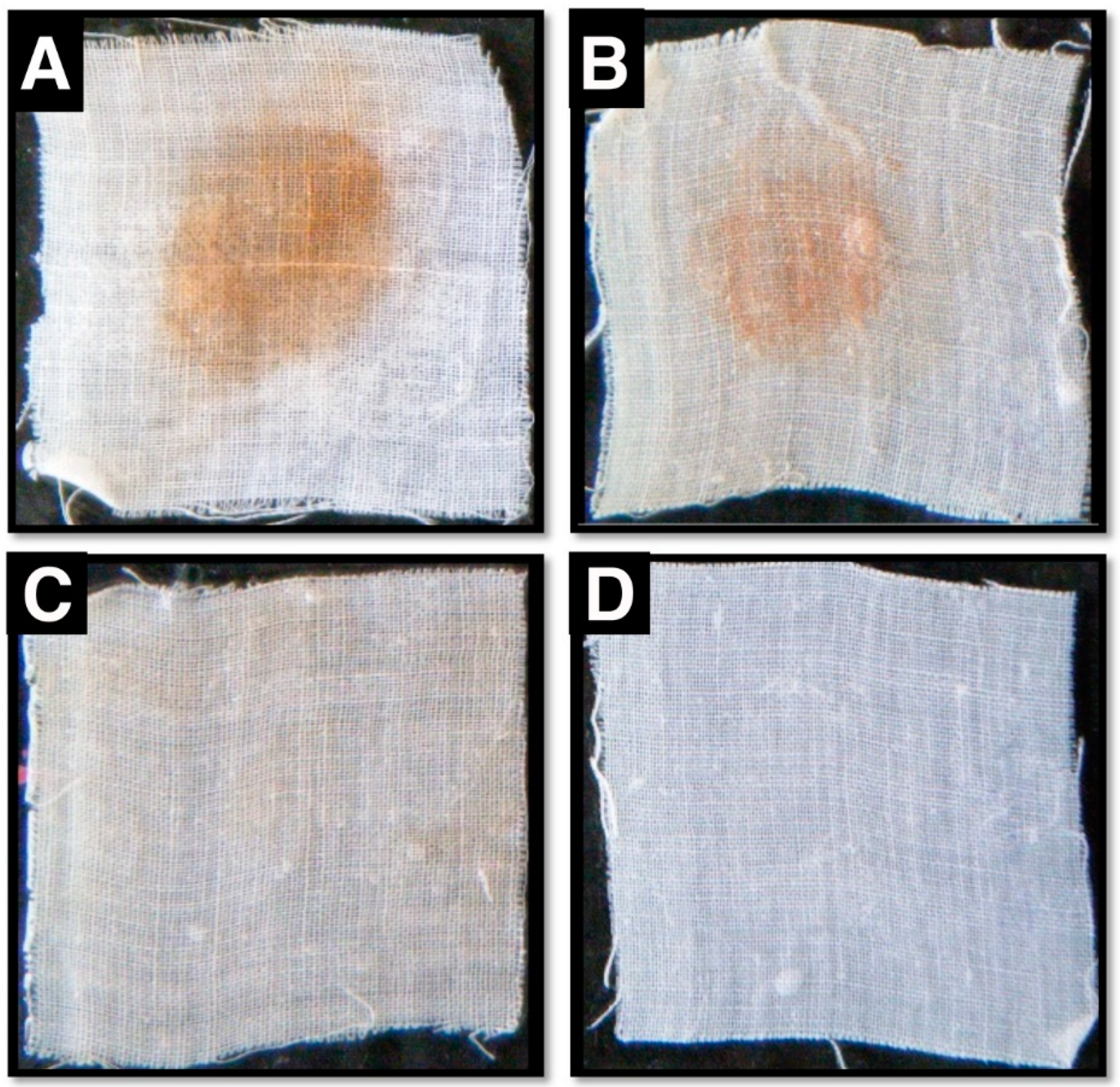
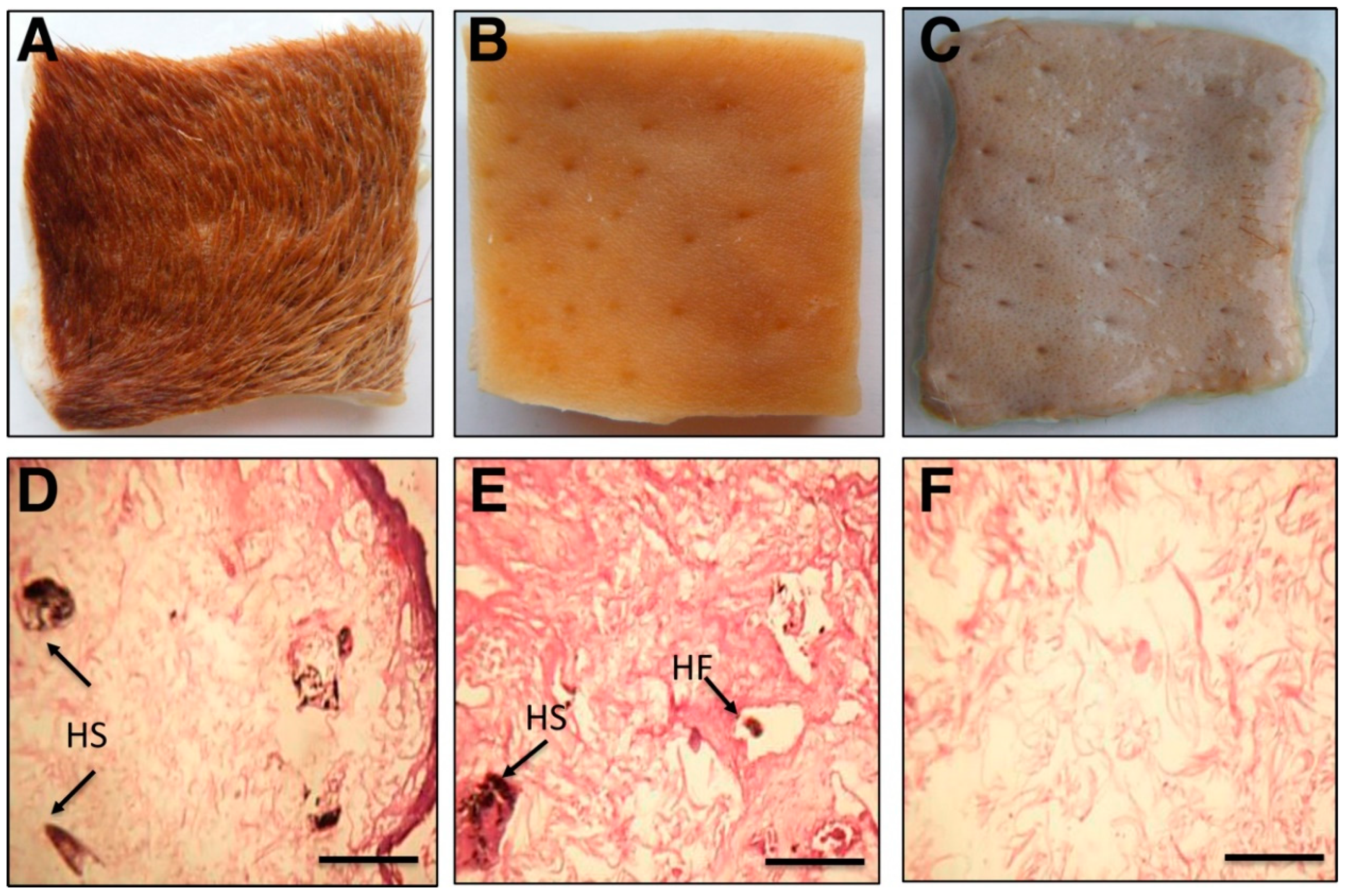
2.9. Blood Stain Removal and Cow Skin Dehairing Efficacy of Purified Protease
2.10. Statistical Analysis
3. Results
3.1. Screening and Identification of Protease Producing Bacterium
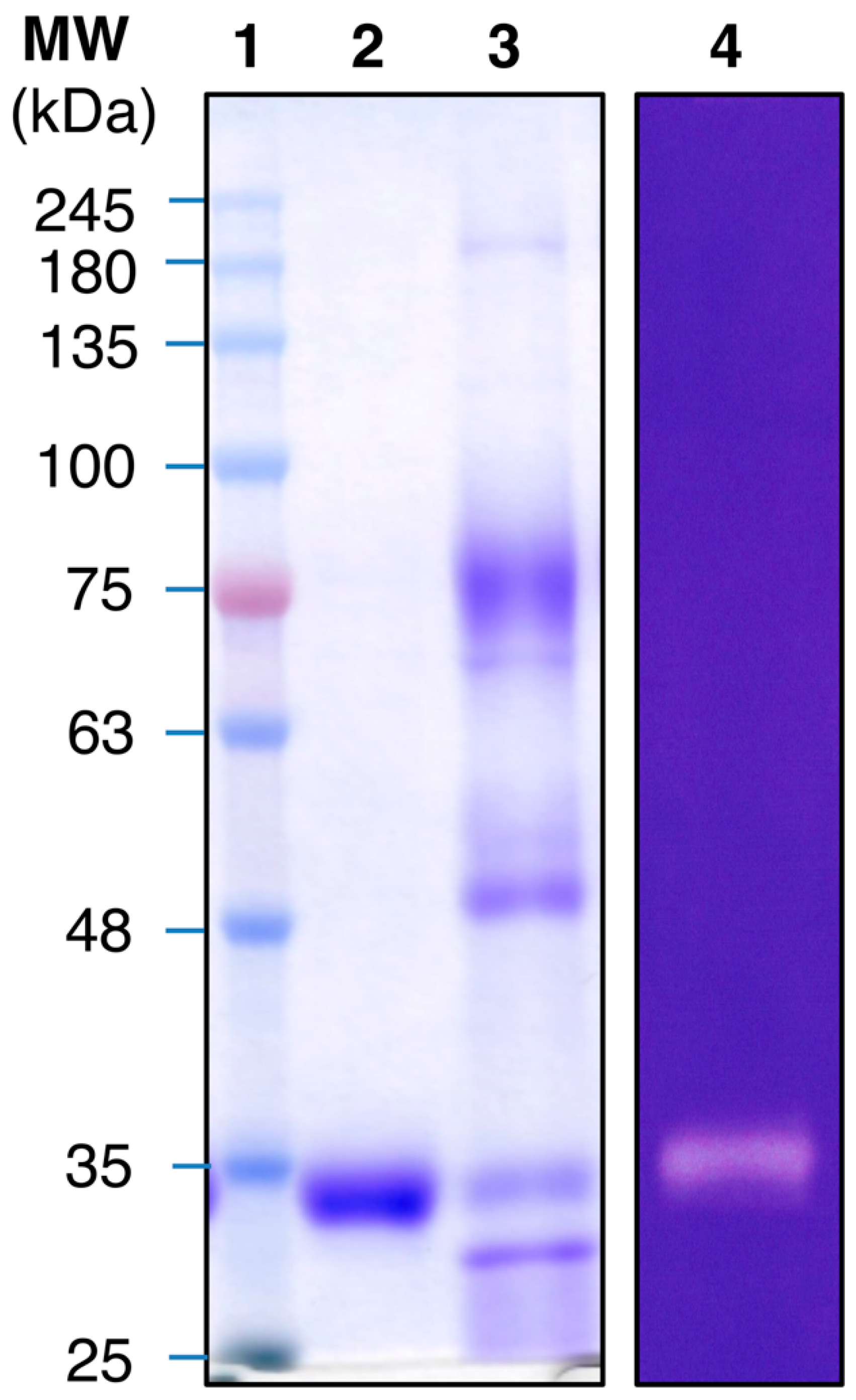
3.2. Protease Purification
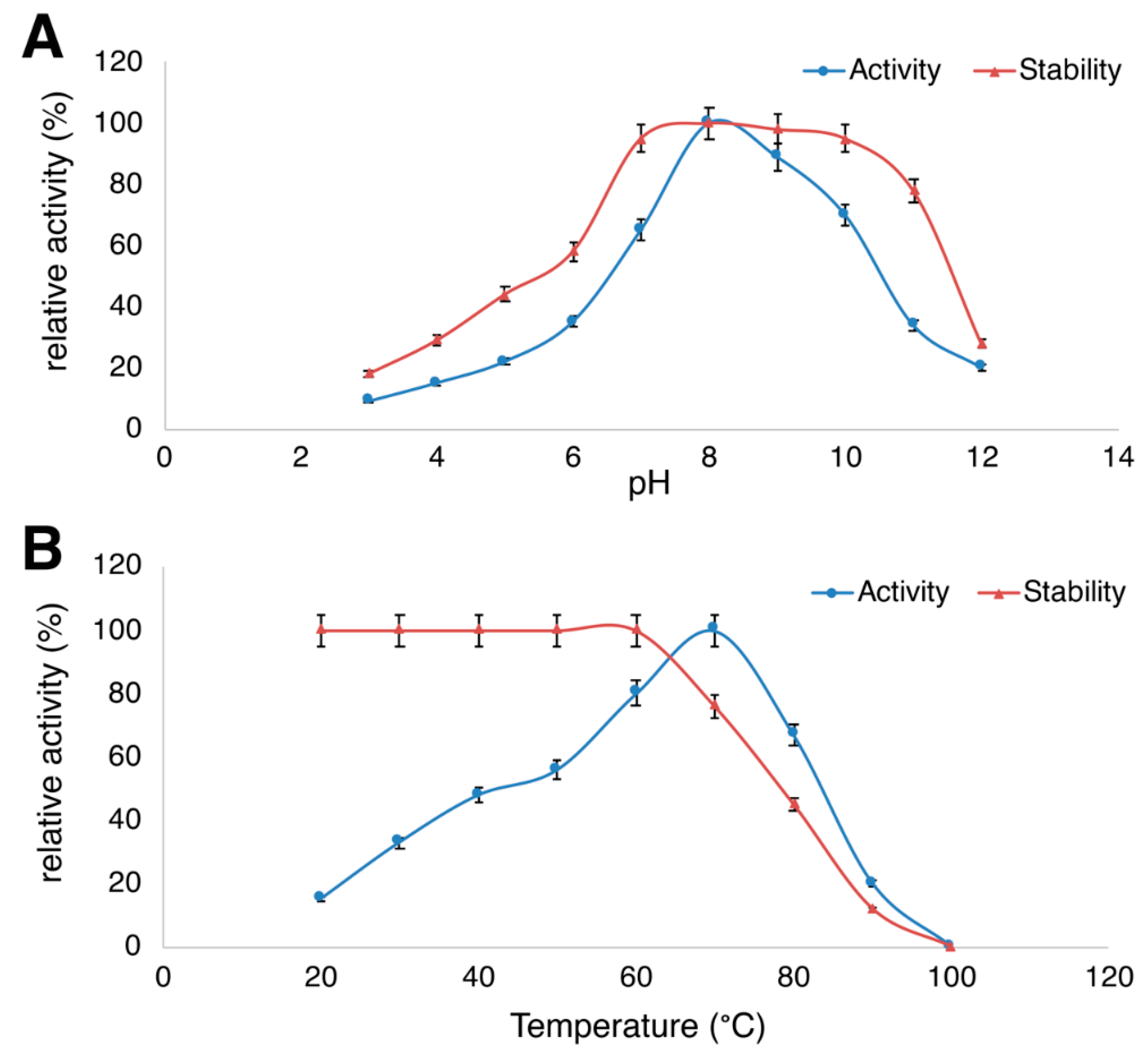
3.3. Temperature- and pH-Dependent Protease Activity
3.4. Protease Substrate Specificity
3.5. Impact of Various Inhibitors, Metal Cations, Solvents, and Surfactants on Purified Protease Activity
3.6. Compatibility of Purified Protease Activity with Various Commercial Detergents and Protease Efficacy During Blood Stain Removal Procedures
3.7. Dehairing of Cow Skin by Chemical Processing or Treatment with Purified Protease
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Razzaq, A.; Shamsi, S.; Ali, A.; Ali, Q.; Sajjad, M.; Malik, A.; Ashraf, M. Microbial Proteases Applications. Front. Bioeng. Biotechnol. 2019, 7, 110. [Google Scholar] [CrossRef]
- Si, J.-B.; Jang, E.-J.; Charalampopoulos, D.; Wee, Y.-J. Purification and Characterization of Microbial Protease Produced Extracellularly from Bacillus subtilis FBL-1. Biotechnol. Bioprocess Eng. 2018, 23, 176–182. [Google Scholar] [CrossRef]
- Yu, P.; Wang, X.; Huang, X.; Ren, Q.; Yan, T. Purification and characterization of a propanol-tolerant neutral protease from Bacillus sp. ZG20. Prep. Biochem. Biotechnol. 2019, 49, 718–726. [Google Scholar] [CrossRef]
- Chen, X.; Zhou, C.; Xue, Y.; Shi, J.; Ma, Y. Cloning, expression, and characterization of an alkaline protease, AprV, from Vibrio sp. DA1-1. Bioprocess Biosyst. Eng. 2018, 41, 1437–1447. [Google Scholar] [CrossRef]
- Contesini, F.J.; Melo, R.R.; Sato, H.H. An overview of Bacillus proteases: From production to application. Crit. Rev. Biotechnol. 2018, 38, 321–334. [Google Scholar] [CrossRef]
- Gao, X.; Yin, Y.; Yan, J.; Zhang, J.; Ma, H.; Zhou, C. Separation, biochemical characterization and salt-tolerant mechanisms of alkaline protease from Aspergillus oryzae. J. Sci. Food Agric. 2019, 99, 3359–3366. [Google Scholar] [CrossRef]
- Huang, Q.; Li, Q.; Chen, A.S.; Kang, C. West Nile virus protease activity in detergent solutions and application for affinity tag removal. Anal. Biochem. 2013, 435, 44–46. [Google Scholar] [CrossRef]
- Kim, J.Y. Isolation of Protease-producing Yeast, Pichia farinose CO-2 and Characterization of Its Extracellular Enzyme. J. Korean Soc. Appl. Biol. Chem. 2010, 53, 133–141. [Google Scholar] [CrossRef]
- Rajkumar, R.; Jayappriyan, K.R.; Rengasamy, R. Purification and characterization of a protease produced by Bacillus megaterium RRM2: Application in detergent and dehairing industries. J. Basic Microbiol. 2011, 51, 614–624. [Google Scholar] [CrossRef]
- Senthilvelan, T.; Kanagaraj, J.; Mandal, A.B. Application of enzymes for dehairing of skins: Cleaner leather processing. Clean Technol. Environ. Policy 2012, 14, 889–897. [Google Scholar] [CrossRef]
- Zhou, C.; Qin, H.; Chen, X.; Zhang, Y.; Xue, Y.; Ma, Y. A novel alkaline protease from alkaliphilic Idiomarina sp. C9-1 with potential application for eco-friendly enzymatic dehairing in the leather industry. Sci. Rep. 2018, 8, 16467. [Google Scholar] [CrossRef] [PubMed]
- Oztas Gulmus, E.; Gormez, A. Characterization and biotechnological application of protease from thermophilic Thermomonas haemolytica. Arch. Microbiol. 2020, 202, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Suthindhiran, K.; Jayasri, M.A.; Dipali, D.; Prasar, A. Screening and characterization of protease producing actinomycetes from marine saltern. J. Basic Microbiol. 2014, 54, 1098–1109. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Arulmani, M.; Aparanjini, K.; Vasanthi, K.; Arumugam, P.; Arivuchelvi, M.; Kalaichelvan, P.T. Purification and partial characterization of serine protease from thermostable alkalophilic Bacillus laterosporus-AK1. World J. Microbiol. Biotechnol. 2007, 23, 475–481. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, R.M.; Manavalan, T.; Ramesh, J.; Thangavelu, K.P.; Heese, K. Improvement of Saccharification and Delignification Efficiency of Trichoderma reesei Rut-C30 by Genetic Bioengineering. Microorganisms 2020, 8, 159. [Google Scholar] [CrossRef]
- Clesceri, L.S.; Greenberg, A.E.; Trussell, R.R.; American Public Health Association; American Water Works Association; Water Pollution Control Federation. Standard Methods for the Examination of Water and Wastewater; American Public Health Association: Washington, DC, USA, 1989. [Google Scholar]
- Asker, M.M.S.; Mahmoud, M.G.; El Shebwy, K.; Abd el Aziz, M.S. Purification and characterization of two thermostable protease fractions from Bacillus megaterium. J. Genet. Eng. Biotechnol. 2013, 11, 103–109. [Google Scholar] [CrossRef]
- Yossana, S.; Reungsangb, A.; Yasudac, M. Purification and characterization of alkaline protease from Bacillus megaterium isolated from Thai fish sauce fermentation process. Sci. Asia 2006, 32, 377–383. [Google Scholar] [CrossRef]
- Yang, S.; Zhai, L.; Huang, L.; Meng, D.; Li, J.; Hao, Z.; Guan, Z.; Cai, Y.; Liao, X. Mining of alkaline proteases from Bacillus altitudinis W3 for desensitization of milk proteins: Their heterologous expression, purification, and characterization. Int. J. Biol. Macromol. 2020, 153, 1220–1230. [Google Scholar] [CrossRef]
- Annamalai, N.; Rajeswari, M.V.; Balasubramanian, T. Extraction, purification and application of thermostable and halostable alkaline protease from Bacillus alveayuensis CAS 5 using marine wastes. Food Bioprod. Process. Trans. Inst. Chem. Eng. Part C 2014, 92, 335–342. [Google Scholar] [CrossRef]
- Bhari, R.; Kaur, M.; Singh, R.S. Thermostable and halotolerant keratinase from Bacillus aerius NSMk2 with remarkable dehairing and laundary applications. J. Basic Microbiol. 2019, 59, 555–568. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xu, A.; Wan, Y.; Li, Q. Purification and characterization of a new metallo-neutral protease for beer brewing from Bacillus amyloliquefaciens SYB-001. Appl. Biochem. Biotechnol. 2013, 170, 2021–2033. [Google Scholar] [CrossRef] [PubMed]
- Tekin, A.; Uzuner, U.; Sezen, K. Homology modeling and heterologous expression of highly alkaline subtilisin-like serine protease from Bacillus halodurans C-125. Biotechnol. Lett. 2020. [Google Scholar] [CrossRef] [PubMed]
- Hammami, A.; Hamdi, M.; Abdelhedi, O.; Jridi, M.; Nasri, M.; Bayoudh, A. Surfactant- and oxidant-stable alkaline proteases from Bacillus invictae: Characterization and potential applications in chitin extraction and as a detergent additive. Int. J. Biol. Macromol. 2017, 96, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Anbu, P. Characterization of solvent stable extracellular protease from Bacillus koreensis (BK-P21A). Int. J. Biol. Macromol. 2013, 56, 162–168. [Google Scholar] [CrossRef]
- Mhamdi, S.; Bkhairia, I.; Nasri, R.; Mechichi, T.; Nasri, M.; Kamoun, A.S. Evaluation of the biotechnological potential of a novel purified protease BS1 from Bacillus safensis S406 on the chitin extraction and detergent formulation. Int. J. Biol. Macromol. 2017, 104, 739–747. [Google Scholar] [CrossRef]
- Jalkute, C.B.; Waghmare, S.R.; Nadaf, N.H.; Dhanavade, M.J.; Jadhav, D.B.; Pendhari, S.I.; Patil, R.S.; Sonawane, K.D. Purification and characterization of SDS stable protease from Bacillus safensis strain CK. Biocatal. Agric. Biotechnol. 2017, 10, 91–95. [Google Scholar] [CrossRef]
- Briki, S.; Hamdi, O.; Landoulsi, A. Enzymatic dehairing of goat skins using alkaline protease from Bacillus sp. SB12. Protein Expr. Purif. 2016, 121, 9–16. [Google Scholar] [CrossRef]
- Jain, D.; Pancha, I.; Mishra, S.K.; Shrivastav, A.; Mishra, S. Purification and characterization of haloalkaline thermoactive, solvent stable and SDS-induced protease from Bacillus sp.: A potential additive for laundry detergents. Bioresour. Technol. 2012, 115, 228–236. [Google Scholar] [CrossRef]
- Omidinia, E. Isolation, Purification and Characterization of a Thermophilic Alkaline Protease from Bacillus subtilis BP-36. J. Sci. Islamic Repub. Iran 2012, 23, 7–13. [Google Scholar]
- Farhadian, S.; Asoodeh, A.; Lagzian, M. Purification, biochemical characterization and structural modeling of a potential htrA-like serine protease from Bacillus subtilis DR8806. J. Mol. Catal. B Enzym. 2015, 115, 51–58. [Google Scholar] [CrossRef]
- Sathishkumar, R.; Ananthan, G.; Arun, J. Production, purification and characterization of alkaline protease by ascidian associated Bacillus subtilis GA CAS8 using agricultural wastes. Biocatal. Agric. Biotechnol. 2015, 4, 214–220. [Google Scholar] [CrossRef]
- Rehman, R.; Ahmed, M.; Siddique, A.; Hasan, F.; Hameed, A.; Jamal, A. Catalytic Role of Thermostable Metalloproteases from Bacillus subtilis KT004404 as Dehairing and Destaining Agent. Appl. Biochem. Biotechnol. 2017, 181, 434–450. [Google Scholar] [CrossRef]
- Yang, H.; Liu, Y.; Ning, Y.; Wang, C.; Zhang, X.; Weng, P.; Wu, Z. Characterization of an Intracellular Alkaline Serine Protease from Bacillus velezensis SW5 with Fibrinolytic Activity. Curr. Microbiol. 2020, 77, 1610–1621. [Google Scholar] [CrossRef]
- Moridshahi, R.; Bahreini, M.; Sharifmoghaddam, M.; Asoodeh, A. Biochemical characterization of an alkaline surfactant-stable keratinase from a new keratinase producer, Bacillus zhangzhouensis. Extremophiles 2020, 24, 693–704. [Google Scholar] [CrossRef]
- Iqbal, I.; Aftab, M.N.; Afzal, M.; Ur-Rehman, A.; Aftab, S.; Zafar, A.; Ud-Din, Z.; Khuharo, A.R.; Iqbal, J.; Ul-Haq, I. Purification and characterization of cloned alkaline protease gene of Geobacillus stearothermophilus. J. Basic Microbiol. 2015, 55, 160–171. [Google Scholar] [CrossRef]
- Anbu, P. Enhanced production and organic solvent stability of a protease from Brevibacillus laterosporus strain PAP04. Braz. J. Med. Biol. Res. 2016, 49, e5178. [Google Scholar] [CrossRef]
- Khandelwal, H.B.; More, S.V.; Kalal, K.M.; Laxman, R.S. Eco-friendly enzymatic dehairing of skins and hides by C. brefeldianus protease. Clean Technol. Environ. Policy 2015, 17, 393–405. [Google Scholar] [CrossRef]
- Ammasi, R.; Victor, J.S.; Chellan, R.; Chellappa, M. Alkaline protease for an efficacious rehydration of skin matrix by a novel Bacillus crolab MTCC 5468 in sustainable leather production: A green approach. Biotechnol. Lett. 2020, 42, 249–267. [Google Scholar] [CrossRef]
| Purification Step | Total Protein (mg) | Total Activity (Units) | Specific Activity (Units/mg Protein) | Purification Fold | Yield (%) |
|---|---|---|---|---|---|
| Culture supernatant | 62 | 22,666 | 366 | 1 | 100 |
| (NH4)2SO4 Fraction 30–80% (w/v) | 25 | 16,250 | 650 | 1.77 | 71.69 |
| DEAE-C | 10 | 7250 | 725 | 1.98 | 31.98 |
| Sephadex G-100 | 6 | 5380 | 897 | 2.45 | 23.73 |
| Substrate (1%) | Relative Activity (%) |
|---|---|
| Azo casein | 85 ± 0.5 |
| Bovine albumin | 80 ± 3 |
| Casein | 100 ± 1 |
| Egg albumin | 49 ± 1.8 |
| Gelatin | 35 ± 1.5 |
| Hemoglobin | 79 ± 1 |
| Chemicals (Concentration) | Relative Activity (%) |
|---|---|
| Control (none) | 100 ± 0.1 |
| Inhibitors (5 mM) | |
| DTT | 67 ± 1.6 |
| EDTA | 90 ± 1.88 |
| Iodoacetate | 85 ± 1.0 |
| 2-mercaptoethanol | 71 ± 0.5 |
| PMSF | 0.0 ± 0.0 |
| Metal cations (1 mM) | |
| Control | 100 ± 0.1 |
| Ca2+ | 120 ± 1.9 |
| Co2+ | 105 ± 2.5 |
| Cu2+ | 95 ± 0.6 |
| Hg2+ | 20 ± 1.0 |
| Mg2+ | 111 ± 0.8 |
| Mn2+ | 123 ± 2 |
| Na+ | 155 ± 1.85 |
| Zn2+ | 85 ± 1.5 |
| Solvents (10%) | |
| Acetone | 100 ± 0.9 |
| DMSO | 109 ± 4.0 |
| Ethanol | 95 ± 3.0 |
| Methanol | 88 ± 1.4 |
| Isopropanol | 90 ± 0.7 |
| Surfactants 1% (1 h) | |
| Control | 100 ± 0.1 |
| CTAB | 71 ± 1.5 |
| SDS | 69 ± 0.6 |
| Triton X-100 | 109 ± 1.7 |
| Tween-20 | 105 ± 0.9 |
| Detergents (1% w/v) | Relative Activity (%) |
|---|---|
| Ariel | 89 ± 1.2 |
| Challenge | 85 ± 0.2 |
| Henko | 79 ± 0.5 |
| Rin | 93 ± 0.3 |
| Surf Excel | 99 ± 1.0 |
| Tide | 97 ± 0.5 |
| Pollution Loads (PPM) | Chemical Treatment | Enzymatic Treatment | Pollution Reduction (%) |
|---|---|---|---|
| BOD | 2130 ± 50 | 1390 ± 20 | 65.2 |
| COD | 5795 ± 30 | 4500 ± 25 | 77.6 |
| TDS | 17,500 ± 40 | 14,750 ± 50 | 84.2 |
| TSS | 5650 ± 60 | 4950 ± 10 | 87.6 |
| Organism | Molecular Weight (kDa)/pI * | Optimal Activity at pH/Temperature (°C) | Stability at pH/Temperature (°C) | Reference |
|---|---|---|---|---|
| B. megaterium RRM2 | 27/– | 10/60 | 7–11/60 | [9] |
| B. megaterium | 28/– | 7.5/50 | 7.0–8.5/80 | [19] |
| B. megaterium | 27/– | 10/50 | 7.5–9.5/30–45 | [20] |
| B. megaterium | 25/– | 7.5/50 | 7.0–8.5/80 | [19] |
| B. megaterium-TK1 | 33/– | 8/70 | 7–11/20–60 | present study |
| B. altitudinis W3 | 37.90/8.67 | 9.5/55 | 6.5–11.5/50 | [21] |
| B. altitudinis W3 | 37.29/6.15 | 8.5/50 | 6–10.5/40 | [21] |
| B. altitudinis W3 | 34.94/7.61 | 10.5/45 | 10.5–11.5/50 | [21] |
| B. alveayuensis CAS 5 | 33/– | 9/50 | 8–11/80 | [22] |
| B. aerius NSMk2 | 9/– | 8/45 | 6.5–9.5/65 | [23] |
| B. amyloliquefaciens SYB-001 | 36.8/– | 7/50 | 6–10/60 | [24] |
| B. halodurans C-125 | 28.3/9.47 | 12/60 | 12/50 | [25] |
| B. invictae | –/– | 9–11/60 | 6–12/30–50 | [26] |
| B. koreensis BK-P21A | 48/– | 9/60 | 7–10/70 | [27] |
| B. safensis S406 (BS1) | 29/– | 11/60 | 6.0–12.0/45 | [28] |
| B. safensis CK | 40/– | 7/37 | – | [29] |
| B. sp. SB12 | 41/– | 9/37 | 7–11/60 | [30] |
| B. sp. SM2014 | 71/– | 10/60 | 7–12/80 | [31] |
| B. subtilis BP36 | 40/– | 9/60 | 9–11/60 | [32] |
| B. subtilis DR8806 | 37/– | 8/45 | 6–9/60 | [33] |
| B. subtilis GA CAS8 | 41/– | 9/50 | 9–11/60 | [34] |
| B. subtilis KT004404 | 28.2/– | 6/55 | 5–8/65 | [35] |
| B. velezensis SW5 | 34/4.66 | 8/40 | 6–9/20–40 | [36] |
| B. zhangzhouensis | 42/– | 9.5/60 | 7–10.5/30–70 | [37] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manavalan, T.; Manavalan, A.; Ramachandran, S.; Heese, K. Identification of a Novel Thermostable Alkaline Protease from Bacillus megaterium-TK1 for the Detergent and Leather Industry. Biology 2020, 9, 472. https://doi.org/10.3390/biology9120472
Manavalan T, Manavalan A, Ramachandran S, Heese K. Identification of a Novel Thermostable Alkaline Protease from Bacillus megaterium-TK1 for the Detergent and Leather Industry. Biology. 2020; 9(12):472. https://doi.org/10.3390/biology9120472
Chicago/Turabian StyleManavalan, Tamilvendan, Arulmani Manavalan, Shiyamsundar Ramachandran, and Klaus Heese. 2020. "Identification of a Novel Thermostable Alkaline Protease from Bacillus megaterium-TK1 for the Detergent and Leather Industry" Biology 9, no. 12: 472. https://doi.org/10.3390/biology9120472
APA StyleManavalan, T., Manavalan, A., Ramachandran, S., & Heese, K. (2020). Identification of a Novel Thermostable Alkaline Protease from Bacillus megaterium-TK1 for the Detergent and Leather Industry. Biology, 9(12), 472. https://doi.org/10.3390/biology9120472