Targeted Protein Degradation Tools: Overview and Future Perspectives
Abstract
Simple Summary
Abstract
1. Introduction
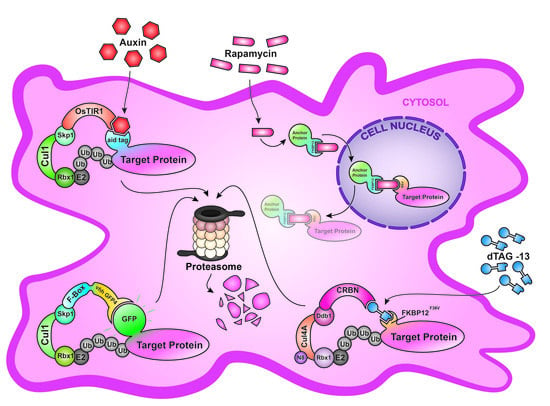
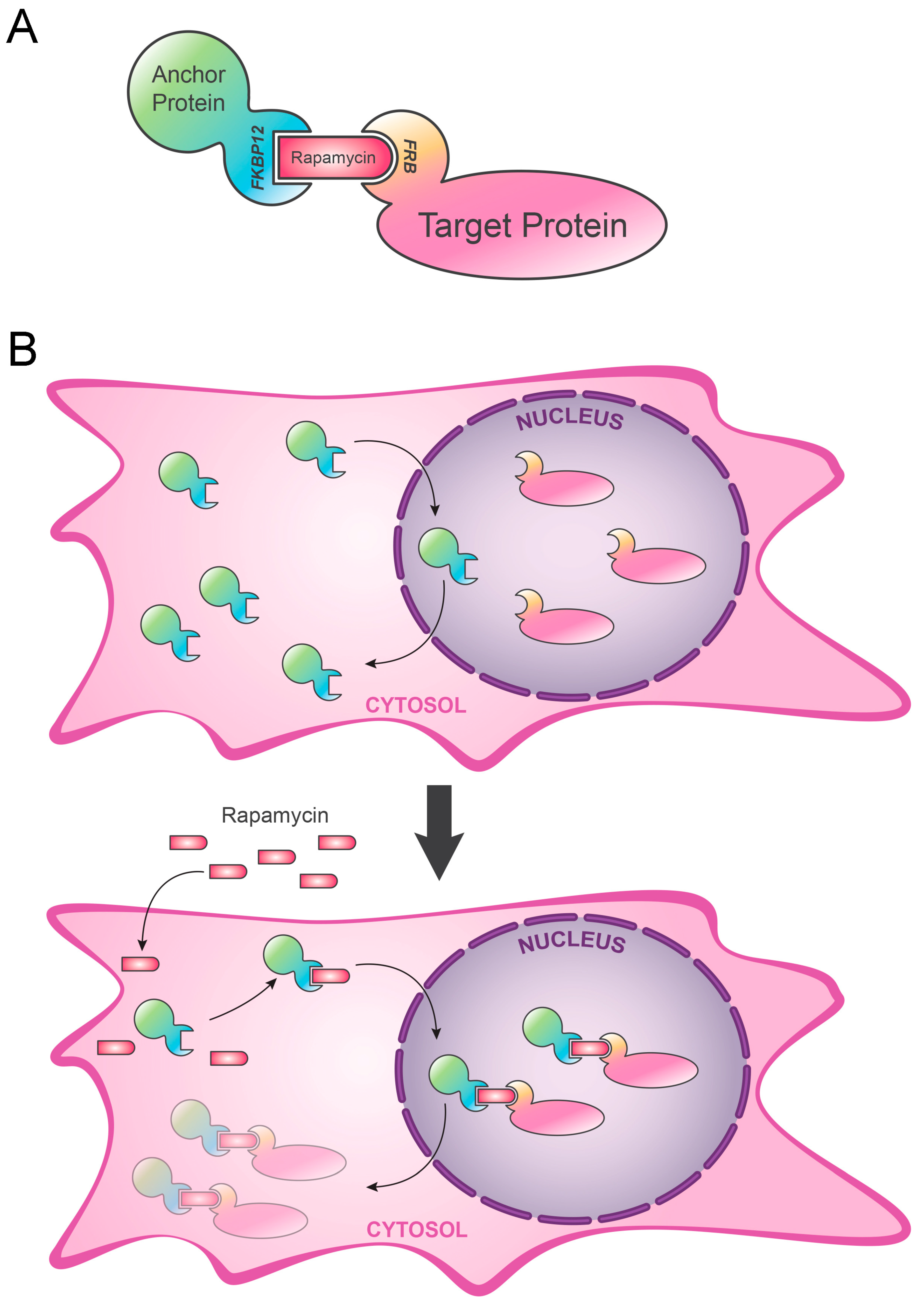
2. Anchor-Away (AA)
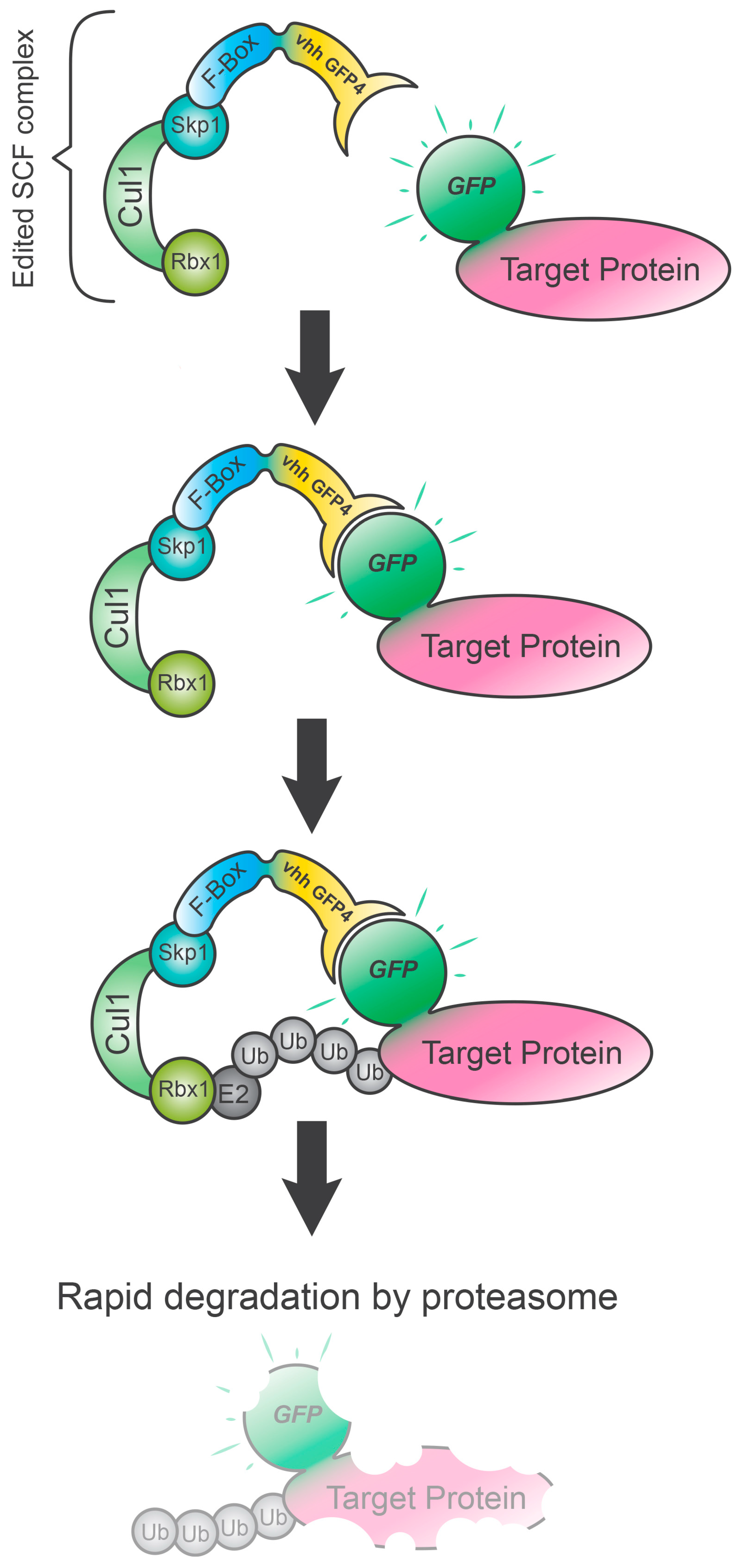
3. deGradFP
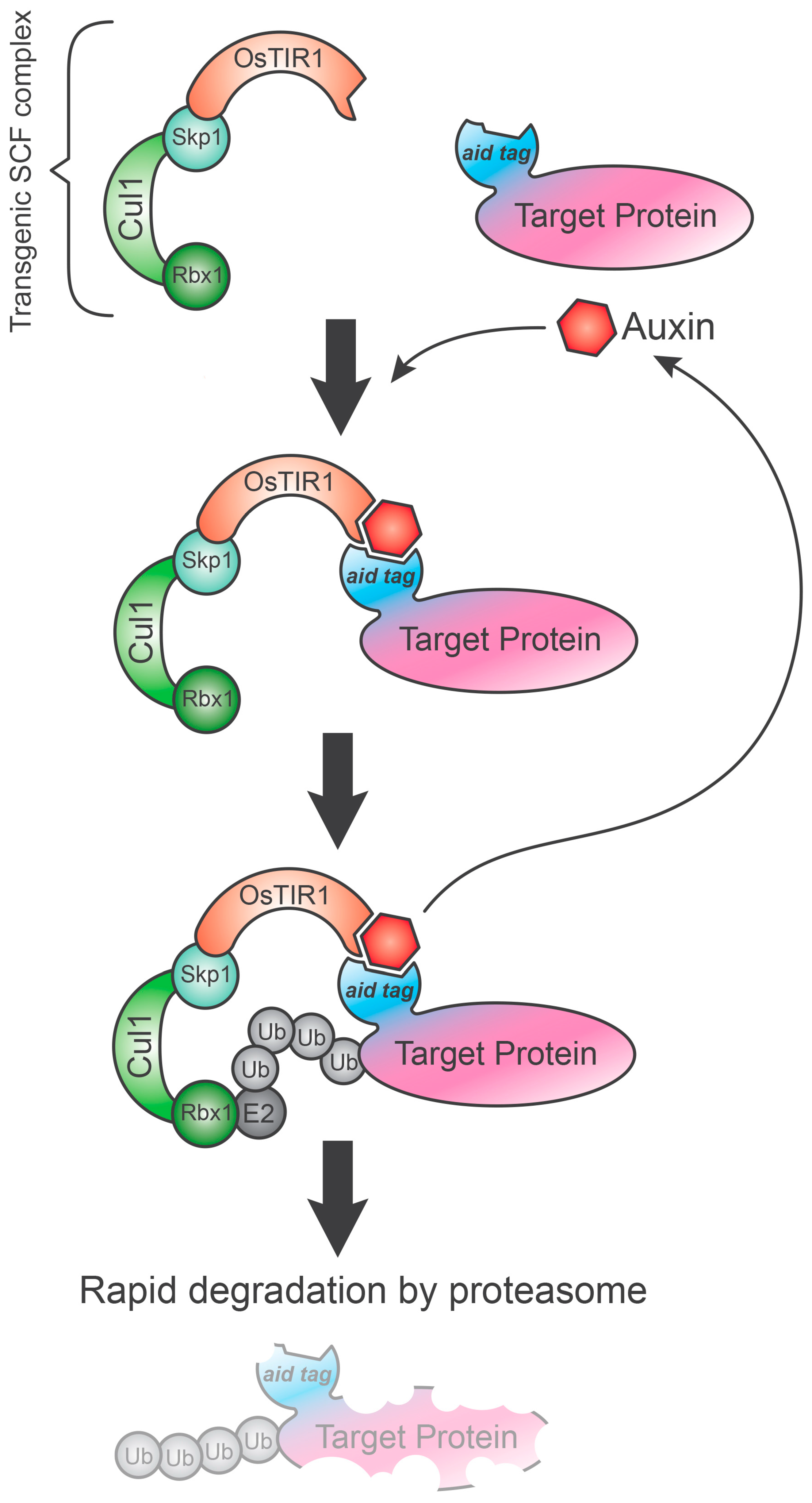
4. Auxin-Inducible Degron System (AID)
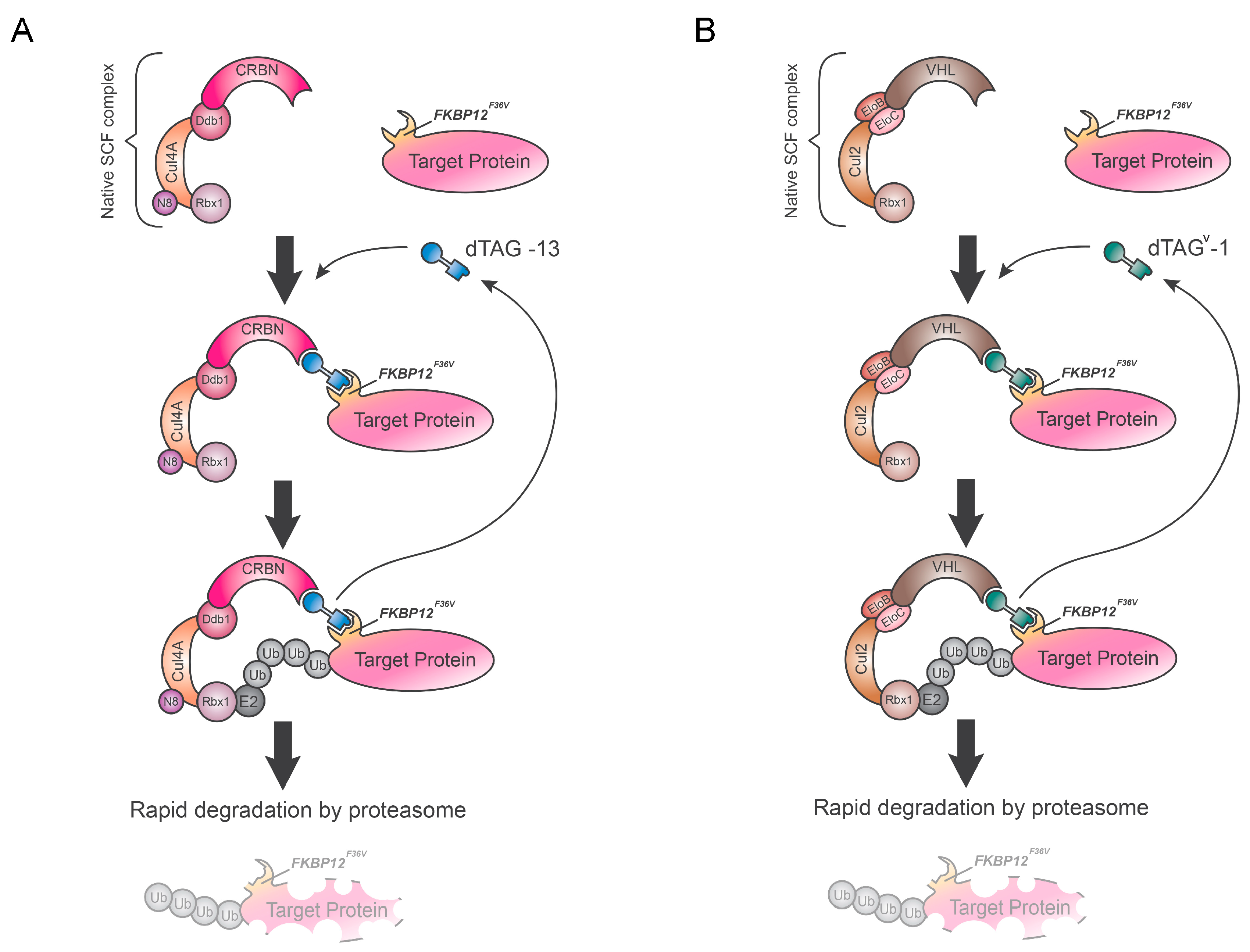
5. Degradation TAG (dTAG) System
6. A Future Perspective: The Nano-Grad System
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Prozzillo, Y.; Delle Monache, F.; Ferreri, D.; Cuticone, S.; Dimitri, P.; Messina, G. The True Story of Yeti, the “Abominable” Heterochromatic Gene of Drosophila melanogaster. Front. Physiol. 2019, 10, 1093. [Google Scholar] [CrossRef] [PubMed]
- Messina, G.; Atterrato, M.T.; Fanti, L.; Giordano, E.; Dimitri, P. Expression of human Cfdp1 gene in Drosophila reveals new insights into the function of the evolutionarily conserved BCNT protein family. Sci. Rep. 2016, 6, 25511. [Google Scholar] [CrossRef] [PubMed]
- Messina, G.; Atterrato, M.T.; Prozzillo, Y.; Piacentini, L.; Losada, A.; Dimitri, P. The human Cranio Facial Development Protein 1 (Cfdp1) gene encodes a protein required for the maintenance of higher-order chromatin organization. Sci. Rep. 2017, 7, 45022. [Google Scholar] [CrossRef] [PubMed]
- Taylor, E.; Alqadri, N.; Dodgson, L.; Mason, D.; Lyulcheva, E.; Messina, G.; Bennett, D. MRL proteins cooperate with activated Ras in glia to drive distinct oncogenic outcomes. Oncogene 2017, 36, 4311–4322. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jackson, A.L.; Bartz, S.R.; Schelter, J.; Kobayashi, S.V.; Burchard, J.; Mao, M.; Li, B.; Cavet, G.; Linsley, P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003, 21, 635–637. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, M.; Zheng, T.; Hou, Y.; Zhang, P.; Tang, T.; Wei, J.; Du, Q. Specificity profiling of CRISPR system reveals greatly enhanced off-target gene editing. Sci. Rep. 2020, 10, 2269. [Google Scholar] [CrossRef]
- Rossi, A.; Kontarakis, Z.; Gerri, C.; Nolte, H.; Holper, S.; Kruger, M.; Stainier, D.Y. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 2015, 524, 230–233. [Google Scholar] [CrossRef]
- Roth, S.; Fulcher, L.J.; Sapkota, G.P. Advances in targeted degradation of endogenous proteins. Cell. Mol. Life Sci. 2019, 76, 2761–2777. [Google Scholar] [CrossRef]
- Haruki, H.; Nishikawa, J.; Laemmli, U.K. The anchor-away technique: Rapid, conditional establishment of yeast mutant phenotypes. Mol. Cell 2008, 31, 925–932. [Google Scholar] [CrossRef]
- Bosch, P.S.; Pepperl, J.; Basler, K. Anchor Away—A Fast, Reliable and Reversible Technique to Inhibit Proteins in Drosophila melanogaster. G3 2020, 10, 1745–1752. [Google Scholar] [CrossRef]
- Samwer, M.; Schneider, M.W.G.; Hoefler, R.; Schmalhorst, P.S.; Jude, J.G.; Zuber, J.; Gerlich, D.W. DNA Cross-Bridging Shapes a Single Nucleus from a Set of Mitotic Chromosomes. Cell 2017, 170, 956–972.e23. [Google Scholar] [CrossRef]
- Chen, J.; Zheng, X.F.; Brown, E.J.; Schreiber, S.L. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc. Natl. Acad. Sci. USA 1995, 92, 4947–4951. [Google Scholar] [CrossRef]
- Caussinus, E.; Kanca, O.; Affolter, M. Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat. Struct. Mol. Biol. 2011, 19, 117–121. [Google Scholar] [CrossRef]
- Caussinus, E.; Affolter, M. deGradFP: A System to Knockdown GFP-Tagged Proteins. Methods Mol. Biol. 2016, 1478, 177–187. [Google Scholar] [CrossRef]
- Caussinus, E.; Kanca, O.; Affolter, M. Protein knockouts in living eukaryotes using deGradFP and green fluorescent protein fusion targets. Curr. Protoc. Protein Sci. 2013, 73, 30.2.1–30.2.13. [Google Scholar] [CrossRef]
- Fedorova, S.A.; Dorogova, N.V. Protein trap: A new Swiss army knife for geneticists? Mol. Biol. Rep. 2020, 47, 1445–1458. [Google Scholar] [CrossRef]
- Shin, Y.J.; Park, S.K.; Jung, Y.J.; Kim, Y.N.; Kim, K.S.; Park, O.K.; Kwon, S.H.; Jeon, S.H.; Trinh le, A.; Fraser, S.E.; et al. Nanobody-targeted E3-ubiquitin ligase complex degrades nuclear proteins. Sci. Rep. 2015, 5, 14269. [Google Scholar] [CrossRef]
- Baudisch, B.; Pfort, I.; Sorge, E.; Conrad, U. Nanobody-Directed Specific Degradation of Proteins by the 26S-Proteasome in Plants. Front. Plant Sci. 2018, 9, 130. [Google Scholar] [CrossRef]
- Yamaguchi, N.; Colak-Champollion, T.; Knaut, H. zGrad is a nanobody-based degron system that inactivates proteins in zebrafish. Elife 2019, 8, e43125. [Google Scholar] [CrossRef]
- Nishimura, K.; Fukagawa, T.; Takisawa, H.; Kakimoto, T.; Kanemaki, M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 2009, 6, 917–922. [Google Scholar] [CrossRef]
- Holland, A.J.; Fachinetti, D.; Han, J.S.; Cleveland, D.W. Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. Proc. Natl. Acad. Sci. USA 2012, 109, E3350–E3357. [Google Scholar] [CrossRef]
- Lambrus, B.G.; Uetake, Y.; Clutario, K.M.; Daggubati, V.; Snyder, M.; Sluder, G.; Holland, A.J. p53 protects against genome instability following centriole duplication failure. J. Cell Biol. 2015, 210, 63–77. [Google Scholar] [CrossRef]
- Kwon, J.Y.; Hong, M.; Choi, M.S.; Kang, S.; Duke, K.; Kim, S.; Lee, S.; Lee, J. Ethanol-response genes and their regulation analyzed by a microarray and comparative genomic approach in the nematode Caenorhabditis elegans. Genomics 2004, 83, 600–614. [Google Scholar] [CrossRef]
- Papagiannakis, A.; de Jonge, J.J.; Zhang, Z.; Heinemann, M. Quantitative characterization of the auxin-inducible degron: A guide for dynamic protein depletion in single yeast cells. Sci. Rep. 2017, 7, 4704. [Google Scholar] [CrossRef]
- Camlin, N.J.; Evans, J.P. Auxin-inducible protein degradation as a novel approach for protein depletion and reverse genetic discoveries in mammalian oocytesdagger. Biol. Reprod. 2019, 101, 704–718. [Google Scholar] [CrossRef]
- Natsume, T.; Kiyomitsu, T.; Saga, Y.; Kanemaki, M.T. Rapid Protein Depletion in Human Cells by Auxin-Inducible Degron Tagging with Short Homology Donors. Cell Rep. 2016, 15, 210–218. [Google Scholar] [CrossRef]
- Muhar, M.; Ebert, A.; Neumann, T.; Umkehrer, C.; Jude, J.; Wieshofer, C.; Rescheneder, P.; Lipp, J.J.; Herzog, V.A.; Reichholf, B.; et al. SLAM-seq defines direct gene-regulatory functions of the BRD4-MYC axis. Science 2018, 360, 800–805. [Google Scholar] [CrossRef]
- Lok, T.M.; Wang, Y.; Xu, W.K.; Xie, S.; Ma, H.T.; Poon, R.Y.C. Mitotic slippage is determined by p31(comet) and the weakening of the spindle-assembly checkpoint. Oncogene 2020, 39, 2819–2834. [Google Scholar] [CrossRef]
- Ito, S.; Goto, H.; Kuniyasu, K.; Shindo, M.; Yamada, M.; Tanaka, K.; Toh, G.T.; Sawa, M.; Inagaki, M.; Bartek, J.; et al. Cdc7 kinase stimulates Aurora B kinase in M-phase. Sci. Rep. 2019, 9, 18622. [Google Scholar] [CrossRef] [PubMed]
- Ng, L.Y.; Ma, H.T.; Liu, J.C.Y.; Huang, X.; Lee, N.; Poon, R.Y.C. Conditional gene inactivation by combining tetracycline-mediated transcriptional repression and auxin-inducible degron-mediated degradation. Cell Cycle 2019, 18, 238–248. [Google Scholar] [CrossRef]
- Morawska, M.; Ulrich, H.D. An expanded tool kit for the auxin-inducible degron system in budding yeast. Yeast 2013, 30, 341–351. [Google Scholar] [CrossRef]
- Nishimura, K.; Fukagawa, T. An efficient method to generate conditional knockout cell lines for essential genes by combination of auxin-inducible degron tag and CRISPR/Cas9. Chromosome Res. Int. J. Mol. Supramol. Evol. Asp. Chromosome Biol. 2017, 25, 253–260. [Google Scholar] [CrossRef]
- Zasadzinska, E.; Huang, J.; Bailey, A.O.; Guo, L.Y.; Lee, N.S.; Srivastava, S.; Wong, K.A.; French, B.T.; Black, B.E.; Foltz, D.R. Inheritance of CENP-A Nucleosomes during DNA Replication Requires HJURP. Dev. Cell 2018, 47, 348–362. [Google Scholar] [CrossRef]
- Guilfoyle, T.J.; Hagen, G. Auxin response factors. Curr. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef]
- Sathyan, K.M.; McKenna, B.D.; Anderson, W.D.; Duarte, F.M.; Core, L.; Guertin, M.J. An improved auxin-inducible degron system preserves native protein levels and enables rapid and specific protein depletion. Genes Dev. 2019, 33, 1441–1455. [Google Scholar] [CrossRef] [PubMed]
- Yesbolatova, A.; Saito, Y.; Kanemaki, M.T. Constructing Auxin-Inducible Degron Mutants Using an All-in-One Vector. Pharmaceuticals 2020, 13, 103. [Google Scholar] [CrossRef] [PubMed]
- Nabet, B.; Roberts, J.M.; Buckley, D.L.; Paulk, J.; Dastjerdi, S.; Yang, A.; Leggett, A.L.; Erb, M.A.; Lawlor, M.A.; Souza, A.; et al. The dTAG system for immediate and target-specific protein degradation. Nat. Chem. Biol. 2018, 14, 431–441. [Google Scholar] [CrossRef]
- Clackson, T.; Yang, W.; Rozamus, L.W.; Hatada, M.; Amara, J.F.; Rollins, C.T.; Stevenson, L.F.; Magari, S.R.; Wood, S.A.; Courage, N.L.; et al. Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity. Proc. Natl. Acad. Sci. USA 1998, 95, 10437–10442. [Google Scholar] [CrossRef] [PubMed]
- Brand, M.; Winter, G.E. Locus-Specific Knock-In of a Degradable Tag for Target Validation Studies. Methods Mol. Biol. 2019, 1953, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Erb, M.A.; Scott, T.G.; Li, B.E.; Xie, H.; Paulk, J.; Seo, H.S.; Souza, A.; Roberts, J.M.; Dastjerdi, S.; Buckley, D.L.; et al. Transcription control by the ENL YEATS domain in acute leukaemia. Nature 2017, 543, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.T.; Seo, H.S.; Zhang, T.; Wang, Y.; Jiang, B.; Li, Q.; Buckley, D.L.; Nabet, B.; Roberts, J.M.; Paulk, J.; et al. MELK is not necessary for the proliferation of basal-like breast cancer cells. Elife 2017, 6, e26693. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Ball, C.B.; Collins, G.; Hu, Q.; Luse, D.S.; Price, D.H.; Meier, J.L. Human cytomegalovirus IE2 drives transcription initiation from a select subset of late infection viral promoters by host RNA polymerase II. PLoS Pathog. 2020, 16, e1008402. [Google Scholar] [CrossRef] [PubMed]
- Bensimon, A.; Pizzagalli, M.D.; Kartnig, F.; Dvorak, V.; Essletzbichler, P.; Winter, G.E.; Superti-Furga, G. Targeted Degradation of SLC Transporters Reveals Amenability of Multi-Pass Transmembrane Proteins to Ligand-Induced Proteolysis. Cell Chem. Biol. 2020, 27, 728–739. [Google Scholar] [CrossRef]
- Mayor-Ruiz, C.; Winter, G.E. Identification and characterization of cancer vulnerabilities via targeted protein degradation. Drug discovery today. Technologies 2019, 31, 81–90. [Google Scholar] [CrossRef]
- Nabet, B.; Ferguson, F.M.; Seong, B.K.A.; Kuljanin, M.; Leggett, A.L.; Mohardt, M.L.; Robichaud, A.; Conway, A.S.; Buckley, D.L.; Mancias, J.D.; et al. Rapid and direct control of target protein levels with VHL-recruiting dTAG molecules. Nat. Commun. 2020, 11, 4687. [Google Scholar] [CrossRef]
- Messina, G.; Prozzillo, Y.; Monache, F.D.; Santopietro, M.V.; Atterrato, M.T.; Dimitri, P. ATPase SRCAP is a new player in cell division, uncovering molecular aspects of Floating-Harbor syndrome. BioRxiv 2020. [Google Scholar] [CrossRef]
- Messina, G.; Damia, E.; Fanti, L.; Atterrato, M.T.; Celauro, E.; Mariotti, F.R.; Accardo, M.C.; Walther, M.; Verni, F.; Picchioni, D.; et al. Yeti, an essential Drosophila melanogaster gene, encodes a protein required for chromatin organization. J. Cell Sci. 2014, 127, 2577–2588. [Google Scholar] [CrossRef]
- Bery, N.; Keller, L.; Soulie, M.; Gence, R.; Iscache, A.L.; Cherier, J.; Cabantous, S.; Sordet, O.; Lajoie-Mazenc, I.; Pedelacq, J.D.; et al. A Targeted Protein Degradation Cell-Based Screening for Nanobodies Selective toward the Cellular RHOB GTP-Bound Conformation. Cell Chem. Biol. 2019, 26, 1544–1558.e6. [Google Scholar] [CrossRef]
- Clift, D.; McEwan, W.A.; Labzin, L.I.; Konieczny, V.; Mogessie, B.; James, L.C.; Schuh, M. A Method for the Acute and Rapid Degradation of Endogenous Proteins. Cell 2017, 171, 1692–1706. [Google Scholar] [CrossRef]
- Sakamoto, K.M.; Kim, K.B.; Kumagai, A.; Mercurio, F.; Crews, C.M.; Deshaies, R.J. Protacs: Chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. USA 2001, 98, 8554–8559. [Google Scholar] [CrossRef] [PubMed]
- Prozzillo, Y.; Cingolani, A.; Messina, G. Department of Biology and Biotechnology “Charles Darwin”. Sapienza University of Rome: Rome, Italy, Unpublished work. 2020. [Google Scholar]
- Herce, H.D.; Schumacher, D.; Schneider, A.F.L.; Ludwig, A.K.; Mann, F.A.; Fillies, M.; Kasper, M.A.; Reinke, S.; Krause, E.; Leonhardt, H.; et al. Cell-permeable nanobodies for targeted immunolabelling and antigen manipulation in living cells. Nat. Chem. 2017, 9, 762–771. [Google Scholar] [CrossRef] [PubMed]

| System | Tag Size | Degrader (MW g/mol) | Organisms | Advantages | Disadvantages | Reversible/Inducible | Transgenic Elements |
|---|---|---|---|---|---|---|---|
| Anchor-away | 12-kDa | Rapamycin (914) |
|
|
| Yes/Yes | 2 |
| deGradFP | 27-kDa | - |
|
|
| No/No | 2 |
| AID system | 7-kDa | IAA (175) NAA (186) |
|
|
| Yes/Yes | 2 |
| dTAG system | 12-kDa | dTAG-13 (1049) dTAGV-1 (1361) |
|
|
| Yes/Yes | 1 |
| nano-grad | No | N/A | N/A |
|
| Yes/Yes | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prozzillo, Y.; Fattorini, G.; Santopietro, M.V.; Suglia, L.; Ruggiero, A.; Ferreri, D.; Messina, G. Targeted Protein Degradation Tools: Overview and Future Perspectives. Biology 2020, 9, 421. https://doi.org/10.3390/biology9120421
Prozzillo Y, Fattorini G, Santopietro MV, Suglia L, Ruggiero A, Ferreri D, Messina G. Targeted Protein Degradation Tools: Overview and Future Perspectives. Biology. 2020; 9(12):421. https://doi.org/10.3390/biology9120421
Chicago/Turabian StyleProzzillo, Yuri, Gaia Fattorini, Maria Virginia Santopietro, Luigi Suglia, Alessandra Ruggiero, Diego Ferreri, and Giovanni Messina. 2020. "Targeted Protein Degradation Tools: Overview and Future Perspectives" Biology 9, no. 12: 421. https://doi.org/10.3390/biology9120421
APA StyleProzzillo, Y., Fattorini, G., Santopietro, M. V., Suglia, L., Ruggiero, A., Ferreri, D., & Messina, G. (2020). Targeted Protein Degradation Tools: Overview and Future Perspectives. Biology, 9(12), 421. https://doi.org/10.3390/biology9120421